FtbZIP12 Positively Regulates Responses to Osmotic Stress in Tartary Buckwheat
Abstract
:1. Introduction
2. Results
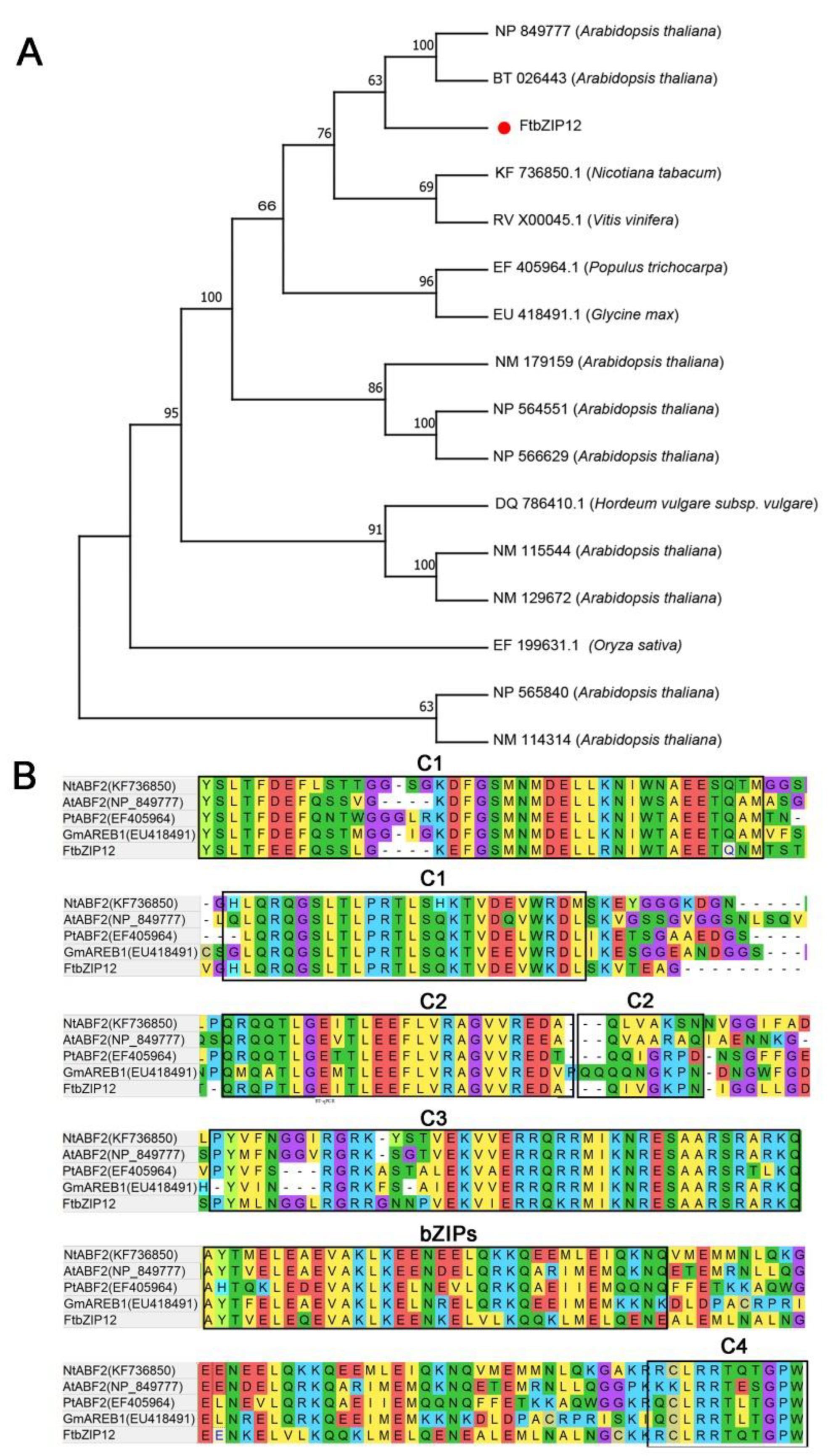
2.1. Isolation and Characterization of FtbZIP12
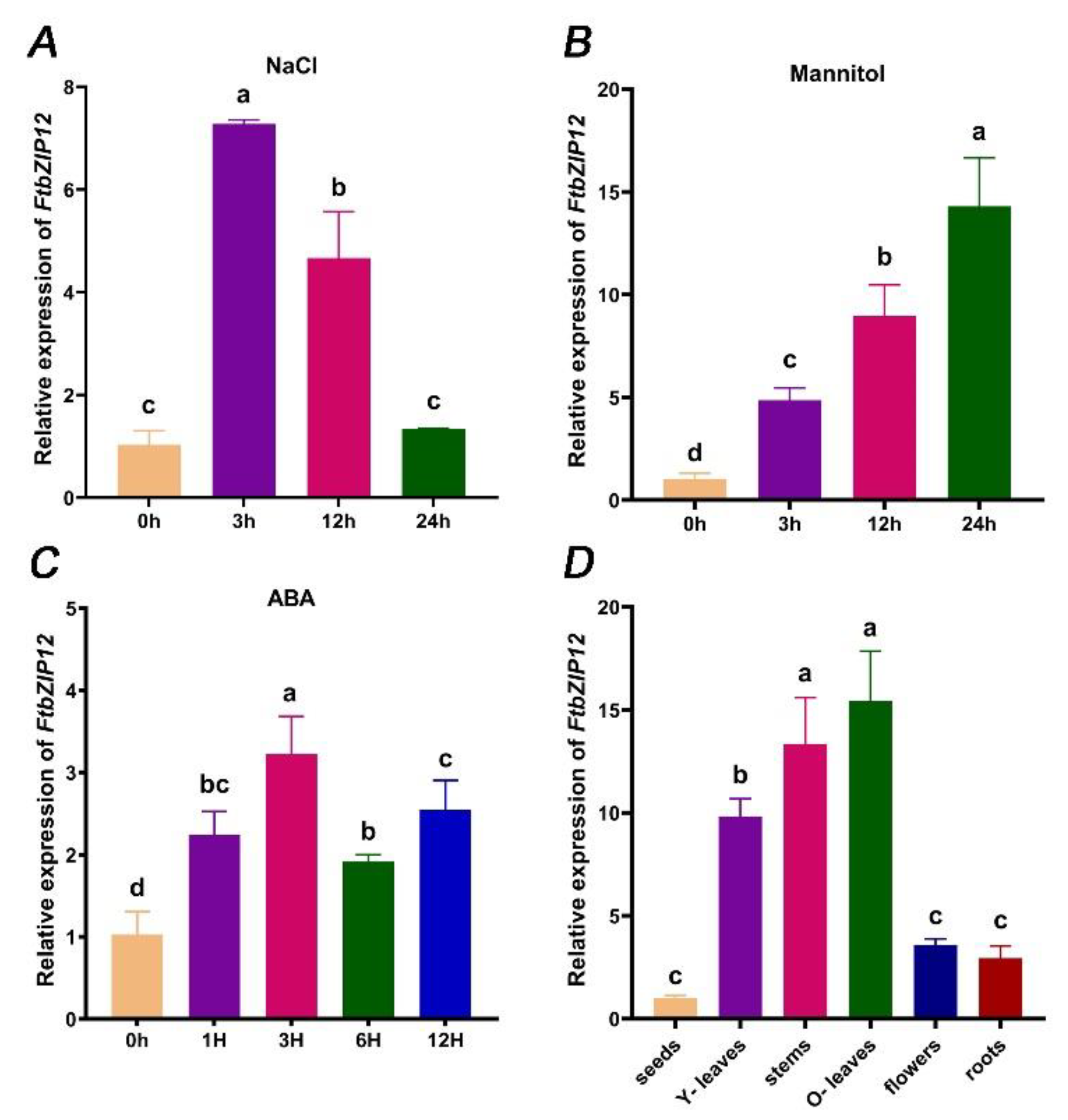
2.2. FtbZIP12 Expression Is Induced by Abiotic Stress Stimuli
2.3. Tissue-Specific Analysis of FtbZIP12
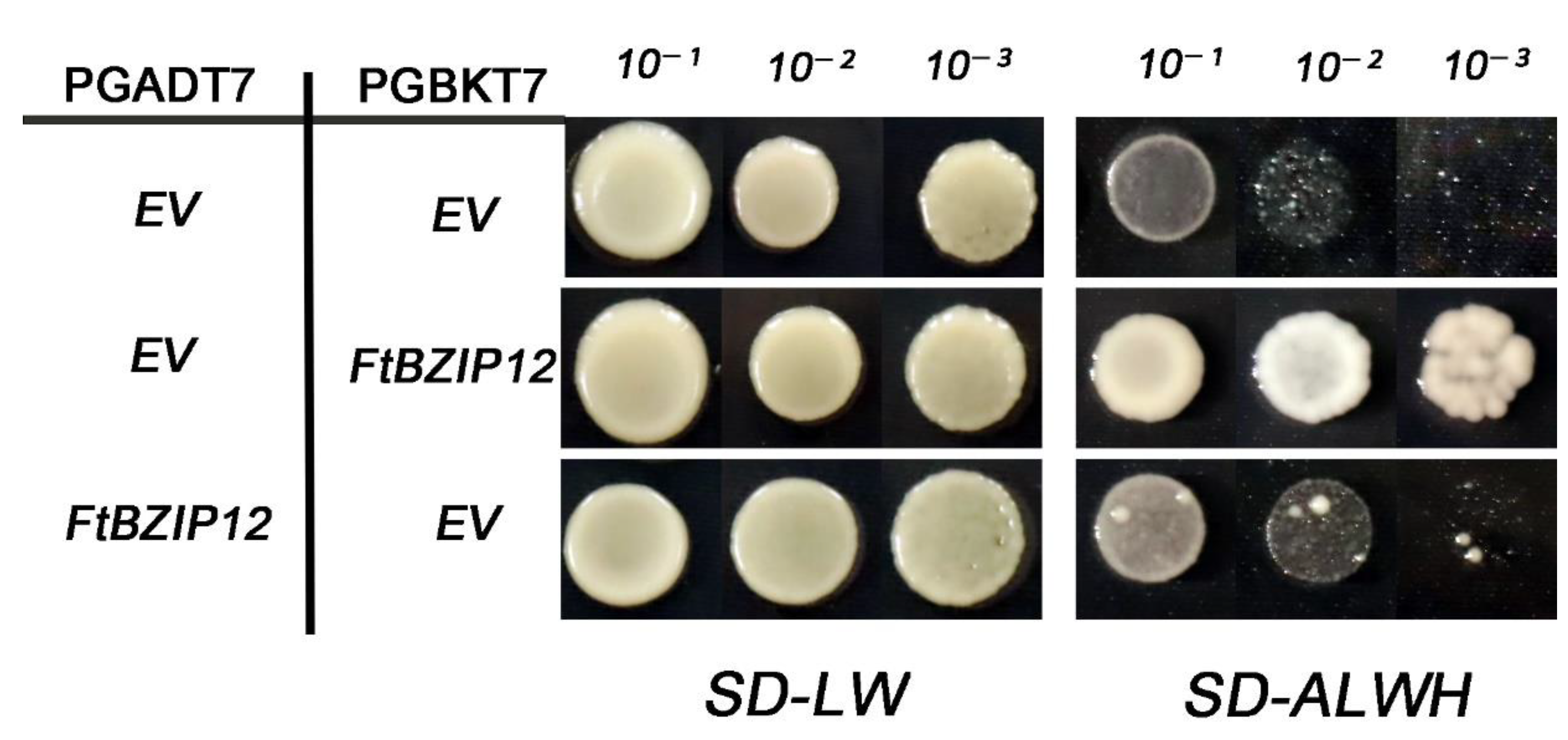
2.4. FtbZIP12 Has Transactivation Activity
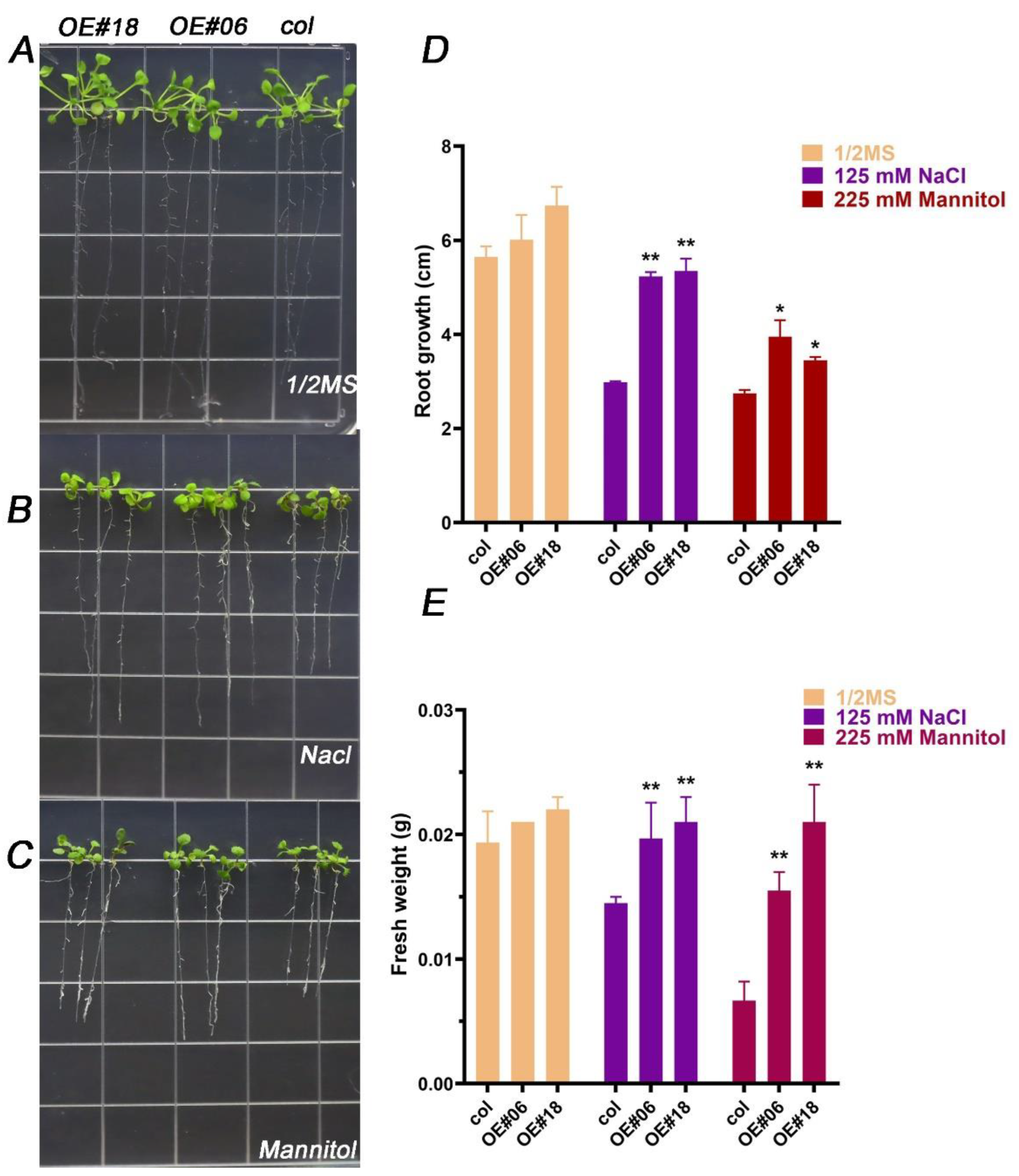
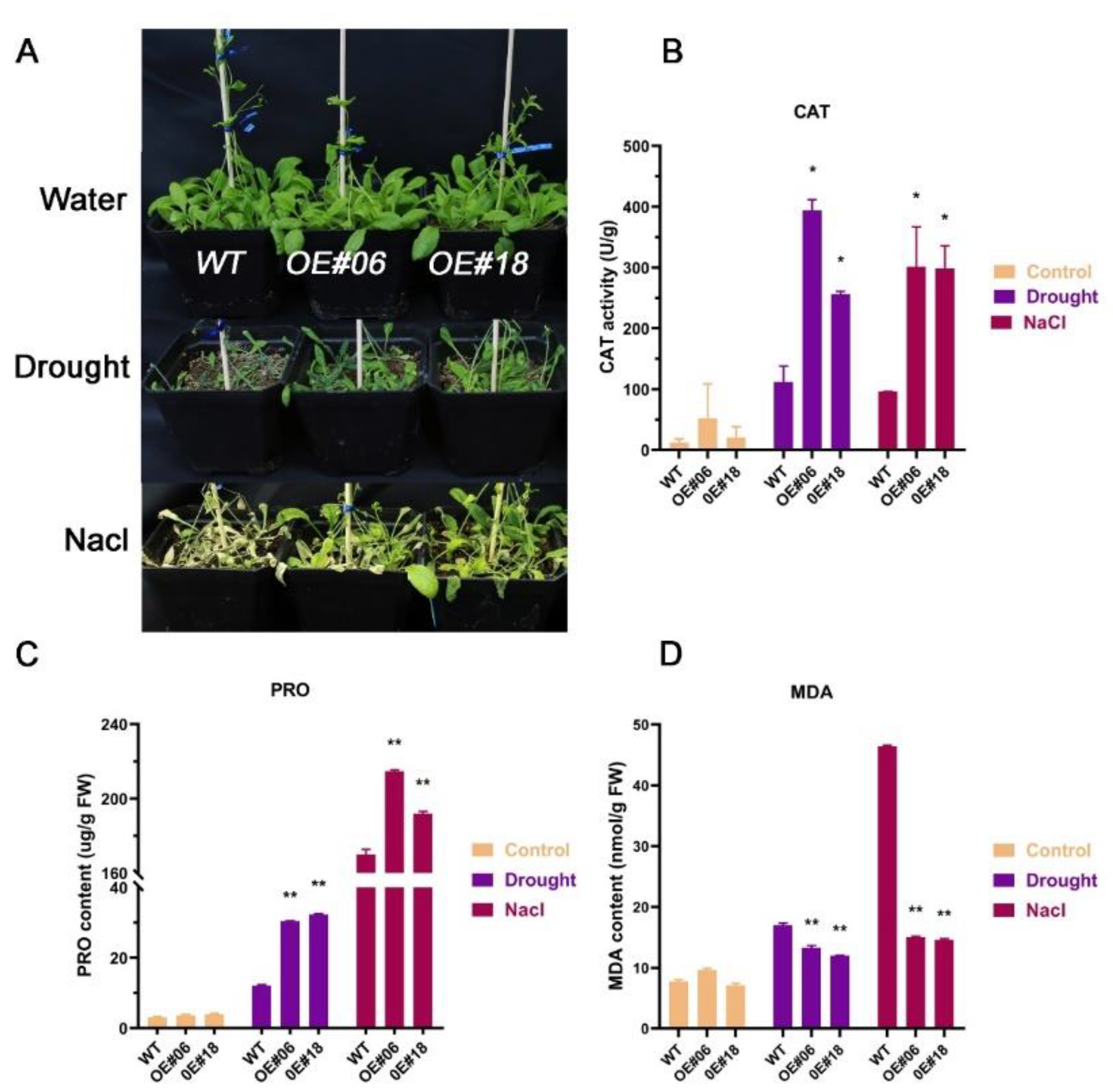
2.5. FtbZIP12 Confers Osmotic Stress Tolerance to Arabidopsis
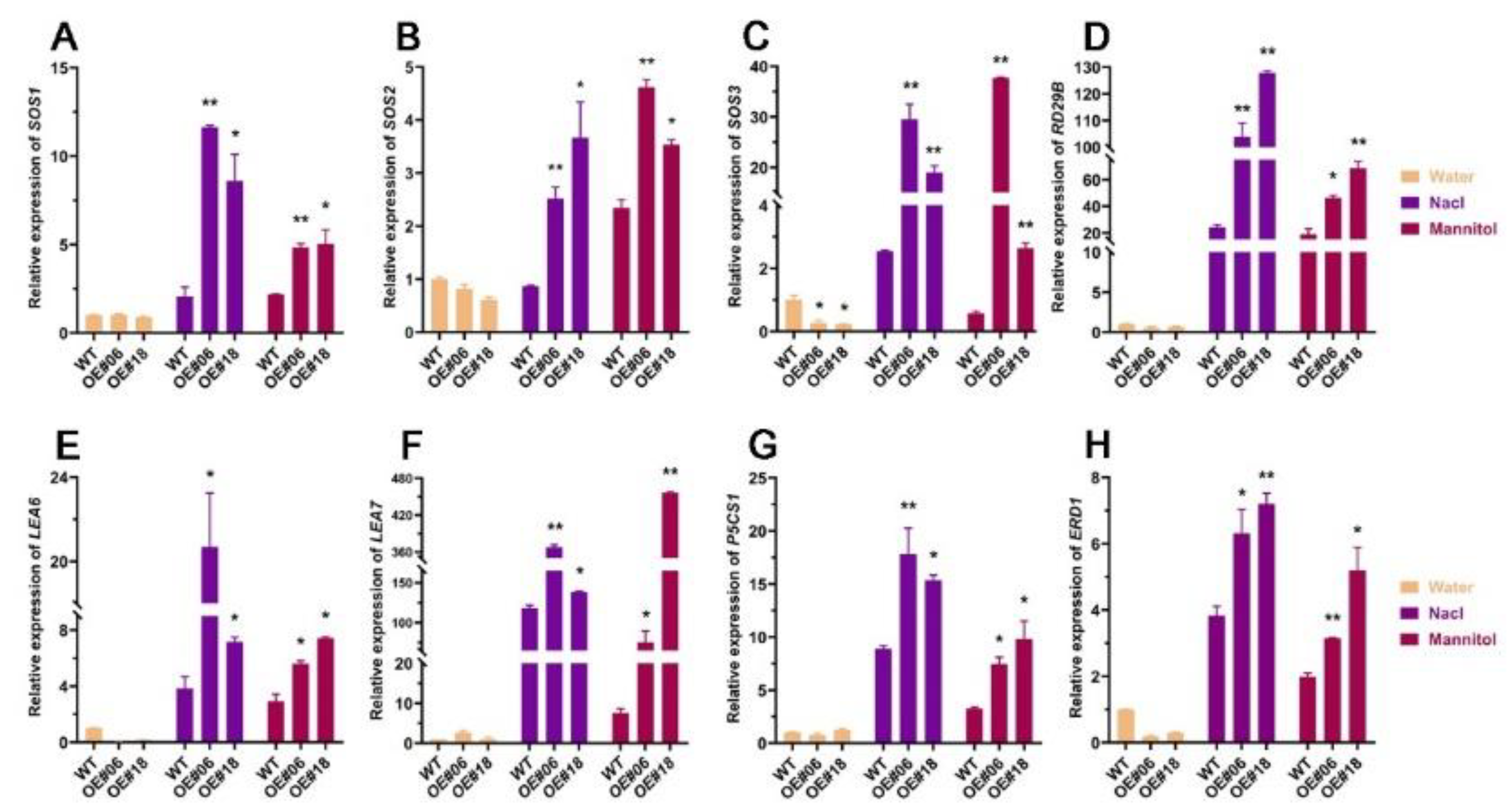
2.6. Analysis of the Changes in the Expression Levels of Genes Downstream of FtbZIP12 Related to Stress
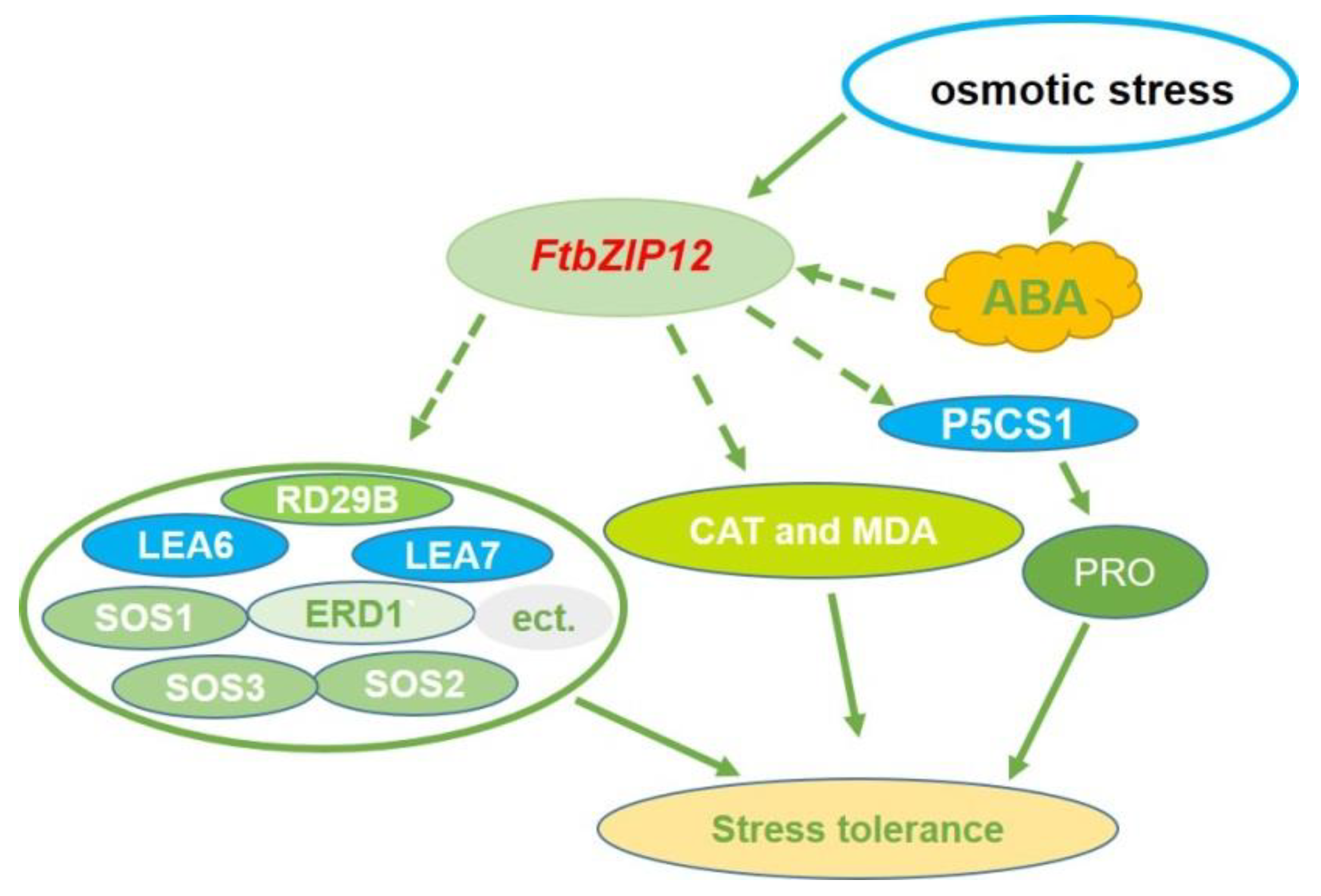
3. Discussion
4. Materials and methods
4.1. Plant Growth and Treatments
4.2. Vector Construction and Plant Transformation
4.3. Analysis of the Transactivation of FtbZIP12
4.4. Stress-Induced Treatment of Buckwheat and Arabidopsis
4.5. Real-Time PCR Analysis
4.6. Stress Tolerance Assays of Transgenic Arabidopsis
4.7. Bioinformatics Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Z.; Wang, Z.; Nie, X.; Qu, M.; Zhao, H.; Ji, X.; Wang, Y. UNFERTILIZED EMBRYO SAC 12 phosphorylation plays a crucial role in conferring salt tolerance. Plant Physiol. 2022, 188, 1385–1401. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 2017, 254, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Hurst, H.C. Transcription factors. 1: bZIP proteins. Protein Profile 1995, 2, 101–168. [Google Scholar] [PubMed]
- Mitchell, P.J.; Tjian, R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 1989, 245, 371–378. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Guo, C.; Peng, J.; Li, C.; Wan, F.; Zhang, S.; Zhou, Y.; Yan, Y.; Qi, L.; Sun, K.; et al. ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol. 2019, 221, 341–355. [Google Scholar] [CrossRef] [Green Version]
- Gao, S.; Gao, J.; Zhu, X.; Song, Y.; Li, Z.; Ren, G.; Zhou, X.; Kuai, B. ABF2, ABF3, and ABF4 Promote ABA-Mediated Chlorophyll Degradation and Leaf Senescence by Transcriptional Activation of Chlorophyll Catabolic Genes and Senescence Associated Genes in Arabidopsis. Mol. Plant. 2016, 9, 1272–1285. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, J.; Lee, H.; Lee, J.; Kim, H.; Ryu, H. ABSCISIC ACID-INSENSITIVE 3 is involved in brassinosteroid-mediated regulation of flowering in plants. Plant Physiol. Biochem. 2019, 139, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Chen, F.; Zhou, W.; Luo, X.; Dai, Y.; Shuai, H.; Yang, W. ABI4 regulates the floral transition independently of ABI5 and ABI3. Mol. Biol. Rep. 2018, 45, 2727–2731. [Google Scholar] [CrossRef]
- Shu, K.; Chen, Q.; Wu, Y.; Liu, R.; Zhang, H.; Wang, S.; Tang, S.; Yang, W.; Xie, Q. ABSCISIC ACID-INSENSITIVE 4 negatively regulates flowering through directly promoting Arabidopsis FLOWERING LOCUS C transcription. J. Exp. Bot. 2016, 67, 195–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amir Hossain, M.; Lee, Y.; Cho, J.I.; Ahn, C.H.; Lee, S.K.; Jeon, J.S.; Kang, H.; Lee, C.H.; An, G.; Park, P.B. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol. 2010, 72, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, C.; Liu, J.; Lv, Y.; Yu, C.; Li, H.; Zhao, T.; Liu, B. The OsABF1 transcription factor improves drought tolerance by activating the transcription of COR413-TM1 in rice. J. Exp. Bot. 2017, 68, 4695–4707. [Google Scholar] [CrossRef] [Green Version]
- Fukumoto, T.; Kano, A.; Ohtani, K.; Inoue, M.; Yoshihara, A.; Izumori, K.; Tajima, S.; Shigematsu, Y.; Tanaka, K.; Ohkouchi, T.; et al. Phosphorylation of D-allose by hexokinase involved in regulation of OsABF1 expression for growth inhibition in Oryza sativa L. Planta 2013, 237, 1379–1391. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Zhao, T.; Gomez, A.; Li, C.; Yu, C.; Li, H.; Lin, J.; Yang, Y.; Liu, B.; et al. A Drought-Inducible Transcription Factor Delays Reproductive Timing in Rice. Plant Physiol. 2016, 171, 334–343. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Yuan, F.; Wang, M.; Guo, A.; Zhang, Y.; Xie, C.G. Molecular characterization of an ABA insensitive 5 orthologue in Brassica oleracea. Biochem. Biophys. Res. Commun. 2012, 430, 1140–1146. [Google Scholar] [CrossRef]
- Liu, J.; Chen, X.; Wang, S.; Wang, Y.; Ouyang, Y.; Yao, Y.; Li, R.; Fu, S.; Hu, X.; Guo, J. MeABL5, an ABA Insensitive 5-Like Basic Leucine Zipper Transcription Factor, Positively Regulates MeCWINV3 in Cassava (Manihot esculenta Crantz). Front. Plant Sci. 2019, 10, 772. [Google Scholar] [CrossRef]
- Zou, L.; Wu, D.; Ren, G.; Hu, Y.; Peng, L.; Zhao, J.; Garcia-Perez, P.; Carpena, M.; Prieto, M.A.; Cao, H.; et al. Bioactive compounds, health benefits, and industrial applications of Tartary buckwheat (Fagopyrum tataricum). Crit. Rev. Food Sci. Nutr. 2021, 19, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.J.; Zhou, Y.X.; Yan, J.; Zhou, M.L.; Woo, S.H.; Weng, W.F.; Cheng, J.P.; Zhang, K.X. Tartary Buckwheat: An Under-utilized Edible and Medicinal Herb for Food and Nutritional Security. Food Rev. Int. 2020, 38, 440–454. [Google Scholar] [CrossRef]
- Giupponi, L.; Borgonovo, G.; Panseri, S.; Giorgi, A. Multidisciplinary study of a little known landrace of Fagopyrum tataricum Gaertn. of Valtellina (Italian Alps). Genet. Resour. Crop Evol. 2019, 66, 783–796. [Google Scholar] [CrossRef]
- Aubert, L.; Konrádová, D.; Barris, S.; Quinet, M. Different drought resistance mechanisms between two buckwheat species Fagopyrum esculentum and Fagopyrum tataricum. Plant Physiol. 2020, 172, 577–586. [Google Scholar] [CrossRef]
- Liu, M.; Wen, Y.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification, phylogeny, evolutionary expansion and expression analyses of bZIP transcription factor family in tartaty buckwheat. BMC Genom. 2019, 20(1), 483. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Wu, Q.; Wang, A.; Lv, B.; Dong, Q.; Yao, Y.; Wu, Q.; Zhao, H.; Li, C.; Chen, H.; et al. Tartary buckwheat transcription factor FtbZIP83 improves the drought/salt tolerance of Arabidopsis via an ABA-mediated pathway. Plant Physiol. Biochem. 2019, 144, 312–323. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, H.; Wang, X.; Kang, J.; Lv, B.; Dong, Q.; Li, C.; Chen, H.; Wu, Q. Tartary Buckwheat Transcription Factor FtbZIP5, Regulated by FtSnRK2.6, Can Improve Salt/Drought Resistance in Transgenic arabidopsis. Int. J. Mol. Sci. 2020, 21, 1123. [Google Scholar] [CrossRef] [Green Version]
- Brindha, C.; Vasantha, S.; Raja, A.K.; Tayade, A.S. Characterization of the Salt Overly Sensitive pathway genes in sugarcane under salinity stress. Physiol. Plant 2021, 171, 677–687. [Google Scholar] [CrossRef]
- Gao, S.Q.; Chen, M.; Xu, Z.S.; Zhao, C.P.; Li, L.; Xu, H.J.; Tang, Y.M.; Zhao, X.; Ma, Y.Z. The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol. Biol. 2011, 75, 537–553. [Google Scholar] [CrossRef]
- Miyamoto, K.; Nishizawa, Y.; Minami, E.; Nojiri, H.; Yamane, H.; Okada, K. Overexpression of the bZIP transcription factor OsbZIP79 suppresses the production of diterpenoid phytoalexin in rice cells. J. Plant Physiol. 2015, 173, 19–27. [Google Scholar]
- Liu, J.; Chen, N.; Chen, F.; Cai, B.; Dal Santo, S.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.M. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Xu, Q.; Kong, J.; Peng, X.; Zhou, W.; Chen, L.; Wu, J.; Xiang, Y.; Jiang, H.; Cheng, B. Overexpression of ZmbZIP22 gene alters endosperm starch content and composition in maize and rice. Plant Sci. 2019, 283, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kang, J.Y.; Cho, D.I.; Park, J.H.; Kim, S.Y. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2005, 40, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yama-guchi-Shinozaki, K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 2005, 17, 3470–3488. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Hu, Y.; Wang, H.; Guo, Q.; Chen, Y.; Howe, G.A.; Yu, D. Molecular Mechanism Underlying the Synergetic Effect of Jasmonate on Abscisic Acid Signaling during Seed Germination in Arabidopsis. Plant Cell 2020, 32, 3846–3865. [Google Scholar] [CrossRef]
- Chen, X.; Hall, H.; Simpson, J.P.; Leon-Salas, W.D.; Ready, D.F.; Weake, V.M. Cytochrome b5 protects photoreceptors from light stress-induced lipid peroxidation and retinal degeneration. NPJ Aging Mech. Dis. 2017, 3, 18. [Google Scholar] [CrossRef] [Green Version]
- Sarker, U.; Oba, S. Catalase, superoxide dismutase and ascorbate-glutathione cycle enzymes confer drought tolerance of Amaranthus tricolor. Sci. Rep. 2018, 8, 16496. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Jiao, J.; Zhou, M.; Jin, Z.; Yu, Y.; Liang, M. Physiological and Transcriptional Responses of Industrial Rapeseed (Brassica napus) Seedlings to Drought and Salinity Stress. Int. J. Mol. Sci. 2019, 20, 5604. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Lu, G.; Hao, Y.; Guo, H.; Guo, Y.; Zhao, J.; Cheng, H. ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in transgenic cotton. Planta 2017, 246, 453–469. [Google Scholar] [CrossRef]
- Liu, J.; Chu, J.; Ma, C.; Jiang, Y.; Ma, Y.; Xiong, J.; Cheng, Z. Overexpression of an ABA-dependent grapevine bZIP transcription factor, VvABF2, enhances osmotic stress in Arabidopsis. Plant Cell Rep. 2019, 38, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.Y.; Zhang, S.J.; Ruan, M.Y.; Wang, Y.L.; Wang, C.Y. Analysis and application of RD29 genes in abiotic stress response. Acta Physiol. Plant. 2012, 34, 1239–1250. [Google Scholar] [CrossRef]
- Shrestha, A.; Cudjoe, D.K.; Kamruzzaman, M.; Siddique, S.; Fiorani, F.; Léon, J.; Naz, A.A. Abscisic acid-responsive element binding transcription factors contribute to proline synthesis and stress adaptation in Arabidopsis. J. Plant Physiol. 2021, 261, 153414. [Google Scholar] [CrossRef]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ anti-porter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Lee, B.H.; Wu, S.J.; Zhu, J.K. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.K. The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, L.; Yu, F.; Tian, M.; Liu, G.; Wu, Y.; Wu, Y.; Xia, R.; Pardo, J.M.; Guo, Y.; Xie, Q. ESCRT-I Component VPS23A Sustains Salt Tolerance by Strengthening the SOS Module in Arabidopsis. Mol. Plant 2020, 13, 1134–1148. [Google Scholar] [CrossRef]
- Verslues, P.E.; Bray, E.A. Role of abscisic acid (ABA) and Arabidopsis thaliana ABA-insensitive loci in low water potential-induced ABA and proline accumulation. J. Exp. Bot. 2006, 57, 201–212. [Google Scholar] [CrossRef] [Green Version]
- Verslues, P.E.; Sharma, S. Proline metabolism and its implications for plant-environment interaction. Arab. Book 2010, 8, e0140. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Verslues, P.E. Mechanisms independent of abscisic acid (ABA) or proline feedback have a predominant role in transcriptional regulation of proline metabolism during low water potential and stress recovery. Plant Cell Environ. 2010, 33, 1838–1851. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]

| Primer Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Functions |
|---|---|---|---|
| F3H | GTATAGCCCCCTAAATGCTCG | GGATTGTGAGATTGTTGCCTATCG | Actin |
| AtActin2 | GGAAAGGATCTGTACGGTAAC | TGTGAACGATTCCTGGAC | |
| AtRD29B | GAAACCAAAGATGAGTCGACAC | TTTTTCGTAAACCGGAGTCAAC | Stress-related genes |
| AtLEA6 | GCAGAAGCGAATATGGATATGC | CGGAGGATAAGTCGGATGATAG | |
| AtLEA7 | TCAAGAGTCCAAAGACAAGACA | GTATATTCAGCTGCATCGTGTG | |
| AtERD1 | CTTTCTCTATCAGCACGAAACG | CGGTGCGATATATTGACAATCC | |
| AtP5CS1 | AGCTTGATGACGTTATCGATCT | AGATTCCATCAGCATGACCTAG | |
| AtSOS1 | ATTTTGATGCAGTCAGTGGATG | GCAAGCAGATTCTAGTCTTTCG | |
| AtSOS2 | GCGAACTCAATGGGTTTTAAGT | CTTACGTCTACCATGAAAAGCG | |
| AtSOS3 | CCGGTCCATGAAAAAGTCAAAT | CTCTTTCAATTCTTCTCGCTCG | |
| FtbZIP12 | TGCCTCGAACACTAAGCCAG | ATGGGGACTAATATGAACTTCAAATC | qPCR |
| FtbZIP12 | ATGGGGACTAATATGAACTTCAAATC | TTACCATGGACCCGTCTGT | CDS |
| FtbZIP12 | gacttgaactcggtatctagaATGGGGACTAATATGAACTTCAAATC | cttgatatcgaattcctgcagTTACCATGGACCCGTCTGT | Homologous recombination |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weng, W.; Lu, X.; Zhou, M.; Gao, A.; Yao, X.; Tang, Y.; Wu, W.; Ma, C.; Bai, Q.; Xiong, R.; et al. FtbZIP12 Positively Regulates Responses to Osmotic Stress in Tartary Buckwheat. Int. J. Mol. Sci. 2022, 23, 13072. https://doi.org/10.3390/ijms232113072
Weng W, Lu X, Zhou M, Gao A, Yao X, Tang Y, Wu W, Ma C, Bai Q, Xiong R, et al. FtbZIP12 Positively Regulates Responses to Osmotic Stress in Tartary Buckwheat. International Journal of Molecular Sciences. 2022; 23(21):13072. https://doi.org/10.3390/ijms232113072
Chicago/Turabian StyleWeng, Wenfeng, Xiang Lu, Meiliang Zhou, Anjing Gao, Xin Yao, Yong Tang, Weijiao Wu, Chao Ma, Qing Bai, Ruiqi Xiong, and et al. 2022. "FtbZIP12 Positively Regulates Responses to Osmotic Stress in Tartary Buckwheat" International Journal of Molecular Sciences 23, no. 21: 13072. https://doi.org/10.3390/ijms232113072
APA StyleWeng, W., Lu, X., Zhou, M., Gao, A., Yao, X., Tang, Y., Wu, W., Ma, C., Bai, Q., Xiong, R., & Ruan, J. (2022). FtbZIP12 Positively Regulates Responses to Osmotic Stress in Tartary Buckwheat. International Journal of Molecular Sciences, 23(21), 13072. https://doi.org/10.3390/ijms232113072






