The Cytogenetic Map of the Nile Crocodile (Crocodylus niloticus, Crocodylidae, Reptilia) with Fluorescence In Situ Localization of Major Repetitive DNAs
Abstract
1. Introduction
2. Results
2.1. C. niloticus Karyotype
2.2. Bioinformatics Analysis of C. niloticus Repetitive DNA
2.3. Comparative Analysis of Satellite Sequences
2.4. Chromosomal Distribution of Repetitive Sequences
3. Discussion
4. Materials and Methods
4.1. Cell Line Establishment and Karyotype Analysis
4.2. DNA Extraction, Library Preparation, and Whole-Genome DNA Sequencing
4.3. Repetitive DNA Identification and Characterization
4.4. Fluorescence In Situ Hybridization (FISH) with Repeats
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deakin, J.E.; Ezaz, T. E-Mail Understanding the Evolution of Reptile Chromosomes through Applications of Combined Cytogenetics and Genomics Approaches. Cytogenet. Genome Res. 2019, 157, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Olmo, E. Trends in the evolution of reptilian chromosomes. Integr. Comp. Biol. 2008, 48, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Straková, B.; Rovatsos, M.; Kubička, L.; Kratochvíl, L. Evolution of Sex Determination in Amniotes: Did Stress and Sequential Hermaphroditism Produce Environmental Determination? BioEssays 2020, 42, 2000050. [Google Scholar] [CrossRef] [PubMed]
- Chiari, Y.; Cahais, V.; Galtier, N.; Delsuc, F. Phylogenomic analyses support the position of turtles as the sister group of birds and crocodiles (Archosauria). BMC Biol. 2012, 10, 65. [Google Scholar] [CrossRef]
- Blair Hedges, S.; Poling, L.L. A molecular phylogeny of reptiles. Science 1999, 283, 998–1001. [Google Scholar] [CrossRef]
- Brochu, C.A. Phylogenetic Approaches Toward Crocodylian History. Annu. Rev. Earth Planet. Sci. 2003, 31, 357–397. [Google Scholar] [CrossRef]
- Hekkala, E.; Shirley, M.H.; Amato, G.; Austin, J.D.; Charter, S.; Thorbjarnarson, J.; Vliet, K.A.; Houck, M.L.; Desalle, R.; Blum, M.J. An ancient icon reveals new mysteries: Mummy DNA resurrects a cryptic species within the Nile crocodile. Mol. Ecol. 2011, 20, 4199–4215. [Google Scholar] [CrossRef]
- McAliley, L.R.; Willis, R.E.; Ray, D.A.; White, P.S.; Brochu, C.A.; Densmore, L.D. Are crocodiles really monophyletic?--Evidence for subdivisions from sequence and morphological data. Mol. Phylogenet. Evol. 2006, 39, 16–32. [Google Scholar] [CrossRef]
- Muniz, F.L.; Ximenes, A.M.; Bittencourt, P.S.; Hernández-Rangel, S.M.; Campos, Z.; Hrbek, T.; Farias, I.P. Detecting population structure of Paleosuchus trigonatus (Alligatoridae: Caimaninae) through microsatellites markers developed by next generation sequencing. Mol. Biol. Rep. 2019, 46, 2472–2484. [Google Scholar] [CrossRef]
- Shirley, M.H.; Vliet, K.A.; Carr, A.N.; Austin, J.D. Rigorous approaches to species delimitation have significant implications for African crocodilian systematics and conservation. Proc. Biol. Sci. 2013, 281, 20132483. [Google Scholar] [CrossRef]
- Srikulnath, K.; Thapana, W.; Muangmai, N. Role of Chromosome Changes in Crocodylus Evolution and Diversity. Genom. Inform. 2015, 13, 102. [Google Scholar] [CrossRef]
- Martin, S. Global diversity of crocodiles (Crocodilia, Reptilia) in freshwater. Hydrobiologia 2008, 595, 587–591. [Google Scholar] [CrossRef]
- Uetz, P.; Freed, P.; Aguilar, R.; Hošek, J. The Reptile Database. Available online: http://www.reptile-database.org (accessed on 31 August 2022).
- Cohen, M.M.; Clark, H.F. The somatic chromosomes of five crocodilian species. Cytogenetics 1967, 6, 193–203. [Google Scholar] [CrossRef]
- Cohen, M.M.; Gans, C. The chromosomes of the order Crocodilia. Cytogenetics 1970, 9, 81–105. [Google Scholar] [CrossRef]
- Kasai, F.; O’Brien, P.C.M.; Martin, S.; Ferguson-Smith, M.A. Extensive homology of chicken macrochromosomes in the karyotypes of Trachemys scripta elegans and Crocodylus niloticus revealed by chromosome painting despite long divergence times. Cytogenet. Genome Res. 2012, 136, 303–307. [Google Scholar] [CrossRef]
- Kawagoshi, T.; Nishida, C.; Ota, H.; Kumazawa, Y.; Endo, H.; Matsuda, Y. Molecular structures of centromeric heterochromatin and karyotypic evolution in the Siamese crocodile (Crocodylus siamensis) (Crocodylidae, Crocodylia). Chromosom. Res. 2008, 16, 1119–1132. [Google Scholar] [CrossRef]
- Oliveira, V.C.S.; Altmanová, M.; Viana, P.F.; Ezaz, T.; Bertollo, L.A.C.; Ráb, P.; Liehr, T.; Al-Rikabi, A.; Feldberg, E.; Hatanaka, T.; et al. Revisiting the Karyotypes of Alligators and Caimans (Crocodylia, Alligatoridae) after a Half-Century Delay: Bridging the Gap in the Chromosomal Evolution of Reptiles. Cells 2021, 10, 1397. [Google Scholar] [CrossRef]
- King, M.; Honeycutt, R.; Contreras, N. Chromosomal repatterning in crocodiles: C, G and N-banding and the in situ hybridization of 18S and 26S rRNA cistrons. Genetica 1986, 70, 191–201. [Google Scholar] [CrossRef]
- Uno, Y.; Nishida, C.; Tarui, H.; Ishishita, S.; Takagi, C. Inference of the Protokaryotypes of Amniotes and Tetrapods and the Evolutionary Processes of Microchromosomes from Comparative Gene Mapping. PLoS ONE 2012, 7, 53027. [Google Scholar] [CrossRef]
- Charlesworth, B.; Jarne, P.; Assimacopoulos, S. The distribution of transposable elements within and between chromosomes in a population of Drosophila melanogaster. III. Element abundances in heterochromatin. Genet. Res. 1994, 64, 183–197. [Google Scholar] [CrossRef]
- Elder, J.F.; Turner, B.J. Concerted evolution of repetitive DNA sequences in eukaryotes. Q. Rev. Biol. 1995, 70, 297–320. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Heslop-Harrison, J.S. Genomes, genes and junk: The large-scale organization of plant chromosomes. Trends Plant Sci. 1998, 3, 195–199. [Google Scholar] [CrossRef]
- Biscotti, M.A.; Canapa, A.; Forconi, M.; Olmo, E.; Barucca, M. Transcription of tandemly repetitive DNA: Functional roles. Chromosome Res. 2015, 23, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Miga, K.H. Completing the human genome: The progress and challenge of satellite DNA assembly. Chromosome Res. 2015, 23, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Nishibuchi, G.; Déjardin, J. The molecular basis of the organization of repetitive DNA-containing constitutive heterochromatin in mammals. Chromosome Res. 2017, 25, 77–87. [Google Scholar] [CrossRef]
- Arney, K.L.; Fisher, A.G. Epigenetic aspects of differentiation. J. Cell Sci. 2004, 117, 4355–4363. [Google Scholar] [CrossRef][Green Version]
- Hall, A.E.; Keith, K.C.; Hall, S.E.; Copenhaver, G.P.; Preuss, D. The rapidly evolving field of plant centromeres. Curr. Opin. Plant Biol. 2004, 7, 108–114. [Google Scholar] [CrossRef]
- Bloom, K. Centromere dynamics. Curr. Opin. Genet. Dev. 2007, 17, 151–156. [Google Scholar] [CrossRef]
- Hartley, G.; O’neill, R.J. Centromere Repeats: Hidden Gems of the Genome. Genes 2019, 10, 223. [Google Scholar] [CrossRef]
- Plohl, M.; Meštrović, N.; Mravinac, B. Satellite DNA evolution. Genome Dyn. 2012, 7, 126–152. [Google Scholar] [CrossRef]
- Ferreira, D.; Meles, S.; Escudeiro, A.; Mendes-da-Silva, A.; Adega, F.; Chaves, R. Satellite non-coding RNAs: The emerging players in cells, cellular pathways and cancer. Chromosome Res. 2015, 23, 479–493. [Google Scholar] [CrossRef]
- Grewal, S.I.S.; Elgin, S.C.R. Transcription and RNA interference in the formation of heterochromatin. Nature 2007, 447, 399–406. [Google Scholar] [CrossRef]
- Trofimova, I.; Krasikova, A. Transcription of highly repetitive tandemly organized DNA in amphibians and birds: A historical overview and modern concepts. RNA Biol. 2016, 13, 1246–1257. [Google Scholar] [CrossRef][Green Version]
- Plohl, M.; Luchetti, A.; Meštrović, N.; Mantovani, B. Satellite DNAs between selfishness and functionality: Structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 2008, 409, 72–82. [Google Scholar] [CrossRef]
- Garrido-Ramos, M.A. Satellite DNA: An Evolving Topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef]
- Mahfooz, S.; Singh, P.; Akhter, Y. A comparative study of microsatellites among crocodiles and development of genomic resources for the critically endangered Indian gharial. Genetica 2022, 150, 67–75. [Google Scholar] [CrossRef]
- Lemskaya, N.A.; Kulemzina, A.I.; Beklemisheva, V.R.; Biltueva, L.S.; Proskuryakova, A.A.; Hallenbeck, J.M.; Perelman, P.L.; Graphodatsky, A.S. A combined banding method that allows the reliable identification of chromosomes as well as differentiation of AT- and GC-rich heterochromatin. Chromosome Res. 2018, 26, 307–315. [Google Scholar] [CrossRef]
- Ames, D.; Murphy, N.; Helentjaris, T.; Sun, N.; Chandler, V. Comparative analyses of human single- and multilocus tandem repeats. Genetics 2008, 179, 1693–1704. [Google Scholar] [CrossRef][Green Version]
- Deryusheva, S.; Krasikova, A.; Kulikova, T.; Gaginskaya, E. Tandem 41-bp repeats in chicken and Japanese quail genomes: FISH mapping and transcription analysis on lampbrush chromosomes. Chromosoma 2007, 116, 519–530. [Google Scholar] [CrossRef]
- Kulak, M.; Komissarov, A.; Fillon, V.; Tsukanova, K.; Saifitdinova, A.; Galkina, S. Genome organization of major tandem repeats and their specificity for heterochromatin of macro- and microchromosomes in Japanese quail. Genome 2022, 65, 391–403. [Google Scholar] [CrossRef]
- Shang, W.H.; Hori, T.; Toyoda, A.; Kato, J.; Popendorf, K.; Sakakibara, Y.; Fujiyama, A.; Fukagawa, T. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 2010, 20, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T. Repetitive Elements in Humans. Int. J. Mol. Sci. 2021, 22, 2072. [Google Scholar] [CrossRef] [PubMed]
- Komissarov, A.S.; Gavrilova, E.V.; Demin, S.J.; Ishov, A.M.; Podgornaya, O.I. Tandemly repeated DNA families in the mouse genome. BMC Genom. 2011, 12, 531. [Google Scholar] [CrossRef] [PubMed]
- Biltueva, L.S.; Prokopov, D.Y.; Makunin, A.I.; Komissarov, A.S.; Kudryavtseva, A.V.; Lemskaya, N.A.; Vorobieva, N.V.; Serdyukova, N.A.; Romanenko, S.A.; Gladkikh, O.L.; et al. Genomic Organization and Physical Mapping of Tandemly Arranged Repetitive DNAs in Sterlet (Acipenser ruthenus). Cytogenet. Genome Res. 2017, 152, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Biltueva, L.S.; Prokopov, D.Y.; Romanenko, S.A.; Interesova, E.A.; Schartl, M.; Trifonov, V.A. Chromosome Distribution of Highly Conserved Tandemly Arranged Repetitive DNAs in the Siberian Sturgeon (Acipenser baerii). Genes 2020, 11, 1375. [Google Scholar] [CrossRef]
- Lamelas, L.; Arroyo, M.; Fernández, F.J.; Marchal, J.A.; Sánchez, A. Structural and Evolutionary Relationships in the Giant Sex Chromosomes of Three Microtus Species. Genes 2018, 9, 27. [Google Scholar] [CrossRef]
- Rovatsos, M.; Marchal, J.A.; Giagia-Athanasopoulou, E.; Sánchez, A. Molecular Composition of Heterochromatin and Its Contribution to Chromosome Variation in the Microtus thomasi/ Microtus atticus Species Complex. Genes 2021, 12, 807. [Google Scholar] [CrossRef]
- Vicari, M.R.; Nogaroto, V.; Noleto, R.B.; Cestari, M.M.; Cioffi, M.B.; Almeida, M.C.; Moreira-Filho, O.; Bertollo, L.A.C.; Artoni, R.F. Satellite DNA and chromosomes in Neotropical fishes: Methods, applications and perspectives. J. Fish Biol. 2010, 76, 1094–1116. [Google Scholar] [CrossRef]
- Vozdova, M.; Kubickova, S.; Martínková, N.; Galindo, D.J.; Bernegossi, A.M.; Cernohorska, H.; Kadlcikova, D.; Musilová, P.; Duarte, J.M.; Rubes, J. Satellite DNA in Neotropical Deer Species. Genes 2021, 12, 123. [Google Scholar] [CrossRef]
- Melters, D.P.; Bradnam, K.R.; Young, H.A.; Telis, N.; May, M.R.; Ruby, J.G.; Sebra, R.; Peluso, P.; Eid, J.; Rank, D.; et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013, 14, R10. [Google Scholar] [CrossRef]
- Feliciello, I.; Akrap, I.; Brajkovi, J.; Zlatar, I.; Ugarkovic, D. Satellite DNA as a driver of population divergence in the red flour beetle Tribolium castaneum. Genome Biol. Evol. 2014, 7, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Warburton, P.E.; Hasson, D.; Guillem, F.; Lescale, C.; Jin, X.; Abrusan, G. Analysis of the largest tandemly repeated DNA families in the human genome. BMC Genom. 2008, 9, 533. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Suzuki, T.; Nojiri, T.; Yamagata, T.; Namikawa, T.; Matsuda, Y. Characterization and chromosomal distribution of a novel satellite DNA sequence of Japanese quail (Coturnix coturnix japonica). J. Hered. 2000, 91, 412–415. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Nishida-Umehara, C.; Matsuda, Y. Characterization and chromosomal distribution of novel satellite DNA sequences of the lesser rhea (Pterocnemia pennata) and the greater rhea (Rhea americana). Chromosome Res. 2002, 10, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, Y.; Nishida-Umehara, C.; Tarui, H.; Kuroiwa, A.; Yamada, K.; Isobe, T.; Ando, J.; Fujiwara, A.; Hirao, Y.; Nishimura, O.; et al. Highly conserved linkage homology between birds and turtles: Bird and turtle chromosomes are precise counterparts of each other. Chromosome Res. 2005, 13, 601–615. [Google Scholar] [CrossRef]
- Kuraku, S.; Ishijima, J.; Nishida-Umehara, C.; Agata, K.; Kuratani, S.; Matsuda, Y. cDNA-based gene mapping and GC3 profiling in the soft-shelled turtle suggest a chromosomal size-dependent GC bias shared by sauropsids. Chromosome Res. 2006, 14, 187–202. [Google Scholar] [CrossRef]
- Shibusawa, M.; Nishibori, M.; Nishida-Umehara, C.; Tsudzuki, M.; Masabanda, J.; Griffin, D.K.; Matsuda, Y. Karyotypic evolution in the Galliformes: An examination of the process of karyotypic evolution by comparison of the molecular cytogenetic findings with the molecular phylogeny. Cytogenet. Genome Res. 2004, 106, 111–119. [Google Scholar] [CrossRef]
- Nishida-Umehara, C.; Tsuda, Y.; Ishijima, J.; Ando, J.; Fujiwara, A.; Matsuda, Y.; Griffin, D.K. The molecular basis of chromosome orthologies and sex chromosomal differentiation in palaeognathous birds. Chromosome Res. 2007, 15, 721–734. [Google Scholar] [CrossRef]
- Kumazawa, Y.; Nishida, M. Complete mitochondrial DNA sequences of the green turtle and blue-tailed mole skink: Statistical evidence for archosaurian affinity of turtles. Mol. Biol. Evol. 1999, 16, 784–792. [Google Scholar] [CrossRef]
- Janke, A.; Erpenbeck, D.; Nilsson, M.; Arnason, U. The mitochondrial genomes of the iguana (Iguana iguana) and the caiman (Caiman crocodylus): Implications for amniote phylogeny. Proc. Biol. Sci. 2001, 268, 623–631. [Google Scholar] [CrossRef]
- Rest, J.S.; Ast, J.C.; Austin, C.C.; Waddell, P.J.; Tibbetts, E.A.; Hay, J.M.; Mindell, D.P. Molecular systematics of primary reptilian lineages and the tuatara mitochondrial genome. Mol. Phylogenet. Evol. 2003, 29, 289–297. [Google Scholar] [CrossRef]
- Kumazawa, Y. Mitochondrial genomes from major lizard families suggest their phylogenetic relationships and ancient radiations. Gene 2007, 388, 19–26. [Google Scholar] [CrossRef]
- Yamada, K.; Nishida-Umehara, C.; Matsuda, Y. A new family of satellite DNA sequences as a major component of centromeric heterochromatin in owls (Strigiformes). Chromosoma 2004, 112, 277–287. [Google Scholar] [CrossRef]
- Kohany, O.; Gentles, A.J.; Hankus, L.; Jurka, J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinform. 2006, 7, 474. [Google Scholar] [CrossRef]
- Green, R.E.; Braun, E.L.; Armstrong, J.; Earl, D.; Nguyen, N.; Hickey, G.; Vandewege, M.W.; St John, J.A.; Capella-Gutiérrez, S.; Castoe, T.A.; et al. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 2014, 346, 1254449. [Google Scholar] [CrossRef]
- Novák, P.; Robledillo, L.Á.; Koblížková, A.; Vrbová, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic. Acids Res. 2017, 45, e111. [Google Scholar] [CrossRef]
- Chong, A.Y.; Kojima, K.K.; Jurka, J.; Ray, D.A.; Smit, A.F.A.; Isberg, S.R.; Gongora, J. Evolution and gene capture in ancient endogenous retroviruses—Insights from the crocodilian genomes. Retrovirology 2014, 11, 71. [Google Scholar] [CrossRef]
- Kasai, F.; O’Brien, P.C.M.; Ferguson-Smith, M.A. Reassessment of genome size in turtle and crocodile based on chromosome measurement by flow karyotyping: Close similarity to chicken. Biol. Lett. 2012, 8, 631–635. [Google Scholar] [CrossRef]
- Graphodatsky, A.S.; Yang, F.; Serdukova, N.; Perelman, P.; Zhdanova, N.S.; Ferguson-Smith, M.A. Dog chromosome-specific paints reveal evolutionary inter- and intrachromosomal rearrangements in the American mink and human. Cytogenet. Cell Genet. 2000, 90, 275–278. [Google Scholar] [CrossRef]
- Yang, F.; O’Brien, P.C.M.; Milne, B.S.; Graphodatsky, A.S.; Solanky, N.; Trifonov, V.; Rens, W.; Sargan, D.; Ferguson-Smith, M.A. A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics 1999, 62, 189–202. [Google Scholar] [CrossRef]
- Gladkikh, O.L.; Romanenko, S.A.; Lemskaya, N.A.; Serdyukova, N.A.; O’Brien, P.C.M.; Kovalskaya, J.M.; Smorkatcheva, A.V.; Golenishchev, F.N.; Perelman, P.L.; Trifonov, V.A.; et al. Rapid karyotype evolution in Lasiopodomys involved at least two autosome—Sex chromosome translocations. PLoS ONE 2016, 11, e0167653. [Google Scholar] [CrossRef]
- Seabright, M. A rapid banding technique for human chromosomes. Lancet 1971, 2, 971–972. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225. [Google Scholar] [CrossRef]
- Ijdo, J.W.; Wells, R.A.; Baldini, A.; Reeders, S.T. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991, 19, 4780. [Google Scholar] [CrossRef]
- Maden, B.E.H.; Dent, C.L.; Farrell, T.E.; Garde, J.; McCallum, F.S.; Wakeman, J.A. Clones of human ribosomal DNA containing the complete 18 S-rRNA and 28 S-rRNA genes. Characterization, a detailed map of the human ribosomal transcription unit and diversity among clones. Biochem. J. 1987, 246, 519–527. [Google Scholar] [CrossRef]
- Romanenko, S.A.; Biltueva, L.S.; Serdyukova, N.A.; Kulemzina, A.I.; Beklemisheva, V.R.; Gladkikh, O.L.; Lemskaya, N.A.; Interesova, E.A.; Korentovich, M.A.; Vorobieva, N.V.; et al. Segmental paleotetraploidy revealed in sterlet (Acipenser ruthenus) genome by chromosome painting. Mol. Cytogenet. 2015, 8, 90. [Google Scholar] [CrossRef]
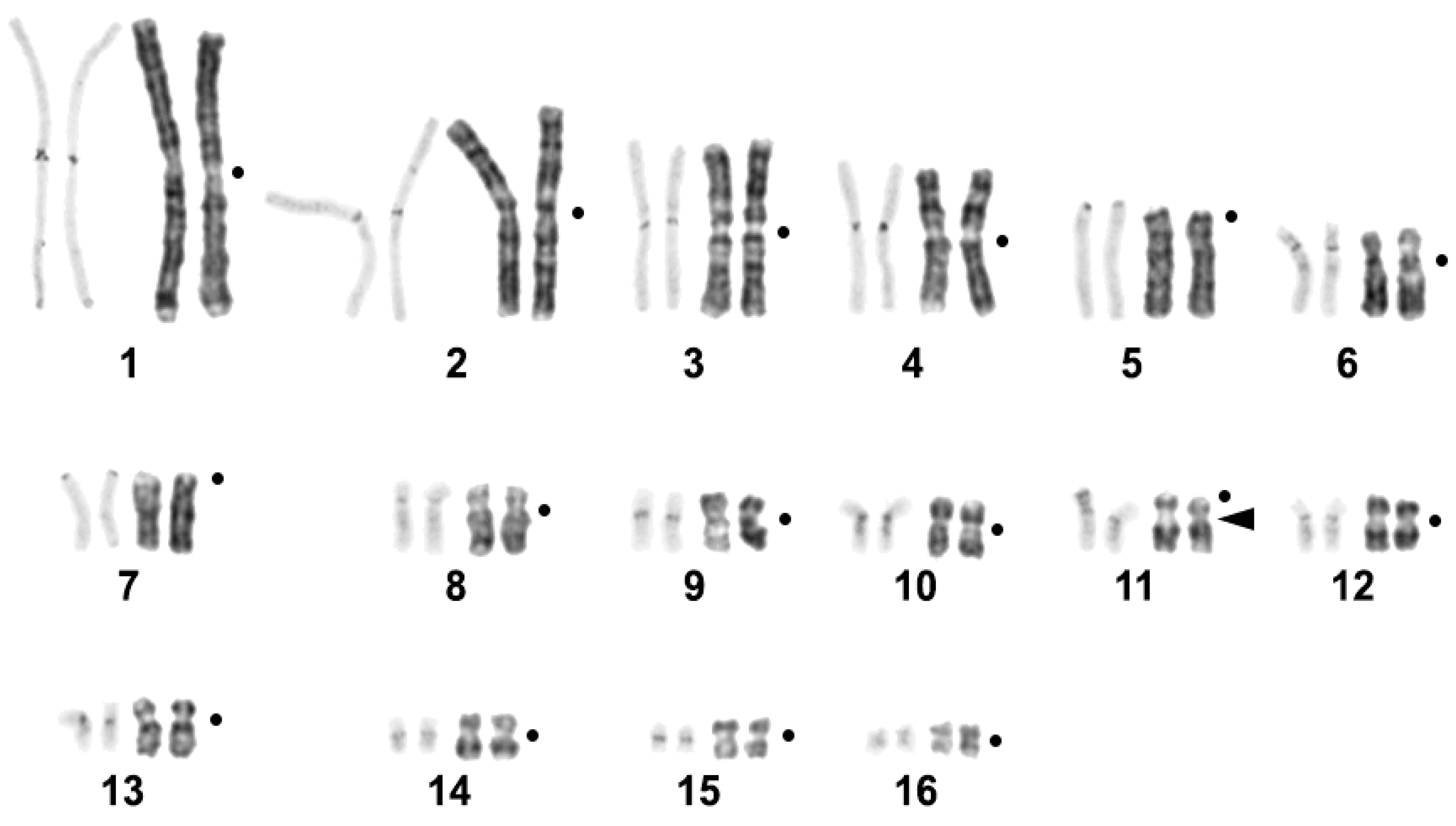
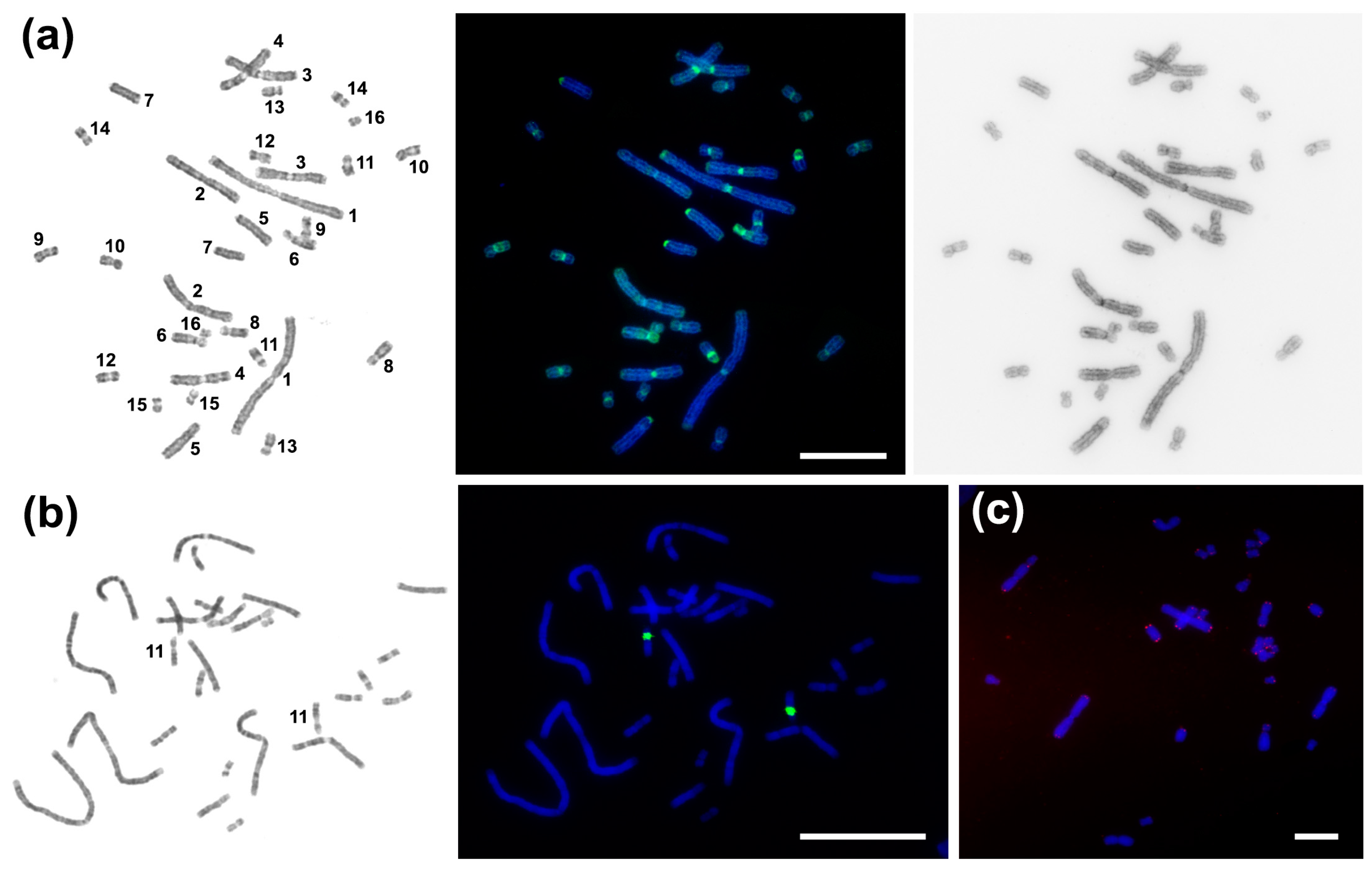
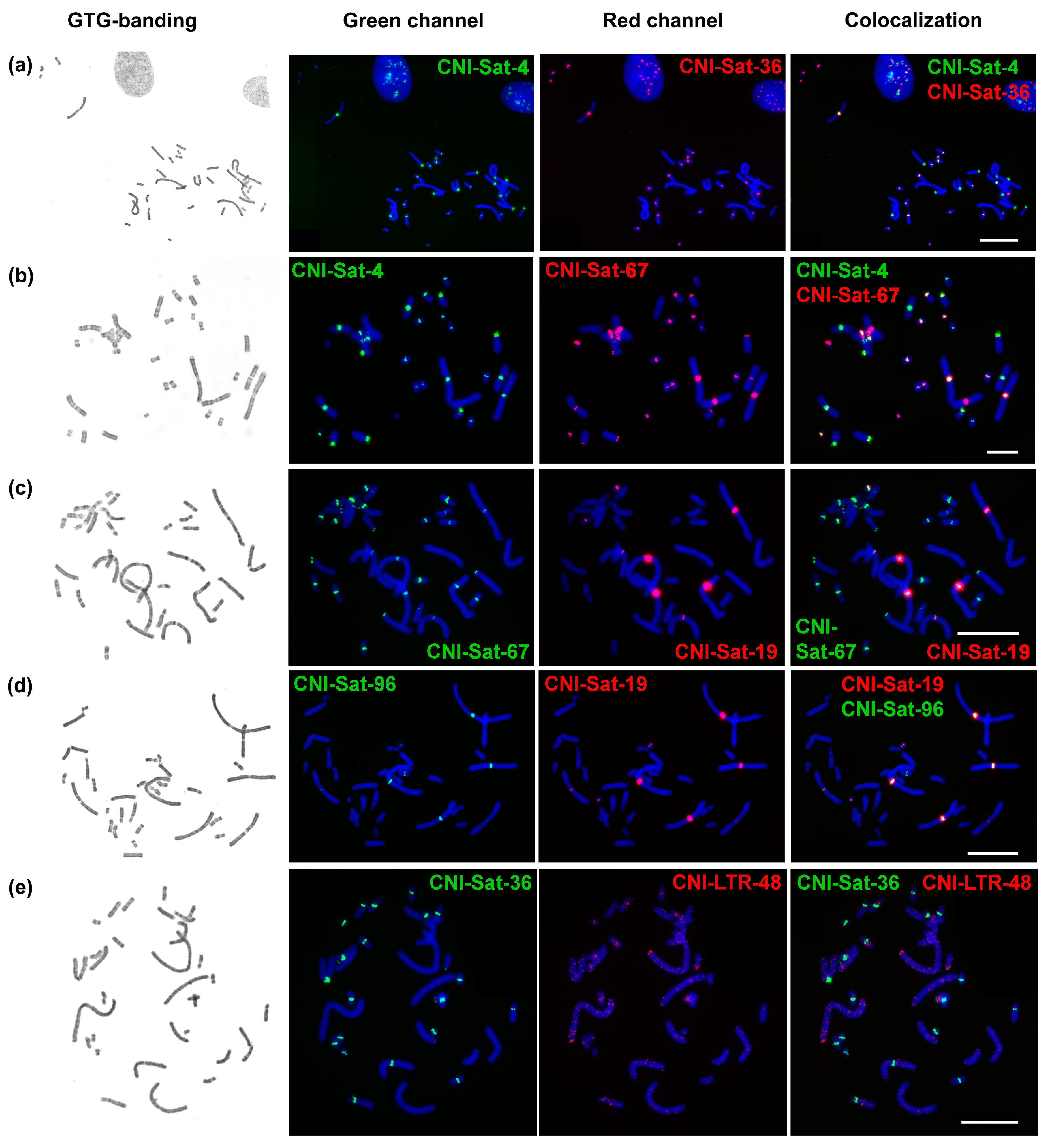
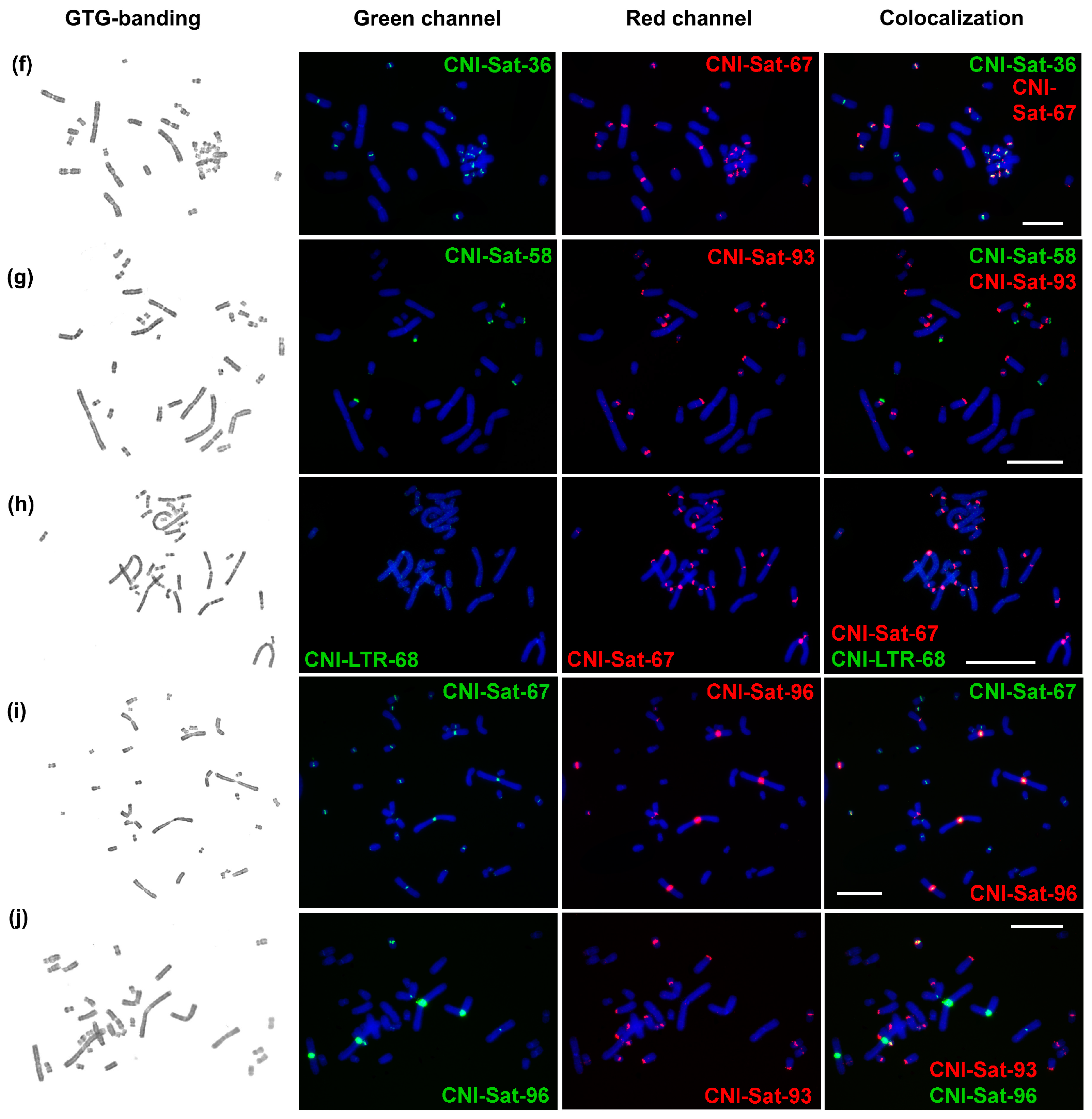

| Repetitive Sequence Name | Genome Proportion | Consensus Length | GC-Content | Accession Number | Homology to Known DNA (Best Hit) | Query Coverage | Percent Identity |
|---|---|---|---|---|---|---|---|
| CNI-Sat-4 | 0.59% | 40 | 65% | OP480173 | Crocodylus siamensis DNA, centromere-specific GC-rich repetitive sequences: clones CSI-HindIII-M02, CSI-HindIII-S40, CSI-HindIII-S07 (GenBank AB434504.1; AB434503.1; AB434500.1) | 100% | 97.5% |
| CNI-Sat-19 | 0.3% | 93 | 40.86% | OP480174 | Crocodylus siamensis DNA, centromere-specific repetitive sequences: clones CSI-DraI-01, CSI-DraI-05 (GenBank AB434505.1; AB434506.1) | 95% | 85.56% |
| CNI-Sat-36 | 0.24% | 101 | 60.4% | OP480175 | - | ||
| CNI-Sat-58 | 0.12% | 112 | 39.29% | OP480176 | Crocodylus porosus, satellite repeat, SAT-2_Crp | ~98% | |
| CNI-Sat-67 | 0.1% | 94 | 44.68% | OP480177 | Crocodylus siamensis DNA, centromere-specific repetitive sequences: clone CSI-DraI-05 (GenBank AB434506.1) | 79% | 90.67% |
| CNI-Sat-93 | 0.05% | 31 | 67.74% | OP480178 | - | - | - |
| CNI-Sat-96 | 0.046% | 162 | 37.65% | OP480179 | Oikopleura dioica, part of Gypsy-9_OD-I | - | 71.7% |
| CNI-LTR-48 | 0.17% | 576 | 40.97% | OP480180 | Crocodylus porosus, ERV1-2B_Crp-LTR | - | 85.64% |
| CNI-LTR-68 | 0.097% | 5414 | 50.68% | OP480181 | Pelodiscus sinensis, ERV1-2_PSi-I | - | 80.68% |
| Satellites Revealed in Crocodylus niloticus Genome | Alligator sinensis (GCA_000455745.1) | Alligator mississippiensis (GCA_000281125.4) | Gavialis gangeticus (GCA_001723915.1) | Crocodylus porosus (GCA_000768395.2) |
|---|---|---|---|---|
| CNI-Sat-4 | 84.62% | 89.47% | 87.18% | 100% |
| CNI-Sat-19 | - | - | 74.68% | 94.62% |
| CNI-Sat-36 | 81.82% | 78.08% | 79.73% | 96.04% |
| CNI-Sat-58 | 83.81% | 84.26% | 88.89% | 97.27% |
| CNI-Sat-67 | - | - | - | 89.77% |
| CNI-Sat-93 | - | - | - | - |
| CNI-Sat-96 | 73.38% | 72.66% | 77.03% | 82.53% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romanenko, S.A.; Prokopov, D.Y.; Proskuryakova, A.A.; Davletshina, G.I.; Tupikin, A.E.; Kasai, F.; Ferguson-Smith, M.A.; Trifonov, V.A. The Cytogenetic Map of the Nile Crocodile (Crocodylus niloticus, Crocodylidae, Reptilia) with Fluorescence In Situ Localization of Major Repetitive DNAs. Int. J. Mol. Sci. 2022, 23, 13063. https://doi.org/10.3390/ijms232113063
Romanenko SA, Prokopov DY, Proskuryakova AA, Davletshina GI, Tupikin AE, Kasai F, Ferguson-Smith MA, Trifonov VA. The Cytogenetic Map of the Nile Crocodile (Crocodylus niloticus, Crocodylidae, Reptilia) with Fluorescence In Situ Localization of Major Repetitive DNAs. International Journal of Molecular Sciences. 2022; 23(21):13063. https://doi.org/10.3390/ijms232113063
Chicago/Turabian StyleRomanenko, Svetlana A., Dmitry Yu. Prokopov, Anastasia A. Proskuryakova, Guzel I. Davletshina, Alexey E. Tupikin, Fumio Kasai, Malcolm A. Ferguson-Smith, and Vladimir A. Trifonov. 2022. "The Cytogenetic Map of the Nile Crocodile (Crocodylus niloticus, Crocodylidae, Reptilia) with Fluorescence In Situ Localization of Major Repetitive DNAs" International Journal of Molecular Sciences 23, no. 21: 13063. https://doi.org/10.3390/ijms232113063
APA StyleRomanenko, S. A., Prokopov, D. Y., Proskuryakova, A. A., Davletshina, G. I., Tupikin, A. E., Kasai, F., Ferguson-Smith, M. A., & Trifonov, V. A. (2022). The Cytogenetic Map of the Nile Crocodile (Crocodylus niloticus, Crocodylidae, Reptilia) with Fluorescence In Situ Localization of Major Repetitive DNAs. International Journal of Molecular Sciences, 23(21), 13063. https://doi.org/10.3390/ijms232113063








