Abstract
Human skin is the largest organ and serves as the first line of defense against environmental factors. The human microbiota is defined as the total microbial community that coexists in the human body, while the microbiome refers to the collective genome of these microorganisms. Skin microbes do not simply reside on the skin but interact with the skin in a variety of ways, significantly affecting the skin barrier function. Here, we discuss recent insights into the symbiotic relationships between the microbiome and the skin barrier in physical, chemical, and innate/adaptive immunological ways. We discuss the gut-skin axis that affects skin barrier function. Finally, we examine the effects of microbiome dysbiosis on skin barrier function and the role of these effects in inflammatory skin diseases, such as acne, atopic dermatitis, and psoriasis. Microbiome cosmetics can help restore skin barrier function and improve these diseases.
1. Introduction
The skin is the outermost surface of the body, with a surface area of about 1 × 8 m2. The primary function of the skin is to form a physical, chemical, and immunological protective barrier between the body and the external environment. The skin is composed of stratified keratinized epithelium, which undergoes terminal differentiation to acquire a strong structure. The normal skin surface is an acidic, high-salt, dry, and aerobic environment, but the follicle-sebaceous units are relatively anaerobic and much richer in lipids [1]. The body is interconnected with a complex community of microbes including bacteria, fungi, and viruses that inhabit the body’s surfaces. These microorganisms and their surrounding environment are the microbiome. The commensal microbiome is essential for maintaining skin barrier function by participating in essential physiological processes that occur in the skin [1,2]. In particular, the microbiome participates in physical, chemical, microbial and both innate and adaptive immunological ways in performing skin barrier functions. These interrelationships are the product of a well-controlled and delicately balanced microbiome. However, continuous exposure to various extrinsic and intrinsic factors affects this balanced system, which can lead to pathophysiological problems, inducing inflammatory skin conditions such as infections, allergic diseases, and autoimmune disorders.
In this review, we organized recent studies on the skin microbiome and highlight the latest insights into the role of the microbiome in forming and strengthening the barrier function of the skin. We also illuminated the relationships between the intestinal microbiome and skin barriers. Finally, we examined the effects of microbiome dysbiosis on skin barrier function and its role in inflammatory skin diseases.
2. Composition of the Skin Microbiome
The skin microbiome is tremendously diverse in organism number and activity. The composition of the microbiome depends on the physiology of the skin, which is related to a microenvironment characterized by moisture, dryness, and sebaceous content. In addition, the skin microbiome undergoes temporal changes with age [3,4,5]. Finally, the microbiome is influenced by environmental factors, such as diet, antibiotic use, and obesity [4,6]. In this section, we discuss the composition of the microbiome of healthy individuals.
2.1. Site-Specific Composition of Microbiome
Recent sequencing studies have extensively mapped the species inhabiting the skin of various body parts. These studies have shown that the skin of each body part, driven by the hair follicle and gland composition resulting in distinct niches for bacterial growth, exhibits different microbe compositions. Sebaceous sites, such as the glabella and back, are dominated by lipophilic Cutibacterium species, which are closely associated with acne vulgaris [4]. Moist sites, such as the bends of the elbows and knees and toe webs, are largely colonized by Staphylococcus and Corynebacterium species and beta-Proteobacteria. Dry sites, such as volar forearm, are mainly colonized by Cutibacterium acnes (C. acnes) and Corynebacterium species [4].
In addition to bacteria, which are the most abundant microbial organisms on the skin, numerous fungal and virus communities inhabit the skin. The species of fungi differ in composition depending on the body part, not the physiological condition of the skin [5,6]. The Malassezia species predominates on the core body and arms, whereas the foot is colonized by a much greater fungal diversity, including Malassezia spp., Aspergillus spp., Cryptococcus spp., Rhodotorula spp., and Epicoccum spp. [5]. In contrast to bacteria and fungi, eukaryotic DNA viral composition, predominantly Polyomaviridae and Papillomaviridae, is specific to the individual rather than physiological state or anatomical site [3].
2.2. Temporal Changes of the Composition of the Microbiome
Skin microbe composition also experiences temporal changes throughout the life cycle. Formation of the skin microbiome is presumed to start in the fetus. The presence of DNA in the microbiome, such as that of Cutibacterium and Staphylococcus species, has been mapped in amniotic fluid [7]. During normal delivery, the neonate’s skin is exposed to the microbes of the birth canal. Next, the neonate’s skin microbiome is influenced by the interaction with the external environment. One study found that a neonate’s skin microbiome is strongly influenced by the mother’s microbiome at birth, and that there are significant differences in both the skin and gut microbiomes between babies born naturally and those born by cesarean section [8,9]. A study by Maria G D. et al. revealed that using multiplexed 16S rRNA gene pyrosequencing, vaginally delivered infants acquired bacterial communities similar to maternal vaginal microbial communities dominated by Lactobacillus, Prevotella, or Sneathia spp., while C-section infants have skin bacterial communities resembling those found on the normal skin surface, dominated by Staphylococcus, Corynebacterium, and Cutibacterium spp. [8]. At puberty, the skin microbiome undergoes a major shift as sex hormones are secreted, driving sebaceous gland maturation and sebum production, transitioning toward a more adult microbiome. Lipophilic Cutibacterium species among bacteria and Malassezia among fungi predominate, with decreasing diversity during this period. Staphylococcus and Streptococcus species, which are more prominent in female children, gradually decrease [10].
After puberty, despite continuous exposure to the external environment and individual factors that are imposed on the skin, the microbial composition of the skin remains relatively stable within an individual over time [3]. The stability of these microbiomes is maintained by mutually beneficial symbiotic relationships between microorganisms and between microorganisms and the host. The relationship between C. acnes and pilosebaceous units play an important role in this stability. The pore structure of the pilosebaceous unit and its anaerobic environment provide a random single-cell bottleneck for C. acnes and enables stable colonization [11].
3. Skin Microbiome and Barrier Function
The skin microbiome and the skin barrier relate symbiotically to affect each other in physical, chemical, and immunological ways. The microbiome also interacts directly with pathogenic microorganisms encountered on the skin surface. In this section, we discuss microbial fortification of the skin barrier (Figure 1).
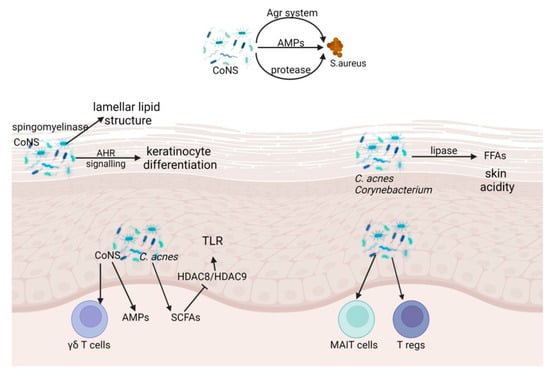
Figure 1.
Microbial, physical, chemical, and innate and adaptive immune barrier function of skin microbiome. CoNS, Coagulase negative Staphylococcus; Agr, auxiliary gene regulator; AMPs, antimicrobial peptides; AHR, aryl hydrocarbonate receptor; C. acnes, Cutibacterium acnes; SCFA, short-chain fatty acids; HDAC, histone deacetylases; MAIT, Mucosal-associated invariant T; T regs, regulatory T cells.
3.1. Physical Barrier
The physical barrier of the skin is the first barricade against external invasions. This barrier is formed by multiple fine layers of epidermal keratinocytes and undergoes a tightly controlled terminal differentiation to form the stratum corneum, which is influenced by the microbiome. Epidermal keratinocytes maintain close physical contingence to each other through a tight junction and adherens intersections, forming a nearly impermeable protective layer against pathogenic microorganisms. In addition, tight junction proteins, such as the zona occludens, can contribute to proliferation and differentiation of keratinocytes in the wound healing process, reconstructing the barrier to microorganisms [12]. Recent studies have shown that using germ free mice, the microbiota is essential for the epithelial barrier integrity and function of the skin barrier. These functions are mediated by the aryl hydrocarbon receptor (AHR) of keratinocytes. Mice lacking AHR are more vulnerable to barrier damage and infection. In skin damage, microbes produce metabolites that activate the AHR in keratinocytes, promoting epithelial differentiation and supporting epithelial integrity [13]. Another role of the microbiome is to secrete the components that make up the lipid structure. Using the mouse model, a study reported that S. epidermidis secretes a sphingomyelinase that helps the host to acquire essential nutrients for the bacteria and produce ceramide, a key component of the epithelial barrier that prevents skin dehydration and aging. In this study, S. epidermidis significantly increases skin ceramide levels and prevents water loss of damaged skin in a manner that is entirely dependent on sphingomyelinase [14].
3.2. Chemical Barrier
The chemical barrier of the skin is organized by numerous lipids and acids secreted by the epidermis and the microbiome. The microbial composition is similar in most areas of the skin but differs among sebaceous, moist and dry areas [2]. Both C. acnes and Corynebacterium, enriched in sebaceous regions, secrete lipases that hydrolyze free fatty acids from triglycerides in sebum [15,16]. Free fatty acids maintain a low pH, inhibiting the growth of pathogenic microorganisms. In addition, free fatty acids not only serve to directly inhibit bacteria, but also enhance skin immunity by stimulating the expression of human β-defensin 2 (hBD-2), one of the most abundant antimicrobial peptides (AMP)s in human skin [17]. The acidity of the stratum corneum is important for both permeability barrier formation and antimicrobial defense. The acidic skin surface creates a chemical environment that is hostile to pathogenic bacterial colonies. In addition, pathogenic microorganisms are directly inhibited by some lipids or free fatty acids. For example, sapienic acid from the stratum corneum can effectively inhibit pathogenic Staphylococcus aureus (S. aureus), but this acid does not have sufficient activity against Staphylococcus and Corynebacterium, which are major components of the skin microbiome [18]. Overall, the composition and function of the microbiome are complementary to the chemical barriers formed by lipids and fatty acids in the skin.
3.3. Innate Immune Barrier
In addition to the aforementioned barriers, the microbiome also stimulates a range of innate immune responses and maintains symbiotic relationships with the skin when the skin barrier is disturbed. For example, S. epidermidis modulates the innate immune system by activating γδ T cells and upregulating perforin-2, an AMP with unique properties against intracellular pathogens [19]. In addition, a specific glycan expressed in S. epidermidis is required for homeostatic T cell activation of S. epidermidis by interaction with C-type lectin in human immune cells [20]. Similarly, Candida albicans, which is a dimorphic fungus causing mucocutaneous and systemic infections, can stimulate T helper (Th) 1 or Th17 immune responses in the skin, protecting against cutaneous or systemic infection [21].
The skin microbiome modulates the production of a variety of innate factors, including interleukin 1a (IL-1α) [22], components of complement C5a receptor [23], and AMPs derived from keratinocytes and sebocytes, to enhance the innate immune system through a variety of mechanisms [24,25,26]. Representatively, AMP LL-37 (the active form of which is cleaved from the protein cathelicidin) is increased in response to the activation of Toll-like receptor (TLR)-2 signaling initiated by small molecules produced by S. epidermidis [27]. Stimulation from the commensal microbiome also induces members of the β-defensin family with bactericidal action against Escherichia coli and S. aureus from the skin [28]. Some functional keratinocytes of the skin appendages, such as sweat glands, sebaceous glands, and hair follicles contain various AMPs associated with a specific intrinsic microenvironment. While dermcidin may be a sweat gland-specific AMP, new evidence has suggested that this protein is also produced by the sebaceous glands of humans and mice [29].
The relationship between the host and the microbiome may vary depending on the oxygen content in site-specific skin areas and the resulting distribution of the microbiome. The facultative anaerobic bacterium C. acnes undergoes fermentation and generates short-chain fatty acids (SCFAs). C. acnes-derived SCFAs inhibit histone deacetylases (HDAC)-8 and -9 and promote the activation of fatty acid receptors, which induce activation of cytokines via the TLR-2 or TLR-3 signaling pathway [30,31]. Additionally, SCFAs can limit biofilm formation by S. epidermidis [32]. Hence, microbial metabolic and inflammatory backgrounds can trigger the unique characteristics of immune responses.
In addition, when the sebaceous gland is exposed to the gram-negative bacterial cell wall component lipopolysaccharide, the gland produces bactericidal molecules and small proline-rich proteins (SPRR)1 and SPRR2. Human SPRR has strong bactericidal activity that directly disturbs the pathogen’s negatively charged membrane [33]. Commensal bacteria can also act on lipids secreted from sebaceous glands and hydrolyze these lipids to free fatty acids (FFAs) [34]. FFAs have a unique antibacterial effect against various gram-positive bacteria and can induce sebaceous cells to upregulate the expression of hBD-2 [17]. Among these, sapienic acid has bactericidal activity against methicillin-resistant S. aureus (MRSA) [35].
3.4. Adaptive Immune Barrier
The relationship between adaptive immunity and microorganisms is especially important in developmental processes. This relationship supports the proposition that much of the complex’s adaptive immune system acts to maintain symbiosis with these microorganisms. Regulatory T cells (T regs) and innate-like/unconventional cells play an important role in this immunity. In a mouse model, exposure to the skin commensal S. epidermidis induced the abrupt accumulation of T regs on neonatal skin [36]. In these experiments, disruption of T reg responses to S. epidermidis in neonates resulted in barrier disruption, increasing inflammatory response upon secondary exposure to commensal microbes. In addition, the accumulation of T regs arises with coordination between hair follicle morphogenesis, which acts as the first reservoir of microbes, and the pathway of chemokine Ccl20-Ccr6 [37].
Mucosal-associated invariant T (MAIT) cells, along with invariant natural killer T cells and γδ T cells, are a type of evolutionarily archaic unconventional T cell that recognizes conserved antigen sets and molecules restricted by major histocompatibility class Ib (MHC-Ib) [38]. As MHC-Ib molecules can present antigens with specific chemical or amino acid sequence motifs, unconventional T cells under their control may be the most effective cells for detection and recognition of antigens and metabolites derived from the microbiome. MAIT cells are activated by MHC class I-like molecules, present in numerous commensal bacteria and yeasts, but not in mammalian cells. The MHC class I-like molecules include a folic acid (vitamin B9) metabolite and riboflavin (vitamin B2) [39]. Within the human skin, MAIT cells comprise a significant portion of lymphocytes, accounting for approximately 2% of CD3+ lymphocytes [40].
Skin microbes do not play as important a role as those of the intestine in optimal seeding of dermal T regs [22]. Even so, the continued action of T regs in the skin modulates responsiveness to symbionts after adulthood, as in the intestine. For example, in experiments using mouse models, commensal-specific T cells produce abnormal type 2 cytokines when there is a specific defect in T reg fitness in the skin. This may be the etiology of several inflammatory skin diseases [41]. A recent study reported that exposure to S. aureus in neonates increased IL-1β in response to S. aureus-associated α-toxin, which actively inhibited the induction of T regs [42]. Collectively, these findings suggest that microbes are critical in the construction and activation of immune cells.
3.5. Microbial Barrier
In addition to microbe-host relationships, microbe–microbe relationships act as a barrier against invasion, colonization, and infection by invading, pathogenic, or opportunistic microbes. The interactions maintain survival and sustain microbes’ niche and access to nutrients. While these relationships are not well understood, regulation of the coagulase-negative Staphylococcus (CoNS) species for S. aureus has been well studied [43]. CoNS species are among the most abundant microbes in the skin microbiome, including S. epidermidis, S. capitis, S. caprae, S. hominis, S. lugdunensis, and S. haemolyticus.
The most representative way in which CoNS inhibits S. aureus is through secretion of AMPs. For example, some strains of S. hominis secrete antibiotics in patients with atopic dermatitis (AD), inhibiting S. aureus colonization [32]. Additionally, S. lugdunensis in the nasal cavity produces lugdunin, a potent anti-S. aureus AMP. Lugdunin activates keratinocytes to release LL-37 and a chemoattractant, CXCL8/MIP-2, resulting in recruitment of neutrophils [44,45]. Phenol-soluble modulin secreted by various CoNS species directly antagonizes S. pyogenes, S. aureus, and C. acnes and enhances antimicrobial activity in cooperation with the host keratinocyte-derived AMP LL-37 [46,47]. Additionally, in a pilosebaceous unit, C. acnes secrete a competitive thiopeptide antibiotic, cutimycin, to maintain its niche against Staphylococcus species colonization [48].
In addition to AMPs, all staphylococci have an inter-microbial communication system called quorum sensing that operates through an auxiliary gene regulator (agr) system [49]. This system is required for S. aureus skin infection, colonization, damage, and inflammation, providing a target for commensal microbes to inhibit the growth and toxicity of pathogenic microbes. S. caprae showed improvement of infection by inhibiting methicillin-resistant S. aureus agr activity [50]. S. hominis and other CoNS interfered with the quorum sensing system of S. aureus, reducing toxin production and tissue damage and inflammation in an AD model [49].
Production of proteases is another effective mechanism by which CoNS species interfere with S. aureus toxicity. A subset of S. epidermidis can produce a serine protease that inhibits biofilm formation and colonization of S. aureus [51]. Corynebacterium accolens release corynebacterial lipase, which restricts the growth of S. pneumoniae [15]. Collectively, the competitive relationship between these microbes plays an important role in maintaining the balance of the skin microbiome.
4. Gut-Skin Axis and Skin Barrier Function
Cumulative evidence has demonstrated that skin and other barrier sites, like intestine, lung, and brain, have a bidirectional connection. Especially, the intestine and skin are both highly innervated and vascularized and interface with the external environment. The immune systems of both organs are consistently activated for homeostatic conditions, called the gut-skin axis [52]. In this regard, skin conditions can affect the intestinal microbiome. For example, exposure to narrow band ultraviolet (NB-UVB) light can inflect the intestinal microbiome. A study reported that after exposing the skin to NB-UVB, fecal microbiota composition analysis using 16S rRNA sequencing significantly increased alpha and beta diversity [53]. In this study, they also revealed that the serum 25(OH)D concentrations, which is associated with exposure to sunlight/UVB light, correlated with the relative abundance of the Lachnospiraceae, Lachnopsira and Fusicatenibacter genera [53]. Also, food allergies can be caused by exposure to epidermal protein in household dust, ultimately leading to immunoglobulin E-mediated mast cell expansion in the intestine [54]. Recent studies reported that scratching in a mouse model caused keratinocytes to release IL-33. This led to type 2 innate lymphoid cell secretion of IL-4 to activate mast cells of the small intestine that increases intestinal permeability and food anaphylaxis [55]. Another study suggested that the skin–gut correlation depends on the intestinal stromal fibroblast. In a mouse model of inflammatory bowel disease, wounding of the skin activated hyaluronan catabolism, resulting in alteration of colon fibroblast function, and subsequently increased inflammation in the colon by the production of AMPs and alteration of the fecal microbiome [56].
The gut microbiome can also have a significant impact on the overall homeostasis of the skin. The integrity of the intestinal barrier, along with the action of intestinal mucus, immune cells, immunoglobulin A, and AMPs, prevents the entrance of bacteria into the bloodstream, ultimately maintaining skin homeostasis. Through its influence on the signaling pathways that coordinate this process that is essential to skin homeostasis, the gut microbiome impacts integumentary health [57]. According to recent studies, the gut microbiome appears to be involved in skin barrier function by controlling the modulatory effect on systemic immunity. Retinoid acid include: Polysaccharide A, a metabolite of the gut microbe Bacteroides fragilis; Faecalibacterium prausnitzii; and Clostridium species that activate the accumulation of T regs [58]. Another important metabolite, SCFA, is also produced by gut microbes; this metabolite executes anti-inflammatory activities in the skin. Segmented filamentous bacteria stimulate pro-inflammatory Th17 and Th1 cells [59,60]. Additionally, a study with a psoriasis mouse model found that enhanced Th17 inflammation is promoted by the intestinal microbiome [61]. Furthermore, the gut microbiome and its metabolites may directly metastasize and affect the skin barrier directly. O’Neill et al. reported that gut microbes and their metabolites can be absorbed systemically when the intestinal barrier is disrupted in psoriatic patients and reach the skin and affect skin barrier function [57].
The constructive effects of intestinal bacteria supplementation on skin barrier function have been suggested in previous studies. In a study by Levkovich et al., mice receiving Lactobacillus reuteri supplementation showed increased dermal thickness, increased hair follicle formation, and increased sebocyte production compared to the control group [62]. In human clinical studies conducted by Ogawa et al. and Guéniche et al., oral supplementation with Lactobacillus induced a significant reduction in transepidermal water loss (TEWL), an indicator of skin barrier function. The reduction of TEWL has been shown to be due to an increase in circulating transforming growth factor beta (TGF-β), a cytokine important for maintaining skin barrier function [63,64].
The intestinal microbiome also contributes to repair damaged skin barrier function through both innate and adaptive immunity processes [65,66]. For example, several studies using an atopic dermatitis mouse model showed that oral supplementation with Lactobacillus increased general skin barrier function, including decrease in severity of disease, subsequent TEWL, and tumor necrosis factor alpha (TNF-α) release [67,68]. The gut microbiome may also have an effect on wound healing. Supplementation with lactobacillus decreased the time required for wound healing by triggering oxytocin to activate host T regs and rapidly remove neutrophils in wound sites [69].
5. Inflammatory Skin Disorders and Microbiome
5.1. Atopic Dermatitis (AD)
The microbial composition in skin with AD lesions is characterized by an increased abundance of S. aureus and reduced diversity compared with healthy skin and skin without lesions. Patients with more severe AD have decreased alpha- and beta-diversity on the skin. These decreases in diversity can also be seen with acute exacerbations of AD, with reductions in the genera Streptococcus, Corynebacterium, and Cutibacterium and the phylum Proteobacteria toward the genus Staphylococcus in general and S. aureus in particular [70]. S. aureus can overwhelm the symbiotic microbiome and induce exacerbation of AD [71]. S. aureus interferes with the host immune response, directly damage the skin barrier, and impair adaptive immunity. This is the result of alpha toxin production that causes IL-1R-mediated inflammation and limits the accumulation of S. aureus-specific T regs [42]. Therefore, changed microbiome metabolites in atopic skin can cause inflammation and maintain and worsen barrier dysfunction. In contrast, CoNS is usually dominant in the non-lesion skin of AD [72]. Nakatsuji et al. have shown that CoNS strains isolated from the skin of healthy individuals have a better ability to kill S. aureus compared to CoNS strains isolated from AD skin [73]. At least 10 proteases from S. aureus in AD lesional skin can facilitate dissolution and penetration through the stratum corneum. Additionally, S. aureus strains isolated from AD patients possess different surface proteins compared to common strains. For instance, Proinflammatory staphylococcal lipoproteins induce thymic stromal lymphopoietin expression in a TLR2/TLR6-dependent manner. These proteins affect skin adhesion and induce imbalanced Th1/Th2 adaptive immune responses via Langerhans cells [74].
Emerging studies have demonstrated that the gut–skin axis can be associated with allergic diseases including AD, supporting the “microbiome hypothesis” [75]. A metagenomic analysis of fecal samples from patients with AD showed a considerable decrease in Faecalibacterium prausnitzii species and SCFAs produced by Faecalibacterium prausnitzii compared with the controls. Data indicated the presence of a possible positive feedback loop. The altered gut microbiome allows penetration of poorly digested food, microbes, and toxins, and is associated with epithelial barrier dysfunction in AD tis patients. The altered microbiome triggers Th2 inflammation, resulting in further disruption of skin barrier functions [76]. Overgrowth of multiple Malassezia spp. (Malassezia furfur and Malassezia sympodialis) may cause AD pathogenesis [77]. A high proportion of AD patients present with a positive reaction to Malassezia allergens. There is increased levels of Malassezia-specific IgE in AD patients and a correlation of AD severity [78]. In particular, M. sympodialis can release extracellular vesicles, inducing IL-4 and TNF-α. M. sympodialis can also induce leukotrienes in IgE-sensitized mast cells and enhance IgE-mediated degranulation of mast cells.
Recently, probiotics and prebiotics have been studied as innovative treatments for skin diseases such as AD [79,80,81]. The results of the studies are still controversial. While some studies suggest improved symptoms, quality of life, and the clinical severity of AD, others have not been able to confirm these findings. The impact of topical treatment on the skin microbiome and consequent influence on AD have been explored in multiple studies. Studies have shown that long-term use of emollients increases the proportion of S. salivarius in infants at a high risk of AD [82]. Emollient-mediated microbiome changes can play a role in correcting the imbalance of the skin microbiome associated with AD and its prevention. In addition, several studies have shown that the use of microbiome-supplemented emollients can improve the symptoms and severity of AD compared to normal emollients. The strains tested in these studies are Vitreoscilla filiformis [83], Aquaphilus dolomiae [84], Lactobacillus reuteri [85], and Roseomonas mucosa [86]; these species increase the proportion of CoNS in the skin and inhibit colonization by S. aureus. The benefits of their use are decreased in pathogenic microbes, reduced production of toxic metabolites, and increased homeostasis of the skin barrier function.
5.2. Psoriasis
As mentioned in the previous section, the microbiome plays an important role in the Th17 immune response. The Th17 inflammatory pathway is key in the pathogenesis of psoriasis, suggesting that the microbiome plays an important role in the pathogenesis of psoriasis. However, previous studies on the microbiome in psoriasis have had inconsistent results, and difficulty arises in drawing a conclusion about the relationship between psoriasis and the microbiome [87]. Diverse studies of the microbiome of patients with psoriasis have reported controversial results on trends in Firmicutes, Actinobacteria, and Proteobacteria. These studies did not draw convincing conclusions on the diversity of the microbial community in skin affected by psoriasis lesions compared to that of healthy skin. In psoriatic lesions, abnormal colonization of S. aureus and a decrease in S. epidermidis abundance were consistently observed, although overt infection is rare [88,89]. Chang et al. reported that mice colonized with S. aureus demonstrated a strong Th17 polarization compared to mice colonized with S. epidermis [90]. Their results suggest that S. aureus can lead to Th17 inflammation observed in psoriasis.
The association between gut dysbiosis and psoriasis is relatively well understood. Host- and microbiome-derived factors lead to gut dysbiosis, resulting in a low-grade chronic inflammatory process [91]. For example, changes in gut microbiome composition can cause changes in levels of SCFAs, produced by Firmicutes and Bacteroidetes. It enhances Th17 inflammation, promoting psoriasis-like skin inflammation. Additionally, these factors cause a decrease in T reg levels in psoriasis patients, resulting in an imbalance between effector T cells and suppressor T cells [92]. Phenol, a metabolite of aromatic amino acids from a disturbed intestinal environment and considered a bioactive toxin, affects abnormal skin keratinocyte differentiation through unknown mechanisms [93]. Further, in a study with a psoriasis mouse model, C. albicans exposure enhanced Th 17 inflammation through IL-17 production by αβ T cells sensitized by Candida [94].
5.3. Acne
While C. acnes is a major commensal organism in the normal skin flora, this bacterium contributes to the pathogenesis of acne [95]. Previous metagenomics studies have found similar proliferation levels of C. acnes between acne patients and healthy individuals, with a slightly higher level reported in healthy subjects [96,97]. Instead, loss of microbial diversity and of balance between C. acnes phylotypes has been reported in acne patients; these may be important etiological factors in the pathogenesis of acne [95]. Moreover, the severity of acne is associated with a loss of diversity of C. acnes phylotypes rather than proliferation of C. acnes [98]. Loss of diversity is characterized by colonization of certain strains of C. acnes, particularly acne-associated phylotype IA1, which is enhanced by a hyper-seborrheic environment [99]. Loss of C. acnes phylotype diversity can activate the innate immune system and skin inflammation in acne. Indeed, monoculture of phylogenetic IA1 has been shown to induce activation of the innate immune system [100]. Additionally, C. acnes strains can differentially modulate the CD4+ T-cell responses, activating Th 17 cells and inducing production of interferon gamma [101].
A recent study reported the importance of the relationship of S. epidermidis and C. acnes in maintaining normal skin barrier function [102,103]. Succinic acid, a metabolite of S. epidermidis, inhibits the growth and colonization of C. acnes and the inflammatory response induced by C. acnes [104]. In addition, S. epidermidis can inhibit the C. acnes-induced keratinocyte production of IL-6 and TNF-α [103].
In the previous section, we noted that the gut microbiome can affect skin barrier function. This phenomenon is also seen in acne patients. Previous studies showed that the gut microbiome composition was different between moderate to severe acne patients and controls. The patients had less abundant Actinobacteria and more abundant Proteobacteria [105]. In another study, the ratio of Bacteroidetes to Firmicutes was higher in acne patients; this was interpreted as an effect of the Western diet [106].
As in AD, research on treatment and management using probiotics and microbiome cosmetics in acne is ongoing. In one study, applying S. thermophilus to the skin increased the production of ceramides, a major skin lipid with anti-inflammatory and antibacterial effects against C. acnes [107,108]. Another study showed that application of emollients with E. faecalis reduced the severity of the disease in patients with mild to moderate acne [109].
The possible mechanisms of the microbiome on each inflammatory skin disease is summarized in Table 1.

Table 1.
Inflammatory skin disorders and microbiome.
6. Conclusions
As academic interest in the microbiome increases, research on the effect of the microbiome on skin barrier function is being actively conducted. In this review, we discussed the interaction between the microbiome and skin barrier function. Microbiome dysbiosis can cause damage to the skin barrier, which is associated with inflammatory skin diseases. Recently, studies on the positive effects of oral or cosmetic microbiome supplementation on skin diseases have been conducted. Microbiome supplementation may help restore skin barrier function and improve disease, which may open new directions for the treatment of inflammatory skin diseases.
Author Contributions
H.-J.L. analyzed the data and wrote the manuscript. M.K. reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Belkaid, Y.; Segre, J.A. Dialogue between skin microbiota and immunity. Science 2014, 346, 954–959. [Google Scholar] [CrossRef] [PubMed]
- Dréno, B.; Araviiskaia, E.; Berardesca, E.; Gontijo, G.; Sanchez Viera, M.; Xiang, L.F.; Martin, R.; Bieber, T. Microbiome in healthy skin, update for dermatologists. J. Eur. Acad. Dermatol. Venereol. JEADV 2016, 30, 2038–2047. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Byrd, A.L.; Park, M.; Kong, H.H.; Segre, J.A. Temporal Stability of the Human Skin Microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; Green, E.D.; et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Findley, K.; Oh, J.; Yang, J.; Conlan, S.; Deming, C.; Meyer, J.A.; Schoenfeld, D.; Nomicos, E.; Park, M.; Kong, H.H.; et al. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013, 498, 367–370. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Deming, C.; Conlan, S.; Kong, H.H.; Segre, J.A. Biogeography and individuality shape function in the human skin metagenome. Nature 2014, 514, 59–64. [Google Scholar] [CrossRef]
- Stinson, L.F.; Boyce, M.C.; Payne, M.S.; Keelan, J.A. The Not-so-Sterile Womb: Evidence That the Human Fetus Is Exposed to Bacteria Prior to Birth. Front. Microbiol. 2019, 10, 1124. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Kalbermatter, C.; Fernandez Trigo, N.; Christensen, S.; Ganal-Vonarburg, S.C. Maternal Microbiota, Early Life Colonization and Breast Milk Drive Immune Development in the Newborn. Front. Immunol. 2021, 12, 683022. [Google Scholar] [CrossRef]
- Park, J.; Schwardt, N.H.; Jo, J.H.; Zhang, Z.; Pillai, V.; Phang, S.; Brady, S.M.; Portillo, J.A.; MacGibeny, M.A.; Liang, H.; et al. Shifts in the Skin Bacterial and Fungal Communities of Healthy Children Transitioning through Puberty. J. Investig. Dermatol. 2022, 142, 212–219. [Google Scholar] [CrossRef]
- Conwill, A.; Kuan, A.C.; Damerla, R.; Poret, A.J.; Baker, J.S.; Tripp, A.D.; Alm, E.J.; Lieberman, T.D. Anatomy promotes neutral coexistence of strains in the human skin microbiome. Cell Host Microbe 2022, 30, 171–182.e177. [Google Scholar] [CrossRef]
- Volksdorf, T.; Heilmann, J.; Eming, S.A.; Schawjinski, K.; Zorn-Kruppa, M.; Ueck, C.; Vidal, Y.S.S.; Windhorst, S.; Jücker, M.; Moll, I.; et al. Tight Junction Proteins Claudin-1 and Occludin Are Important for Cutaneous Wound Healing. Am. J. Pathol. 2017, 187, 1301–1312. [Google Scholar] [CrossRef]
- Uberoi, A.; Bartow-McKenney, C.; Zheng, Q.; Flowers, L.; Campbell, A.; Knight, S.A.B.; Chan, N.; Wei, M.; Lovins, V.; Bugayev, J.; et al. Commensal microbiota regulates skin barrier function and repair via signaling through the aryl hydrocarbon receptor. Cell Host Microbe 2021, 29, 1235–1248.e1238. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Hunt, R.L.; Villaruz, A.E.; Fisher, E.L.; Liu, R.; Liu, Q.; Cheung, G.Y.C.; Li, M.; Otto, M. Commensal Staphylococcus epidermidis contributes to skin barrier homeostasis by generating protective ceramides. Cell Host Microbe 2022, 30, 301–313.e309. [Google Scholar] [CrossRef] [PubMed]
- Bomar, L.; Brugger, S.D.; Yost, B.H.; Davies, S.S.; Lemon, K.P. Corynebacterium accolens Releases Antipneumococcal Free Fatty Acids from Human Nostril and Skin Surface Triacylglycerols. mBio 2016, 7, e01725-15. [Google Scholar] [CrossRef] [PubMed]
- Ridaura, V.K.; Bouladoux, N.; Claesen, J.; Chen, Y.E.; Byrd, A.L.; Constantinides, M.G.; Merrill, E.D.; Tamoutounour, S.; Fischbach, M.A.; Belkaid, Y. Contextual control of skin immunity and inflammation by Corynebacterium. J. Exp. Med. 2018, 215, 785–799. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Kao, M.C.; Zhang, L.; Zouboulis, C.C.; Gallo, R.L.; Huang, C.M. Sebum free fatty acids enhance the innate immune defense of human sebocytes by upregulating beta-defensin-2 expression. J. Investig. Dermatol. 2010, 130, 985–994. [Google Scholar] [CrossRef]
- Moran, J.C.; Alorabi, J.A.; Horsburgh, M.J. Comparative Transcriptomics Reveals Discrete Survival Responses of S. aureus and S. epidermidis to Sapienic Acid. Front. Microbiol. 2017, 8, 33. [Google Scholar] [CrossRef]
- Pastar, I.; O’Neill, K.; Padula, L.; Head, C.R.; Burgess, J.L.; Chen, V.; Garcia, D.; Stojadinovic, O.; Hower, S.; Plano, G.V.; et al. Staphylococcus epidermidis Boosts Innate Immune Response by Activation of Gamma Delta T Cells and Induction of Perforin-2 in Human Skin. Front. Immunol. 2020, 11, 550946. [Google Scholar] [CrossRef]
- Chen, Y.E.; Bouladoux, N.; Hurabielle, C.; Mattke, A.M.; Belkaid, Y.; Fischbach, M.A. Decoding commensal-host communication through genetic engineering of Staphylococcus epidermidis. bioRxiv 2019, 664656. [Google Scholar] [CrossRef]
- Kashem, S.W.; Igyarto, B.Z.; Gerami-Nejad, M.; Kumamoto, Y.; Mohammed, J.A.; Jarrett, E.; Drummond, R.A.; Zurawski, S.M.; Zurawski, G.; Berman, J.; et al. Candida albicans morphology and dendritic cell subsets determine T helper cell differentiation. Immunity 2015, 42, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Naik, S.; Bouladoux, N.; Wilhelm, C.; Molloy, M.J.; Salcedo, R.; Kastenmuller, W.; Deming, C.; Quinones, M.; Koo, L.; Conlan, S.; et al. Compartmentalized control of skin immunity by resident commensals. Science 2012, 337, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Chehoud, C.; Rafail, S.; Tyldsley, A.S.; Seykora, J.T.; Lambris, J.D.; Grice, E.A. Complement modulates the cutaneous microbiome and inflammatory milieu. Proc. Natl. Acad. Sci. USA 2013, 110, 15061–15066. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Braff, M.H.; Zaiou, M.; Fierer, J.; Nizet, V.; Gallo, R.L. Keratinocyte production of cathelicidin provides direct activity against bacterial skin pathogens. Infect. Immun. 2005, 73, 6771–6781. [Google Scholar] [CrossRef]
- Lai, Y.; Cogen, A.L.; Radek, K.A.; Park, H.J.; Macleod, D.T.; Leichtle, A.; Ryan, A.F.; Di Nardo, A.; Gallo, R.L. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J. Investig. Dermatol. 2010, 130, 2211–2221. [Google Scholar] [CrossRef]
- Simanski, M.; Erkens, A.S.; Rademacher, F.; Harder, J. Staphylococcus epidermidis-induced Interleukin-1 Beta and Human Beta-defensin-2 Expression in Human Keratinocytes is Regulated by the Host Molecule A20 (TNFAIP3). Acta Derm. Venereol. 2019, 99, 181–187. [Google Scholar] [CrossRef]
- Dahlhoff, M.; Zouboulis, C.C.; Schneider, M.R. Expression of dermcidin in sebocytes supports a role for sebum in the constitutive innate defense of human skin. J. Dermatol. Sci. 2016, 81, 124–126. [Google Scholar] [CrossRef]
- Sanford, J.A.; Zhang, L.J.; Williams, M.R.; Gangoiti, J.A.; Huang, C.M.; Gallo, R.L. Inhibition of HDAC8 and HDAC9 by microbial short-chain fatty acids breaks immune tolerance of the epidermis to TLR ligands. Sci. Immunol. 2016, 1, eaah4609. [Google Scholar] [CrossRef]
- Sanford, J.A.; O’Neill, A.M.; Zouboulis, C.C.; Gallo, R.L. Short-Chain Fatty Acids from Cutibacterium acnes Activate Both a Canonical and Epigenetic Inflammatory Response in Human Sebocytes. J. Immunol. 2019, 202, 1767–1776. [Google Scholar] [CrossRef]
- Nakamura, K.; O’Neill, A.M.; Williams, M.R.; Cau, L.; Nakatsuji, T.; Horswill, A.R.; Gallo, R.L. Short chain fatty acids produced by Cutibacterium acnes inhibit biofilm formation by Staphylococcus epidermidis. Sci. Rep. 2020, 10, 21237. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Hu, Z.; Lone, A.G.; Artami, M.; Edwards, M.; Zouboulis, C.C.; Stein, M.; Harris-Tryon, T.A. Small proline-rich proteins (SPRRs) are epidermally produced antimicrobial proteins that defend the cutaneous barrier by direct bacterial membrane disruption. eLife 2022, 11, e76729. [Google Scholar] [CrossRef] [PubMed]
- Marples, R.R.; Downing, D.T.; Kligman, A.M. Control of free fatty acids in human surface lipids by Corynebacterium acnes. J. Investig. Dermatol. 1971, 56, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Drake, D.R.; Brogden, K.A.; Dawson, D.V.; Wertz, P.W. Thematic review series: Skin lipids. Antimicrobial lipids at the skin surface. J. Lipid Res. 2008, 49, 4–11. [Google Scholar] [CrossRef]
- Scharschmidt, T.C.; Vasquez, K.S.; Truong, H.A.; Gearty, S.V.; Pauli, M.L.; Nosbaum, A.; Gratz, I.K.; Otto, M.; Moon, J.J.; Liese, J.; et al. A Wave of Regulatory T Cells into Neonatal Skin Mediates Tolerance to Commensal Microbes. Immunity 2015, 43, 1011–1021. [Google Scholar] [CrossRef]
- Scharschmidt, T.C.; Vasquez, K.S.; Pauli, M.L.; Leitner, E.G.; Chu, K.; Truong, H.A.; Lowe, M.M.; Sanchez Rodriguez, R.; Ali, N.; Laszik, Z.G.; et al. Commensal Microbes and Hair Follicle Morphogenesis Coordinately Drive Treg Migration into Neonatal Skin. Cell Host Microbe 2017, 21, 467–477.e465. [Google Scholar] [CrossRef]
- Ansaldo, E.; Farley, T.K.; Belkaid, Y. Control of Immunity by the Microbiota. Annu. Rev. Immunol. 2021, 39, 449–479. [Google Scholar] [CrossRef]
- Kjer-Nielsen, L.; Patel, O.; Corbett, A.J.; Le Nours, J.; Meehan, B.; Liu, L.; Bhati, M.; Chen, Z.; Kostenko, L.; Reantragoon, R.; et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 2012, 491, 717–723. [Google Scholar] [CrossRef]
- Li, J.; Reantragoon, R.; Kostenko, L.; Corbett, A.J.; Varigos, G.; Carbone, F.R. The frequency of mucosal-associated invariant T cells is selectively increased in dermatitis herpetiformis. Australas. J. Dermatol. 2017, 58, 200–204. [Google Scholar] [CrossRef]
- Harrison, O.J.; Linehan, J.L.; Shih, H.Y.; Bouladoux, N.; Han, S.J.; Smelkinson, M.; Sen, S.K.; Byrd, A.L.; Enamorado, M.; Yao, C.; et al. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science 2019, 363, eaat6280. [Google Scholar] [CrossRef]
- Leech, J.M.; Dhariwala, M.O.; Lowe, M.M.; Chu, K.; Merana, G.R.; Cornuot, C.; Weckel, A.; Ma, J.M.; Leitner, E.G.; Gonzalez, J.R.; et al. Toxin-Triggered Interleukin-1 Receptor Signaling Enables Early-Life Discrimination of Pathogenic versus Commensal Skin Bacteria. Cell Host Microbe 2019, 26, 795–809.e795. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.N.; Rea, M.C.; O’Connor, P.M.; Hill, C.; Ross, R.P. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 2019, 95, fiy241. [Google Scholar] [CrossRef] [PubMed]
- Zipperer, A.; Konnerth, M.C.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M.; et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Bitschar, K.; Sauer, B.; Focken, J.; Dehmer, H.; Moos, S.; Konnerth, M.; Schilling, N.A.; Grond, S.; Kalbacher, H.; Kurschus, F.C.; et al. Lugdunin amplifies innate immune responses in the skin in synergy with host- and microbiota-derived factors. Nat. Commun. 2019, 10, 2730. [Google Scholar] [CrossRef]
- Cogen, A.L.; Yamasaki, K.; Sanchez, K.M.; Dorschner, R.A.; Lai, Y.; MacLeod, D.T.; Torpey, J.W.; Otto, M.; Nizet, V.; Kim, J.E.; et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Investig. Dermatol. 2010, 130, 192–200. [Google Scholar] [CrossRef]
- O’Neill, A.M.; Nakatsuji, T.; Hayachi, A.; Williams, M.R.; Mills, R.H.; Gonzalez, D.J.; Gallo, R.L. Identification of a Human Skin Commensal Bacterium that Selectively Kills Cutibacterium acnes. J. Investig. Dermatol. 2020, 140, 1619–1628.e1612. [Google Scholar] [CrossRef] [PubMed]
- Claesen, J.; Spagnolo, J.B.; Ramos, S.F.; Kurita, K.L.; Byrd, A.L.; Aksenov, A.A.; Melnik, A.V.; Wong, W.R.; Wang, S.; Hernandez, R.D.; et al. A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci. Transl. Med. 2020, 12, eaay5445. [Google Scholar] [CrossRef]
- Williams, M.R.; Costa, S.K.; Zaramela, L.S.; Khalil, S.; Todd, D.A.; Winter, H.L.; Sanford, J.A.; O’Neill, A.M.; Liggins, M.C.; Nakatsuji, T.; et al. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci. Transl. Med. 2019, 11, eaat8329. [Google Scholar] [CrossRef]
- Paharik, A.E.; Parlet, C.P.; Chung, N.; Todd, D.A.; Rodriguez, E.I.; Van Dyke, M.J.; Cech, N.B.; Horswill, A.R. Coagulase-Negative Staphylococcal Strain Prevents Staphylococcus aureus Colonization and Skin Infection by Blocking Quorum Sensing. Cell Host Microbe 2017, 22, 746–756.e745. [Google Scholar] [CrossRef]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010, 465, 346–349. [Google Scholar] [CrossRef] [PubMed]
- De Pessemier, B.; Grine, L.; Debaere, M.; Maes, A.; Paetzold, B.; Callewaert, C. Gut-Skin Axis: Current Knowledge of the Interrelationship between Microbial Dysbiosis and Skin Conditions. Microorganisms 2021, 9, 353. [Google Scholar] [CrossRef]
- Bosman, E.S.; Albert, A.Y.; Lui, H.; Dutz, J.P.; Vallance, B.A. Skin Exposure to Narrow Band Ultraviolet (UVB) Light Modulates the Human Intestinal Microbiome. Front. Microbiol. 2019, 10, 2410. [Google Scholar] [CrossRef]
- Brough, H.A.; Liu, A.H.; Sicherer, S.; Makinson, K.; Douiri, A.; Brown, S.J.; Stephens, A.C.; Irwin McLean, W.H.; Turcanu, V.; Wood, R.A.; et al. Atopic dermatitis increases the effect of exposure to peanut antigen in dust on peanut sensitization and likely peanut allergy. J. Allergy Clin. Immunol. 2015, 135, 164–170. [Google Scholar] [CrossRef]
- Leyva-Castillo, J.M.; Galand, C.; Kam, C.; Burton, O.; Gurish, M.; Musser, M.A.; Goldsmith, J.D.; Hait, E.; Nurko, S.; Brombacher, F.; et al. Mechanical Skin Injury Promotes Food Anaphylaxis by Driving Intestinal Mast Cell Expansion. Immunity 2019, 50, 1262–1275.e1264. [Google Scholar] [CrossRef] [PubMed]
- Dokoshi, T.; Seidman, J.S.; Cavagnero, K.J.; Li, F.; Liggins, M.C.; Taylor, B.C.; Olvera, J.; Knight, R.; Chang, J.T.; Salzman, N.H.; et al. Skin inflammation activates intestinal stromal fibroblasts and promotes colitis. J. Clin. Investig. 2021, 131, e147614. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, C.A.; Monteleone, G.; McLaughlin, J.T.; Paus, R. The gut-skin axis in health and disease: A paradigm with therapeutic implications. BioEssays News Rev. Mol. Cell. Dev. Biol. 2016, 38, 1167–1176. [Google Scholar] [CrossRef]
- Forbes, J.D.; Van Domselaar, G.; Bernstein, C.N. The Gut Microbiota in Immune-Mediated Inflammatory Diseases. Front. Microbiol. 2016, 7, 1081. [Google Scholar] [CrossRef]
- Meijer, K.; de Vos, P.; Priebe, M.G. Butyrate and other short-chain fatty acids as modulators of immunity: What relevance for health? Curr. Opin. Clin. Nutr. Metab. Care 2010, 13, 715–721. [Google Scholar] [CrossRef]
- Samuelson, D.R.; Welsh, D.A.; Shellito, J.E. Regulation of lung immunity and host defense by the intestinal microbiota. Front. Microbiol. 2015, 6, 1085. [Google Scholar] [CrossRef] [PubMed]
- Zákostelská, Z.; Málková, J.; Klimešová, K.; Rossmann, P.; Hornová, M.; Novosádová, I.; Stehlíková, Z.; Kostovčík, M.; Hudcovic, T.; Štepánková, R.; et al. Intestinal Microbiota Promotes Psoriasis-Like Skin Inflammation by Enhancing Th17 Response. PLoS ONE 2016, 11, e0159539. [Google Scholar] [CrossRef] [PubMed]
- Levkovich, T.; Poutahidis, T.; Smillie, C.; Varian, B.J.; Ibrahim, Y.M.; Lakritz, J.R.; Alm, E.J.; Erdman, S.E. Probiotic bacteria induce a ‘glow of health’. PLoS ONE 2013, 8, e53867. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Saiki, A.; Matsui, Y.; Tsuchimoto, N.; Nakakita, Y.; Takata, Y.; Nakamura, T. Effects of oral intake of heat-killed Lactobacillus brevis SBC8803 (SBL88™) on dry skin conditions: A randomized, double-blind, placebo-controlled study. Exp. Ther. Med. 2016, 12, 3863–3872. [Google Scholar] [CrossRef]
- Gueniche, A.; Philippe, D.; Bastien, P.; Reuteler, G.; Blum, S.; Castiel-Higounenc, I.; Breton, L.; Benyacoub, J. Randomised double-blind placebo-controlled study of the effect of Lactobacillus paracasei NCC 2461 on skin reactivity. Benef. Microbes 2014, 5, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Wu, C.S.; Chao, Y.H.; Lin, C.C.; Tsai, H.Y.; Li, Y.R.; Chen, Y.Z.; Tsai, W.H.; Chen, Y.K. Lactobacillus pentosus GMNL-77 inhibits skin lesions in imiquimod-induced psoriasis-like mice. J. Food Drug Anal. 2017, 25, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Udayanga, K.G.; Totsuka, N.; Weinberg, J.B.; Núñez, G.; Shibuya, A. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE2. Cell Host Microbe 2014, 15, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Philippe, D.; Blum, S.; Benyacoub, J. Oral Lactobacillus paracasei improves skin barrier function recovery and reduces local skin inflammation. Eur. J. Dermatol. EJD 2011, 21, 279–280. [Google Scholar] [CrossRef]
- Baba, H.; Masuyama, A.; Yoshimura, C.; Aoyama, Y.; Takano, T.; Ohki, K. Oral intake of Lactobacillus helveticus-fermented milk whey decreased transepidermal water loss and prevented the onset of sodium dodecylsulfate-induced dermatitis in mice. Biosci. Biotechnol. Biochem. 2010, 74, 18–23. [Google Scholar] [CrossRef]
- Poutahidis, T.; Kearney, S.M.; Levkovich, T.; Qi, P.; Varian, B.J.; Lakritz, J.R.; Ibrahim, Y.M.; Chatzigiagkos, A.; Alm, E.J.; Erdman, S.E. Microbial symbionts accelerate wound healing via the neuropeptide hormone oxytocin. PLoS ONE 2013, 8, e78898. [Google Scholar] [CrossRef]
- Edslev, S.M.; Agner, T.; Andersen, P.S. Skin Microbiome in Atopic Dermatitis. Acta Derm. Venereol. 2020, 100, adv00164. [Google Scholar] [CrossRef]
- Byrd, A.L.; Deming, C.; Cassidy, S.K.B.; Harrison, O.J.; Ng, W.I.; Conlan, S.; Belkaid, Y.; Segre, J.A.; Kong, H.H. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl. Med. 2017, 9, eaal4651. [Google Scholar] [CrossRef]
- Baurecht, H.; Rühlemann, M.C.; Rodríguez, E.; Thielking, F.; Harder, I.; Erkens, A.S.; Stölzl, D.; Ellinghaus, E.; Hotze, M.; Lieb, W.; et al. Epidermal lipid composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration. J. Allergy Clin. Immunol. 2018, 141, 1668–1676.e1616. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, K.; Moriwaki, M.; Miyake, R.; Hide, M. Staphylococcus aureus in atopic dermatitis: Strain-specific cell wall proteins and skin immunity. Allergol. Int. 2019, 68, 309–315. [Google Scholar] [CrossRef]
- Johnson, C.C.; Ownby, D.R. The infant gut bacterial microbiota and risk of pediatric asthma and allergic diseases. Transl. Res. J. Lab. Clin. Med. 2017, 179, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Yoo, Y.; Hwang, J.; Na, Y.C.; Kim, H.S. Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis. J. Allergy Clin. Immunol. 2016, 137, 852–860. [Google Scholar] [CrossRef]
- Nowicka, D.; Nawrot, U. Contribution of Malassezia spp. to the development of atopic dermatitis. Mycoses 2019, 62, 588–596. [Google Scholar] [CrossRef]
- Zhang, E.; Tanaka, T.; Tajima, M.; Tsuboi, R.; Kato, H.; Nishikawa, A.; Sugita, T. Anti-Malassezia-Specific IgE Antibodies Production in Japanese Patients with Head and Neck Atopic Dermatitis: Relationship between the Level of Specific IgE Antibody and the Colonization Frequency of Cutaneous Malassezia Species and Clinical Severity. J. Allergy 2011, 2011, 645670. [Google Scholar] [CrossRef]
- Notay, M.; Foolad, N.; Vaughn, A.R.; Sivamani, R.K. Probiotics, Prebiotics, and Synbiotics for the Treatment and Prevention of Adult Dermatological Diseases. Am. J. Clin. Dermatol. 2017, 18, 721–732. [Google Scholar] [CrossRef]
- Huang, R.; Ning, H.; Shen, M.; Li, J.; Zhang, J.; Chen, X. Probiotics for the Treatment of Atopic Dermatitis in Children: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Front. Cell. Infect. Microbiol. 2017, 7, 392. [Google Scholar] [CrossRef]
- Jiang, W.; Ni, B.; Liu, Z.; Liu, X.; Xie, W.; Wu, I.X.Y.; Li, X. The Role of Probiotics in the Prevention and Treatment of Atopic Dermatitis in Children: An Updated Systematic Review and Meta-Analysis of Randomized Controlled Trials. Paediatr. Drugs 2020, 22, 535–549. [Google Scholar] [CrossRef] [PubMed]
- Glatz, M.; Jo, J.H.; Kennedy, E.A.; Polley, E.C.; Segre, J.A.; Simpson, E.L.; Kong, H.H. Emollient use alters skin barrier and microbes in infants at risk for developing atopic dermatitis. PLoS ONE 2018, 13, e0192443. [Google Scholar] [CrossRef] [PubMed]
- Seité, S.; Zelenkova, H.; Martin, R. Clinical efficacy of emollients in atopic dermatitis patients-relationship with the skin microbiota modification. Clin. Cosmet. Investig. Dermatol. 2017, 10, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Deleuran, M.; Georgescu, V.; Jean-Decoster, C. An Emollient Containing Aquaphilus dolomiae Extract is Effective in the Management of Xerosis and Pruritus: An International, Real-World Study. Dermatol. Ther. 2020, 10, 1013–1029. [Google Scholar] [CrossRef]
- Butler, É.; Lundqvist, C.; Axelsson, J. Lactobacillus reuteri DSM 17938 as a Novel Topical Cosmetic Ingredient: A Proof of Concept Clinical Study in Adults with Atopic Dermatitis. Microorganisms 2020, 8, 1026. [Google Scholar] [CrossRef]
- Myles, I.A.; Earland, N.J.; Anderson, E.D.; Moore, I.N.; Kieh, M.D.; Williams, K.W.; Saleem, A.; Fontecilla, N.M.; Welch, P.A.; Darnell, D.A.; et al. First-in-human topical microbiome transplantation with Roseomonas mucosa for atopic dermatitis. JCI Insight 2018, 3. [Google Scholar] [CrossRef]
- Chen, L.; Li, J.; Zhu, W.; Kuang, Y.; Liu, T.; Zhang, W.; Chen, X.; Peng, C. Skin and Gut Microbiome in Psoriasis: Gaining Insight Into the Pathophysiology of It and Finding Novel Therapeutic Strategies. Front. Microbiol. 2020, 11, 589726. [Google Scholar] [CrossRef]
- Liu, S.H.; Yu, H.Y.; Chang, Y.C.; Chung-Yee Hui, R.; Huang, Y.C.; Huang, Y.H. Host characteristics and dynamics of Staphylococcus aureus colonization in patients with moderate-to-severe psoriasis before and after treatment: A prospective cohort study. J. Am. Acad. Dermatol. 2019, 81, 605–607. [Google Scholar] [CrossRef]
- Ng, C.Y.; Huang, Y.H.; Chu, C.F.; Wu, T.C.; Liu, S.H. Risks for Staphylococcus aureus colonization in patients with psoriasis: A systematic review and meta-analysis. Br. J. Dermatol. 2017, 177, 967–977. [Google Scholar] [CrossRef]
- Chang, H.W.; Yan, D.; Singh, R.; Liu, J.; Lu, X.; Ucmak, D.; Lee, K.; Afifi, L.; Fadrosh, D.; Leech, J.; et al. Alteration of the cutaneous microbiome in psoriasis and potential role in Th17 polarization. Microbiome 2018, 6, 154. [Google Scholar] [CrossRef]
- Orsmond, A.; Bereza-Malcolm, L.; Lynch, T.; March, L.; Xue, M. Skin Barrier Dysregulation in Psoriasis. Int. J. Mol. Sci. 2021, 22, 10841. [Google Scholar] [CrossRef] [PubMed]
- Komine, M. Recent Advances in Psoriasis Research; the Clue to Mysterious Relation to Gut Microbiome. Int. J. Mol. Sci. 2020, 21, 2582. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, K.; Masuoka, N.; Kano, M.; Iizuka, R. Bifidobacterium fermented milk and galacto-oligosaccharides lead to improved skin health by decreasing phenols production by gut microbiota. Benef. Microbes 2014, 5, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Hurabielle, C.; Link, V.M.; Bouladoux, N.; Han, S.J.; Merrill, E.D.; Lightfoot, Y.L.; Seto, N.; Bleck, C.K.E.; Smelkinson, M.; Harrison, O.J.; et al. Immunity to commensal skin fungi promotes psoriasiform skin inflammation. Proc. Natl. Acad. Sci. USA 2020, 117, 16465–16474. [Google Scholar] [CrossRef] [PubMed]
- Fitz-Gibbon, S.; Tomida, S.; Chiu, B.H.; Nguyen, L.; Du, C.; Liu, M.; Elashoff, D.; Erfe, M.C.; Loncaric, A.; Kim, J.; et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Investig. Dermatol. 2013, 133, 2152–2160. [Google Scholar] [CrossRef] [PubMed]
- Barnard, E.; Shi, B.; Kang, D.; Craft, N.; Li, H. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci. Rep. 2016, 6, 39491. [Google Scholar] [CrossRef]
- Omer, H.; McDowell, A.; Alexeyev, O.A. Understanding the role of Propionibacterium acnes in acne vulgaris: The critical importance of skin sampling methodologies. Clin. Dermatol. 2017, 35, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Dagnelie, M.A.; Montassier, E.; Khammari, A.; Mounier, C.; Corvec, S.; Dréno, B. Inflammatory skin is associated with changes in the skin microbiota composition on the back of severe acne patients. Exp. Dermatol. 2019, 28, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Dréno, B.; Pécastaings, S.; Corvec, S.; Veraldi, S.; Khammari, A.; Roques, C. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: A brief look at the latest updates. J. Eur. Acad. Dermatol. Venereol. JEADV 2018, 32 (Suppl. S2), 5–14. [Google Scholar] [CrossRef]
- Dagnelie, M.A.; Corvec, S.; Saint-Jean, M.; Nguyen, J.M.; Khammari, A.; Dréno, B. Cutibacterium acnes phylotypes diversity loss: A trigger for skin inflammatory process. J. Eur. Acad. Dermatol. Venereol. JEADV 2019, 33, 2340–2348. [Google Scholar] [CrossRef]
- Agak, G.W.; Kao, S.; Ouyang, K.; Qin, M.; Moon, D.; Butt, A.; Kim, J. Phenotype and Antimicrobial Activity of Th17 Cells Induced by Propionibacterium acnes Strains Associated with Healthy and Acne Skin. J. Investig. Dermatol. 2018, 138, 316–324. [Google Scholar] [CrossRef]
- Dreno, B.; Martin, R.; Moyal, D.; Henley, J.B.; Khammari, A.; Seité, S. Skin microbiome and acne vulgaris: Staphylococcus, a new actor in acne. Exp. Dermatol. 2017, 26, 798–803. [Google Scholar] [CrossRef] [PubMed]
- Skabytska, Y.; Biedermann, T. Staphylococcus epidermidis Sets Things Right Again. J. Investig. Dermatol. 2016, 136, 559–560. [Google Scholar] [CrossRef] [PubMed]
- Claudel, J.P.; Auffret, N.; Leccia, M.T.; Poli, F.; Corvec, S.; Dréno, B. Staphylococcus epidermidis: A Potential New Player in the Physiopathology of Acne? Dermatology 2019, 235, 287–294. [Google Scholar] [CrossRef]
- Yan, H.M.; Zhao, H.J.; Guo, D.Y.; Zhu, P.Q.; Zhang, C.L.; Jiang, W. Gut microbiota alterations in moderate to severe acne vulgaris patients. J. Dermatol. 2018, 45, 1166–1171. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wang, H.; Zhou, J.; Mou, Y.; Wang, G.; Xiong, X. Patients with Acne Vulgaris Have a Distinct Gut Microbiota in Comparison with Healthy Controls. Acta Derm. Venereol. 2018, 98, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Di Marzio, L.; Cinque, B.; De Simone, C.; Cifone, M.G. Effect of the lactic acid bacterium Streptococcus thermophilus on ceramide levels in human keratinocytes in vitro and stratum corneum in vivo. J. Investig. Dermatol. 1999, 113, 98–106. [Google Scholar] [CrossRef]
- Bowe, W.P.; Logan, A.C. Acne vulgaris, probiotics and the gut-brain-skin axis-back to the future? Gut Pathog. 2011, 3, 1. [Google Scholar] [CrossRef]
- Kang, B.S.; Seo, J.G.; Lee, G.S.; Kim, J.H.; Kim, S.Y.; Han, Y.W.; Kang, H.; Kim, H.O.; Rhee, J.H.; Chung, M.J.; et al. Antimicrobial activity of enterocins from Enterococcus faecalis SL-5 against Propionibacterium acnes, the causative agent in acne vulgaris, and its therapeutic effect. J. Microbiol. 2009, 47, 101–109. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).