Induced Pluripotent Stem Cell (iPSC) Lines from a Family with Resistant Epileptic Encephalopathy Caused by Compound Heterozygous Mutations in SZT2 Gene
Abstract
1. Introduction
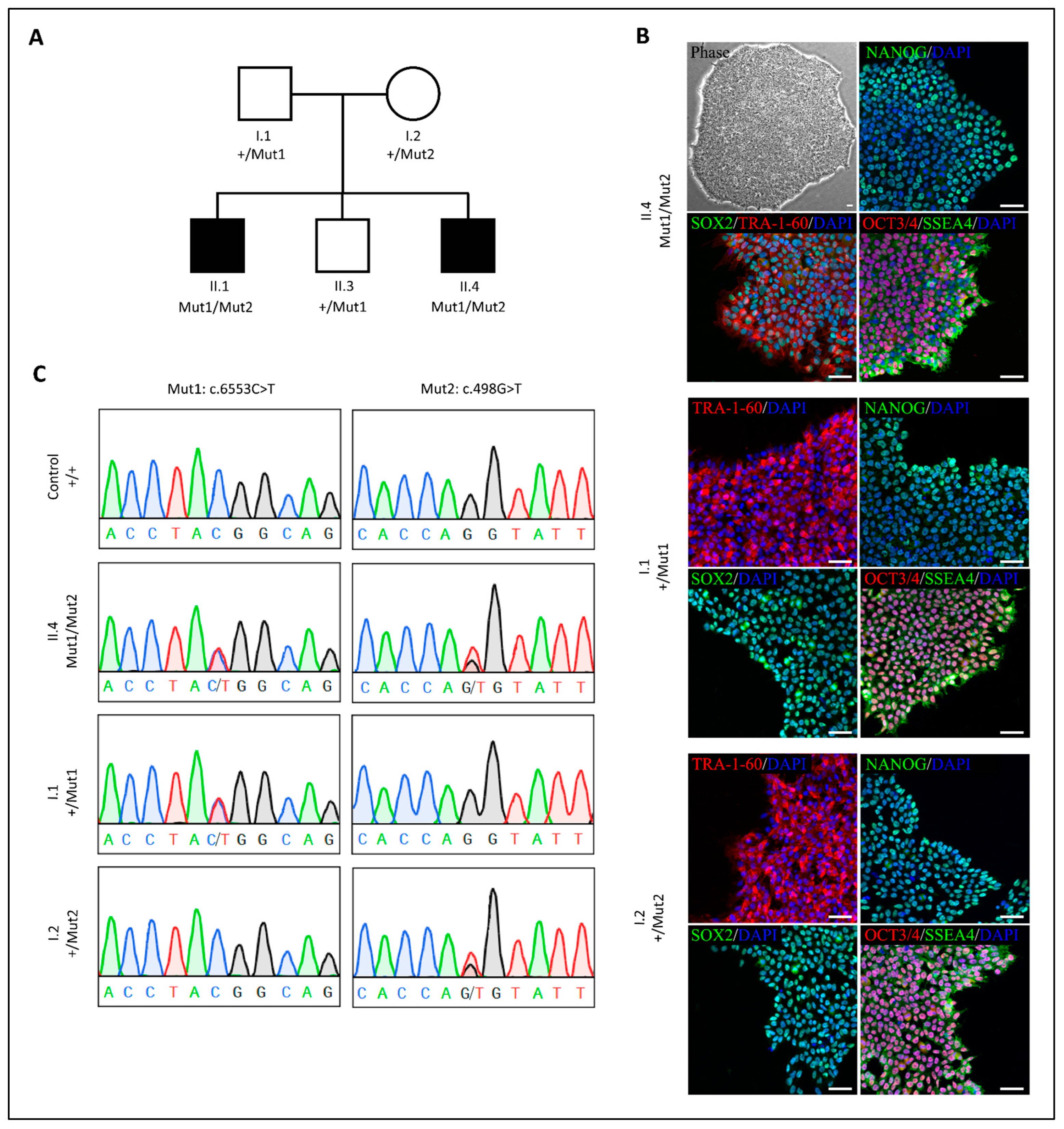
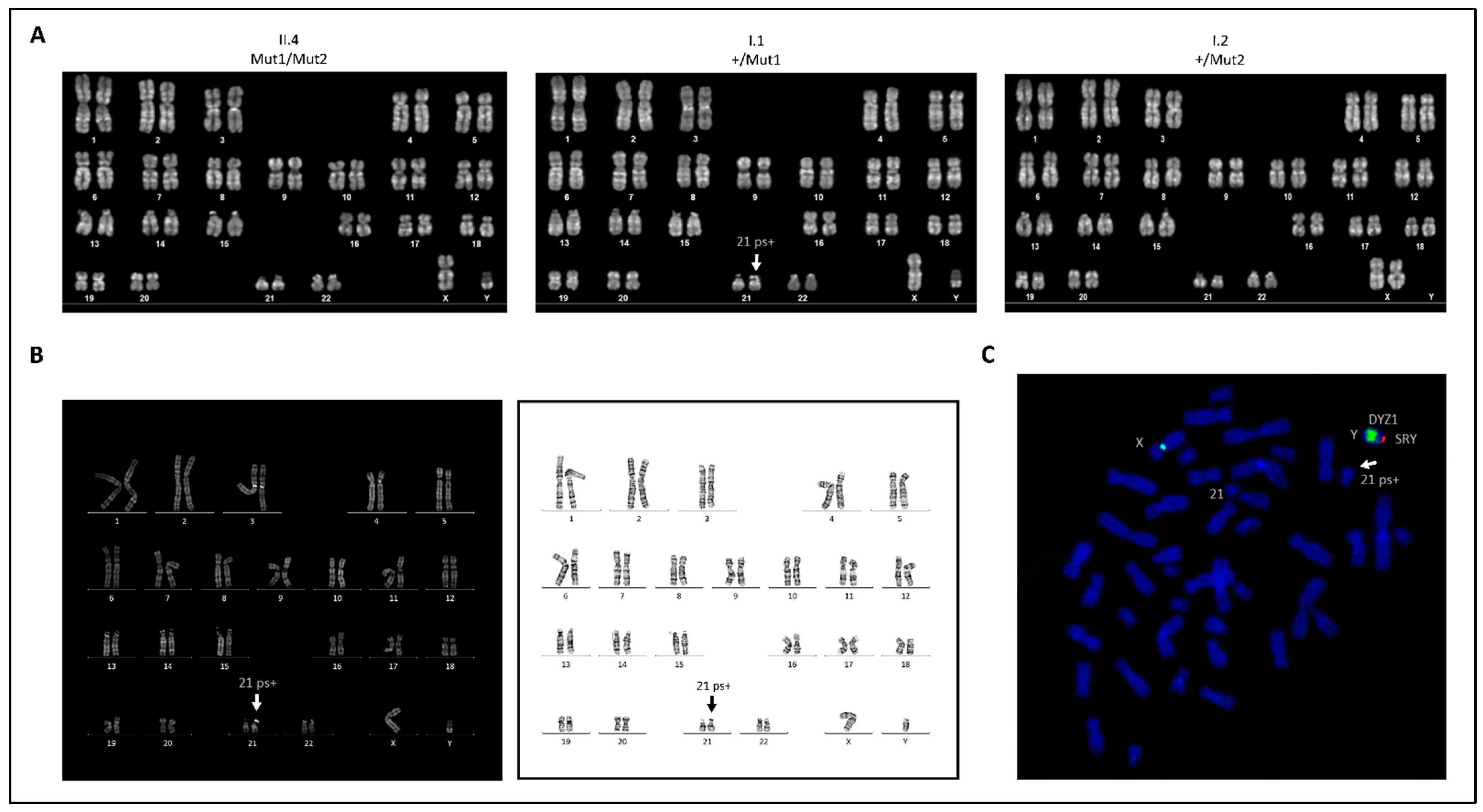
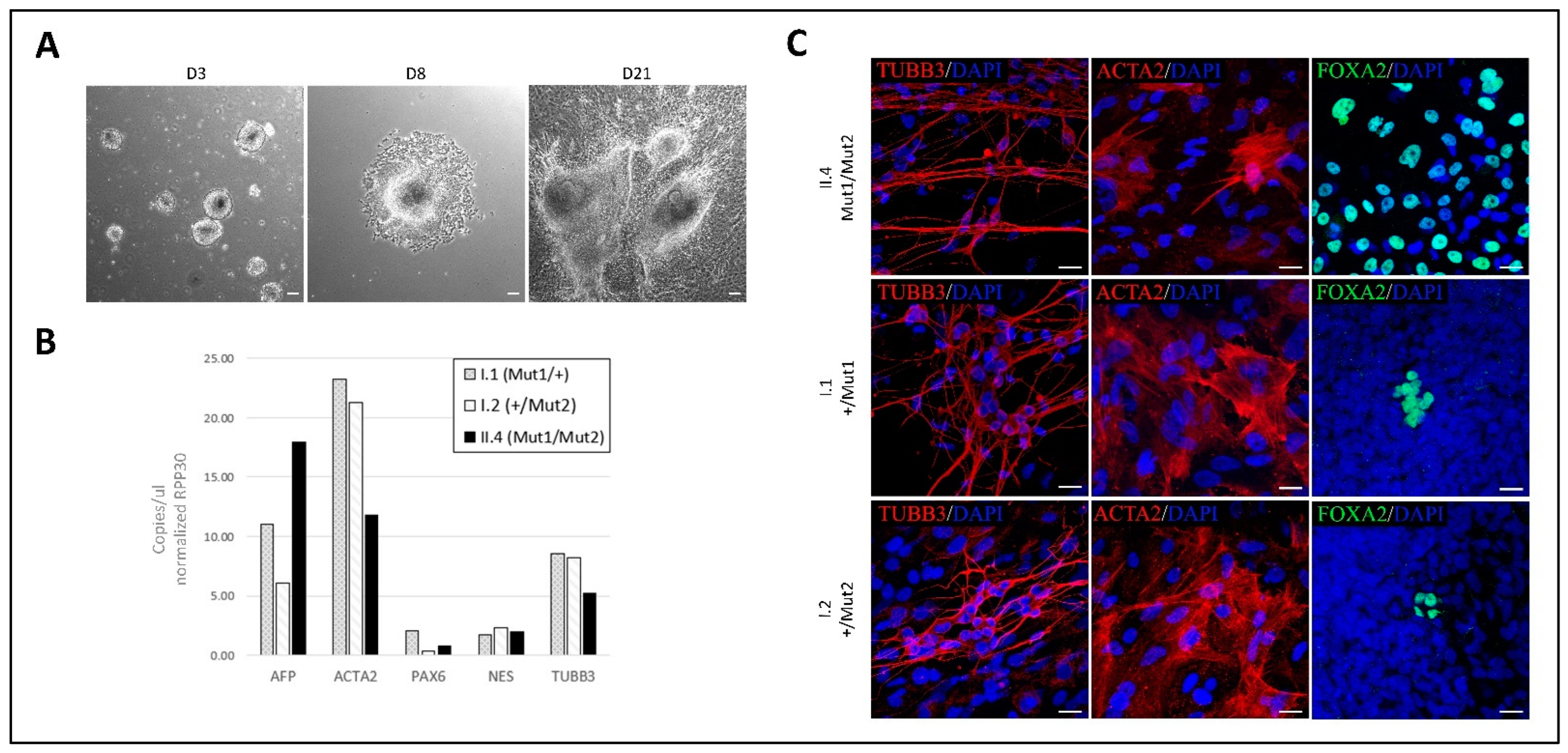
2. Results
3. Discussion
4. Materials and Methods
4.1. PBMCs Collection and Reprogramming to hiPSCs with Sendai Virus Particles
4.2. In Vitro Embryo Bodies Differentiation Assay
4.3. RNA Isolation, Retrotranscription and ddPCR
4.4. Immunofluorescence Assay
4.5. Karyotyping
4.6. PCR and Sequencing
4.7. Mycoplasma Test
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Toutzaris, D.; Lewerenz, J.; Albrecht, P.; Jensen, L.T.; Letz, J.; Geerts, A.; Golz, S.; Methner, A. A novel giant peroxisomal superoxide dismutase motif-containing protein. Free Radic. Biol. Med. 2010, 48, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Frankel, W.N.; Yang, Y.; Mahaffey, C.L.; Beyer, B.J.; O’Brien, T.P. Szt2, a novel gene for seizure threshold in mice. Genes Brain Behav. 2009, 8, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, R.L.; Chantranupong, L.; Wyant, G.A.; Gu, X.; Orozco, J.M.; Shen, K.; Condon, K.J.; Petri, S.; Kedir, J.; Scaria, S.M.; et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 2017, 543, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Yin, N.; Li, M.O. SZT2 dictates GATOR control of mTORC1 signalling. Nature 2017, 543, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Cattelani, C.; Lesiak, D.; Liebscher, G.; Singer, I.I.; Stasyk, T.; Wallnöfer, M.H.; Heberle, A.M.; Corti, C.; Hess, M.W.; Pfaller, K.; et al. The SZT2 interactome unravels new functions of the KICSTOR complex. Cells 2021, 10, 2711. [Google Scholar] [CrossRef] [PubMed]
- Basel-Vanagaite, L.; Hershkovitz, T.; Heyman, E.; Raspall-Chaure, M.; Kakar, N.; Smirin-Yosef, P.; Vila-Pueyo, M.; Kornreich, L.; Thiele, H.; Bode, H.; et al. Biallelic SZT2 mutations cause infantile encephalopathy with epilepsy and dysmorphic corpus callosum. Am. J. Hum. Genet. 2013, 93, 524–529. [Google Scholar] [CrossRef]
- Trivisano, M.; Rivera, M.; Terracciano, A.; Ciolfi, A.; Napolitano, A.; Pepi, C.; Calabrese, C.; Digilio, M.C.; Tartaglia, M.; Curatolo, P.; et al. Developmental and epileptic encephalopathy due to SZT2 genomic variants: Emerging features of a syndromic condition. Epilepsy Behav. 2020, 108, 107097. [Google Scholar] [CrossRef]
- Domingues, F.S.; König, E.; Schwienbacher, C.; Volpato, C.B.; Picard, A.; Cantaloni, C.; Mascalzoni, D.; Lackner, P.; Heimbach, A.; Hoffmann, P.; et al. Compound heterozygous SZT2 mutations in two siblings with Early-onset epilepsy, intellectual disability and macrocephaly. Seizure 2019, 66, 81–85. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Fusaki, N.; Ban, H.; Nishiyama, A.; Saeki, K.; Hasegawa, M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 348–362. [Google Scholar] [CrossRef]
- Yang, W.; Mills, J.A.; Sullivan, S.; Liu, Y.; French, D.L.; Gadue, P. iPSC Reprogramming from human peripheral blood using Sendai virus mediated gene transfer. In Stembook; 2008. Available online: www.ncbi.nlm.nih.gov/books/NBK143766 (accessed on 10 February 2021).
- Barber, J.C.K. Atlas of human chromosome heteromorphisms. Hum. Genet. 2005, 117, 404–405. [Google Scholar] [CrossRef]
- Gardner, R.J.M.; Amor, D.J. Gardner and Sutherland’s Chromosome Abnormalities and Genetic Counseling, 5th ed.; Oxford Monographs on Medical Genetics: Oxford, UK, 2018. [Google Scholar]
- Carpenter, M.K.; Rosler, E.; Rao, M.S. Characterization and differentiation of human embryonic stem cells. Cloning Stem Cells 2003, 5, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Gosden, J.R.; Lawrie, S.S.; Gosden, C.M. Satellite DNA sequences in the human acrocentric chromosomes: Information from translocations and heteromorphisms. Am. J. Hum. Genet. 1981, 33, 243–251. [Google Scholar] [PubMed]
- Waye, J.S.; Willard, H.F. Human β satellite DNA: Genomic organization and sequence definition of a class of highly repetitive tandem DNA. Proc. Natl. Acad. Sci. USA 1989, 86, 6250–6254. [Google Scholar] [CrossRef] [PubMed]
- Redaelli, S.; Conconi, D.; Villa, N.; Sala, E.; Crosti, F.; Corti, C.; Catusi, I.; Garzo, M.; Romitti, L.; Martinoli, E.; et al. Instability of short arm of acrocentric chromosomes: Lesson from non-acrocentric satellited chromosomes. Report of 24 Unrelated Cases. Int. J. Mol. Sci. 2020, 21, 3431. [Google Scholar] [CrossRef]
- Cooke, P.; Curtis, D.J. General and specific patterns of acrocentric association in parents of mongol children. Hum. Genet. 1974, 23, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Taysi, K. Satellite association: Giemsa banding studies in parents of Down’s Syndrome patients. Clin. Genet. 1975, 8, 319–323. [Google Scholar] [CrossRef]
- Yip, M.Y.; Fox, D.P. Variation in pattern and frequency of acrocentric association in normal and Trisomy-21 individuals. Hum. Genet. 1981, 59, 14–22. [Google Scholar] [CrossRef]
- Jacobs, P.A.; Mayer, M. The origin of human Trisomy: A study of heteromorphisms and satellite associations. Ann. Hum. Genet. 1981, 45, 357–365. [Google Scholar] [CrossRef]
- Marcatili, M.; Marsoner, F.; D’Agostino, A.; Karnavas, T.; Bottai, D.; Scarone, S.; Conti, L. Establishment of an induced pluripotent stem cell (IPSC) line from a patient with clozapine-responder Schizophrenia. Stem Cell Res. 2016, 17, 630–633. [Google Scholar] [CrossRef][Green Version]
- Marsoner, F.; Marcatili, M.; Karnavas, T.; Bottai, D.; D’Agostino, A.; Scarone, S.; Conti, L. Generation and characterization of an induced pluripotent stem cell (IPSC) line from a patient with clozapine-resistant Schizophrenia. Stem Cell Res. 2016, 17, 661–664. [Google Scholar] [CrossRef] [PubMed]
| Antibody | Species | Dilution | Source | Identifier |
|---|---|---|---|---|
| OCT3/4 1 | Rabbit | 1:200 | Thermo Fisher Scientific | Cat A24867 RRID: AB_2650999 |
| SOX2 1 | Rat | 1:100 | Thermo Fisher Scientific | Cat A24759 RRID: AB_2651000 |
| TRA-1-60 1 | Mouse | 1:100 | Thermo Fisher Scientific | Cat A24868 RRID: AB_2651002 |
| SSEA4 1 | Mouse | 1:100 | Thermo Fisher Scientific | Cat A24866 RRID: AB_2651001 |
| NANOG | Rabbit | 1:400 | Cell Signaling Technology | Cat 4903 RRID: AB_10559205 |
| FOXA2/HNF3β | Goat | 1:100 | R&D Biotechne | Cat AF2400 RRID: AB_2294104 |
| αSMA | Mouse | 1:1000 | Abcam | Cat ab7817 RRID: AB_262054 |
| βIII Tubulin | Mouse | 1:5000 | Biolegend | Cat 801201 RRID: AB_2313773 |
| Alexa Fluor 488 Donkey anti-Rabbit IgG | 1:1000 | Thermo Fisher Scientific | Cat A21206 RRID: AB_2535792 | |
| Alexa Fluor 555 Donkey anti-Mouse IgG | 1:1000 | Thermo Fisher Scientific | Cat A31570 RRID: AB_2536180 | |
| Alexa Fluor 555 Donkey anti-Rabbit 1 | 1:250 | Thermo Fisher Scientific | Cat A24869 RRID: AB_2651006 | |
| Alexa Fluor 488 Goat anti-Mouse IgG3 1 | 1:250 | Thermo Fisher Scientific | Cat A24877 RRID: AB_2651008 | |
| Alexa Fluor 488 Donkey anti-Rat 1 | 1:250 | Thermo Fisher Scientific | Cat A24876 RRID: AB_2651007 | |
| Alexa Fluor 555 Goat anti-Mouse IgM 1 | 1:250 | Thermo Fisher Scientific | Cat A24871 RRID: AB_2651009 |
| Name | Sequence | Target |
|---|---|---|
| SZT2 gDNA M1 F | GGAACTGCAAGCTGACACCA | SZT2 genomic Exon 47 |
| SZT2 gDNA M1 R | TCGGTAGGAAAGAGTAAGTGCG | SZT2 genomic Exon 47 |
| SZT2 gDNA M2 F | TGGAAGCTGCATGTTTCTGC | SZT2 genomic Exon 4 |
| SZT2 gDNA M2 R | AACAGGTTCAAGAAGCCAGCA | SZT2 genomic Exon 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cattelani, C.; Battistella, I.; Di Leva, F.; Fioravanti, G.; Benedicenti, F.; Stanzial, F.; Schwienbacher, C.; Fanelli, F.; Pramstaller, P.P.; Hicks, A.A.; et al. Induced Pluripotent Stem Cell (iPSC) Lines from a Family with Resistant Epileptic Encephalopathy Caused by Compound Heterozygous Mutations in SZT2 Gene. Int. J. Mol. Sci. 2022, 23, 13095. https://doi.org/10.3390/ijms232113095
Cattelani C, Battistella I, Di Leva F, Fioravanti G, Benedicenti F, Stanzial F, Schwienbacher C, Fanelli F, Pramstaller PP, Hicks AA, et al. Induced Pluripotent Stem Cell (iPSC) Lines from a Family with Resistant Epileptic Encephalopathy Caused by Compound Heterozygous Mutations in SZT2 Gene. International Journal of Molecular Sciences. 2022; 23(21):13095. https://doi.org/10.3390/ijms232113095
Chicago/Turabian StyleCattelani, Cecilia, Ingrid Battistella, Francesca Di Leva, Giulia Fioravanti, Francesco Benedicenti, Franco Stanzial, Christine Schwienbacher, Francesca Fanelli, Peter P. Pramstaller, Andrew A. Hicks, and et al. 2022. "Induced Pluripotent Stem Cell (iPSC) Lines from a Family with Resistant Epileptic Encephalopathy Caused by Compound Heterozygous Mutations in SZT2 Gene" International Journal of Molecular Sciences 23, no. 21: 13095. https://doi.org/10.3390/ijms232113095
APA StyleCattelani, C., Battistella, I., Di Leva, F., Fioravanti, G., Benedicenti, F., Stanzial, F., Schwienbacher, C., Fanelli, F., Pramstaller, P. P., Hicks, A. A., Conti, L., & Corti, C. (2022). Induced Pluripotent Stem Cell (iPSC) Lines from a Family with Resistant Epileptic Encephalopathy Caused by Compound Heterozygous Mutations in SZT2 Gene. International Journal of Molecular Sciences, 23(21), 13095. https://doi.org/10.3390/ijms232113095






