Crosstalk between Tumor-Associated Macrophages and MicroRNAs: A Key Role in Tumor Microenvironment
Abstract
1. Introduction
2. Tumor-Associated Macrophages
2.1. Origin of Tumor-Associated Macrophages
2.2. Polarization of Tumor-Associated Macrophages: Double-Edged Sword
2.3. New Research Techniques Bring New Insights into the Complex Role of Tumor-Associated Macrophages
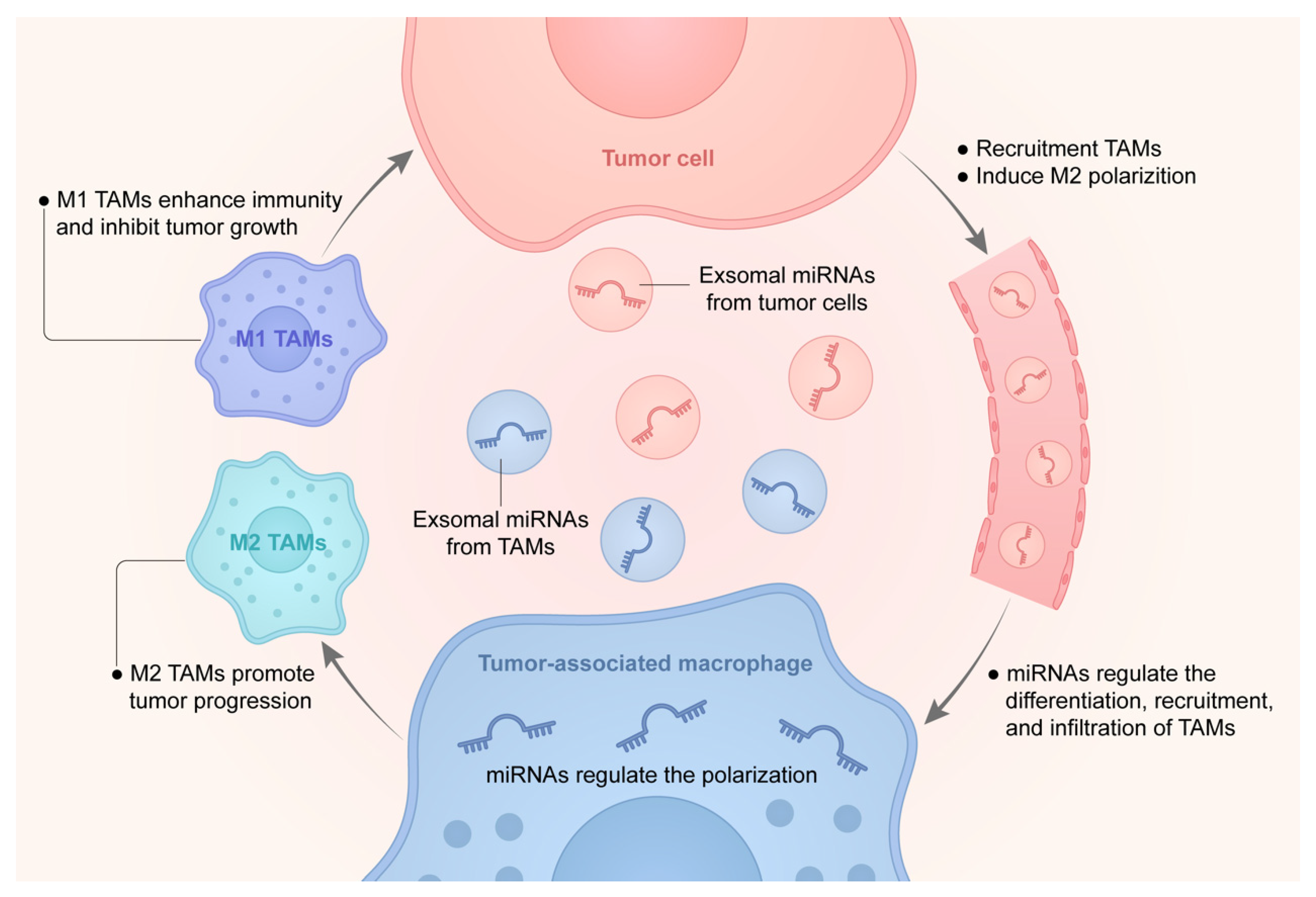
3. Interaction of MicroRNAs with Tumor-Associated Macrophages
3.1. Roles of miRNAs on the Differentiation, Recruitment, and Infiltration of Macrophages
3.2. Regulation of miRNAs on the Polarization of TAMs
3.3. Exosomes Derived miRNAs in the Interaction of TAMs and Cancer Cells
3.4. miRNAs Act as a Bridge for Cancer Cells to Affect the Function of TAMs
4. Clinical Application and Prospect
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Hinshaw, D.C.; Shevde, L.A. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Yu, D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol. Ther. 2021, 221, 107753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef]
- Kumari, S.; Advani, D.; Sharma, S.; Ambasta, R.K.; Kumar, P. Combinatorial therapy in tumor microenvironment: Where do we stand? Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188585. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; Denardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef]
- Sadeghi, R.H.; Monkman, J.; Warkiani, M.E.; Ladwa, R.; O’Byrne, K.; Rezaei, N.; Kulasinghe, A. Understanding the tumor microenvironment for effective immunotherapy. Med. Res. Rev. 2021, 41, 1474–1498. [Google Scholar] [CrossRef]
- Hegde, P.S.; Chen, D.S. Top 10 Challenges in Cancer Immunotherapy. Immunity 2020, 52, 17–35. [Google Scholar] [CrossRef]
- Kirtane, K.; Elmariah, H.; Chung, C.H.; Abate-Daga, D. Adoptive cellular therapy in solid tumor malignancies: Review of the literature and challenges ahead. J. Immunother. Cancer 2021, 9, e002723. [Google Scholar] [CrossRef]
- Mao, X.; Xu, J.; Wang, W.; Liang, C.; Hua, J.; Liu, J.; Zhang, B.; Meng, Q.; Yu, X.; Shi, S. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: New findings and future perspectives. Mol. Cancer 2021, 20, 131. [Google Scholar] [CrossRef]
- Pittet, M.J.; Michielin, O.; Migliorini, D. Clinical relevance of tumour-associated macrophages. Nat. Rev. Clin. Oncol. 2022, 19, 402–421. [Google Scholar] [CrossRef]
- Locati, M.; Curtale, G.; Mantovani, A. Diversity, mechanisms, and significance of macrophage plasticity. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 123–147. [Google Scholar] [CrossRef] [PubMed]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2021, 21, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory mechanism of microRNA expression in cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef] [PubMed]
- Kohlhapp, F.J.; Mitra, A.K.; Lengyel, E.; Peter, M.E. MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene 2015, 34, 5857–5868. [Google Scholar] [CrossRef]
- Ham, S.; Lima, L.G.; Lek, E.; Möller, A. The impact of the cancer microenvironment on macrophage phenotypes. Front. Immunol. 2020, 11, 1308. [Google Scholar] [CrossRef]
- Scott, C.L.; Zheng, F.; De Baetselier, P.; Martens, L.; Saeys, Y.; De Prijck, S.; Lippens, S.; Abels, C.; Schoonooghe, S.; Raes, G.; et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 2016, 7, 10321. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, Z.; Gao, J.; Lin, Z.; Wang, Y.; Shan, P.; Li, M.; Zhou, T.; Li, P. Noncoding RNA-mediated macrophage and cancer cell crosstalk in hepatocellular carcinoma. Mol. Ther. Oncol. 2022, 25, 98–120. [Google Scholar] [CrossRef]
- Mass, E.; Ballesteros, I.; Farlik, M.; Halbritter, F.; Günther, P.; Crozet, L.; Jacome-Galarza, C.E.; Händler, K.; Klughammer, J.; Kobayashi, Y.; et al. Specification of tissue-resident macrophages during organogenesis. Science 2016, 353, aaf4238. [Google Scholar] [CrossRef]
- Loyher, P.; Hamon, P.; Laviron, M.; Meghraoui-Kheddar, A.; Goncalves, E.; Deng, Z.; Torstensson, S.; Bercovici, N.; Baudesson De Chanville, C.; Combadière, B. Macrophages of distinct origins contribute to tumor development in the lung. J. Exp. Med. 2018, 215, 2536–2553. [Google Scholar] [CrossRef]
- Cotechini, T.; Atallah, A.; Grossman, A. Tissue-Resident and recruited macrophages in primary tumor and metastatic microenvironments: Potential targets in cancer therapy. Cells 2021, 10, 960. [Google Scholar] [CrossRef]
- Singhal, S.; Stadanlick, J.; Annunziata, M.J.; Rao, A.S.; Bhojnagarwala, P.S.; O Brien, S.; Moon, E.K.; Cantu, E.; Danet-Desnoyers, G.; Ra, H. Human tumor-associated monocytes/macrophages and their regulation of T cell responses in early-stage lung cancer. Sci. Transl. Med. 2019, 11, eaat1500. [Google Scholar] [CrossRef]
- Lee, H.; Choi, H.; Ha, S.; Lee, K.; Kwon, Y. Recruitment of monocytes/macrophages in different tumor microenvironments. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2013, 1835, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Song, Y.; Du, W.; Gong, L.; Chang, H.; Zou, Z. Tumor-associated macrophages: An accomplice in solid tumor progression. J. Biomed. Sci. 2019, 26, 78. [Google Scholar] [CrossRef] [PubMed]
- Toth, C.A.; Thomas, P. Liver endocytosis and Kupffer cells. Hepatology 1992, 16, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.J.; Barnes, M.; Tang, H.; Pritchard, M.T.; Nagy, L.E. Kupffer cells in the liver. Compr. Physiol. 2013, 3, 785–797. [Google Scholar]
- Morris, D.G.; Huang, X.; Kaminski, N.; Wang, Y.; Shapiro, S.D.; Dolganov, G.; Glick, A.; Sheppard, D. Loss of integrin alpha(v)beta6-mediated TGF-beta activation causes Mmp12-dependent emphysema. Nature 2003, 422, 169–173. [Google Scholar] [CrossRef]
- Snelgrove, R.J.; Goulding, J.; Didierlaurent, A.M.; Lyonga, D.; Vekaria, S.; Edwards, L.; Gwyer, E.; Sedgwick, J.D.; Barclay, A.N.; Hussell, T. A critical function for CD200 in lung immune homeostasis and the severity of influenza infection. Nat. Immunol. 2008, 9, 1074–1083. [Google Scholar] [CrossRef]
- Kumagai, Y.; Takeuchi, O.; Kato, H.; Kumar, H.; Matsui, K.; Morii, E.; Aozasa, K.; Kawai, T.; Akira, S. Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity 2007, 27, 240–252. [Google Scholar] [CrossRef]
- Tate, M.D.; Pickett, D.L.; van Rooijen, N.; Brooks, A.G.; Reading, P.C. Critical role of airway macrophages in modulating disease severity during influenza virus infection of mice. J. Virol. 2010, 84, 7569–7580. [Google Scholar] [CrossRef]
- Ginhoux, F.; Lim, S.; Hoeffel, G.; Low, D.; Huber, T. Origin and differentiation of microglia. Front. Cell. Neurosci. 2013, 7, 45. [Google Scholar] [CrossRef]
- Christofides, A.; Strauss, L.; Yeo, A.; Cao, C.; Charest, A.; Boussiotis, V.A. The complex role of tumor-infiltrating macrophages. Nat. Immunol. 2022, 23, 1148–1156. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Herndon, J.M.; Sojka, D.K.; Kim, K.; Knolhoff, B.L.; Zuo, C.; Cullinan, D.R.; Luo, J.; Bearden, A.R.; Lavine, K.J. Tissue-resident macrophages in pancreatic ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity 2017, 47, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Solinas, G.; Schiarea, S.; Liguori, M.; Fabbri, M.; Pesce, S.; Zammataro, L.; Pasqualini, F.; Nebuloni, M.; Chiabrando, C.; Mantovani, A. Tumor-conditioned macrophages secrete migration-stimulating factor: A new marker for M2-polarization, influencing tumor cell motility. J. Immunol. 2010, 185, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Biswas, S.K.; Galdiero, M.R.; Sica, A.; Locati, M. Macrophage plasticity and polarization in tissue repair and remodelling. J. Pathol. 2013, 229, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Shapouri Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Yin, Y.; Li, N.; Zhu, D.; Zhang, J.; Zhang, C.; Zen, K. Re-polarization of tumor-associated macrophages to pro-inflammatory M1 macrophages by microRNA-155. J. Mol. Cell Biol. 2012, 4, 341–343. [Google Scholar] [CrossRef] [PubMed]
- Flavell, R.A.; Sanjabi, S.; Wrzesinski, S.H.; Licona-Limón, P. The polarization of immune cells in the tumour environment by TGFβ. Nat. Rev. Immunol. 2010, 10, 554–567. [Google Scholar] [CrossRef]
- Han, S.; Wang, W.; Wang, S.; Yang, T.; Zhang, G.; Wang, D.; Ju, R.; Lu, Y.; Wang, H.; Wang, L. Tumor microenvironment remodeling and tumor therapy based on M2-like tumor associated macrophage-targeting nano-complexes. Theranostics 2021, 11, 2892. [Google Scholar] [CrossRef]
- Yang, Q.; Guo, N.; Zhou, Y.; Chen, J.; Wei, Q.; Han, M. The role of tumor-associated macrophages (TAMs) in tumor progression and relevant advance in targeted therapy. Acta Pharm. Sin. B 2020, 10, 2156–2170. [Google Scholar] [CrossRef]
- Li, Y.; Cao, F.; Li, M.; Li, P.; Yu, Y.; Xiang, L.; Xu, T.; Lei, J.; Tai, Y.Y.; Zhu, J. Hydroxychloroquine induced lung cancer suppression by enhancing chemo-sensitization and promoting the transition of M2-TAMs to M1-like macrophages. J. Exp. Clin. Cancer Res. 2018, 37, 1–16. [Google Scholar] [CrossRef]
- Binnemars Postma, K.; Bansal, R.; Storm, G.; Prakash, J. Targeting the Stat6 pathway in tumor-associated macrophages reduces tumor growth and metastatic niche formation in breast cancer. FASEB J. 2018, 32, 969–978. [Google Scholar] [CrossRef] [PubMed]
- Griess, B.; Mir, S.; Datta, K.; Teoh-Fitzgerald, M. Scavenging reactive oxygen species selectively inhibits M2 macrophage polarization and their pro-tumorigenic function in part, via Stat3 suppression. Free Radic. Biol. Med. 2020, 147, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Mazzone, M.; Menga, A.; Castegna, A. Metabolism and TAM functions—It takes two to tango. FEBS J. 2018, 285, 700–716. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, D.; Cang, H.; Guo, B. Crosstalk between cancer and immune cells: Role of tumor-associated macrophages in the tumor microenvironment. Cancer Med. 2019, 8, 4709–4721. [Google Scholar] [CrossRef]
- Wang, L.X.; Zhang, S.X.; Wu, H.J.; Rong, X.L.; Guo, J. M2b macrophage polarization and its roles in diseases. J. Leukoc. Biol. 2019, 106, 345–358. [Google Scholar] [CrossRef]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Duluc, D.; Delneste, Y.; Tan, F.; Moles, M.; Grimaud, L.; Lenoir, J.; Preisser, L.; Anegon, I.; Catala, L.; Ifrah, N. Tumor-associated leukemia inhibitory factor and IL-6 skew monocyte differentiation into tumor-associated macrophage-like cells. Blood J. Am. Soc. Hematol. 2007, 110, 4319–4330. [Google Scholar] [CrossRef]
- Zhou, K.; Cheng, T.; Zhan, J.; Peng, X.; Zhang, Y.; Wen, J.; Chen, X.; Ying, M. Targeting tumor-associated macrophages in the tumor microenvironment. Oncol. Lett. 2020, 20, 234. [Google Scholar] [CrossRef]
- Tsuchimoto, Y.; Asai, A.; Tsuda, Y.; Ito, I.; Nishiguchi, T.; Garcia, M.C.; Suzuki, S.; Kobayashi, M.; Higuchi, K.; Suzuki, F. M2b monocytes provoke bacterial pneumonia and gut bacteria–associated sepsis in alcoholics. J. Immunol. 2015, 195, 5169–5177. [Google Scholar] [CrossRef]
- Myers, K.V.; Amend, S.R.; Pienta, K.J. Targeting Tyro3, Axl and MerTK (TAM receptors): Implications for macrophages in the tumor microenvironment. Mol. Cancer 2019, 18, 94. [Google Scholar] [CrossRef]
- Cheng, H.; Wang, Z.; Fu, L.; Xu, T. Macrophage polarization in the development and progression of ovarian cancers: An overview. Front. Oncol. 2019, 9, 421. [Google Scholar] [CrossRef] [PubMed]
- Boutilier, A.J.; Elsawa, S.F. Macrophage polarization states in the tumor microenvironment. Int. J. Mol. Sci. 2021, 22, 6995. [Google Scholar] [CrossRef]
- Xu, Y.; Lu, J.; Tang, Y.; Xie, W.; Zhang, H.; Wang, B.; Zhang, S.; Hou, W.; Zou, C.; Jiang, P. PINK1 deficiency in gastric cancer compromises mitophagy, promotes the Warburg effect, and facilitates M2 polarization of macrophages. Cancer Lett. 2022, 529, 19–36. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Dai, X.; Zhang, X.; Mao, Z.; Huang, H. Sophoridine suppresses macrophage-mediated immunosuppression through TLR4/IRF3 pathway and subsequently upregulates CD8+ T cytotoxic function against gastric cancer. Biomed. Pharmacother. 2020, 121, 109636. [Google Scholar] [CrossRef] [PubMed]
- Kouketsu, A.; Sato, I.; Oikawa, M.; Shimizu, Y.; Saito, H.; Tashiro, K.; Yamashita, Y.; Takahashi, T.; Kumamoto, H. Regulatory T cells and M2-polarized tumour-associated macrophages are associated with the oncogenesis and progression of oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 2019, 48, 1279–1288. [Google Scholar] [CrossRef]
- Yu, M.; Guan, R.; Hong, W.; Zhou, Y.; Lin, Y.; Jin, H.; Hou, B.; Jian, Z. Prognostic value of tumor-associated macrophages in pancreatic cancer: A meta-analysis. Cancer Manag. Res. 2019, 11, 4041. [Google Scholar] [CrossRef]
- Yagi, T.; Baba, Y.; Okadome, K.; Kiyozumi, Y.; Hiyoshi, Y.; Ishimoto, T.; Iwatsuki, M.; Miyamoto, Y.; Yoshida, N.; Watanabe, M. Tumour-associated macrophages are associated with poor prognosis and programmed death ligand 1 expression in oesophageal cancer. Eur. J. Cancer 2019, 111, 38–49. [Google Scholar] [CrossRef]
- Kumar, S.; Ramesh, A.; Kulkarni, A. Targeting macrophages: A novel avenue for cancer drug discovery. Expert Opin. Drug Dis. 2020, 15, 561–574. [Google Scholar] [CrossRef]
- Cheng, N.; Bai, X.; Shu, Y.; Ahmad, O.; Shen, P. Targeting tumor-associated macrophages as an antitumor strategy. Biochem. Pharmacol. 2021, 183, 114354. [Google Scholar] [CrossRef]
- Qian, B.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Mckay, D.; Pollard, J.W.; Lewis, C.E. Diverse functions of macrophages in different tumor MicroenvironmentsSpatial heterogeneity of TAMs in tumors. Cancer Res. 2018, 78, 5492–5503. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Li, Z.; Gao, R.; Xing, B.; Gao, Y.; Yang, Y.; Qin, S.; Zhang, L.; Ouyang, H.; Du, P. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 2021, 184, 792–809. [Google Scholar] [CrossRef] [PubMed]
- Cassetta, L.; Fragkogianni, S.; Sims, A.H.; Swierczak, A.; Forrester, L.M.; Zhang, H.; Soong, D.Y.; Cotechini, T.; Anur, P.; Lin, E.Y. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell 2019, 35, 588–602. [Google Scholar] [CrossRef]
- Kovaleva, O.V.; Samoilova, D.V.; Shitova, M.S.; Gratchev, A. Tumor associated macrophages in kidney cancer. Anal. Cell. Pathol. 2016, 2016, 9307549. [Google Scholar] [CrossRef]
- Falleni, M.; Savi, F.; Tosi, D.; Agape, E.; Cerri, A.; Moneghini, L.; Bulfamante, G.P. M1 and M2 macrophages’ clinicopathological significance in cutaneous melanoma. Melanoma Res. 2017, 27, 200–210. [Google Scholar] [CrossRef]
- Kainulainen, K.; Takabe, P.; Heikkinen, S.; Aaltonen, N.; de la Motte, C.; Rauhala, L.; Durst, F.C.; Oikari, S.; Hukkanen, T.; Rahunen, E.; et al. M1 macrophages induce protumor inflammation in melanoma cells through TNFR-NF-κB signaling. J. Investig. Dermatol. 2022, 142, 3041–3051.e10. [Google Scholar] [CrossRef]
- Huggins, D.N.; Larue, R.S.; Wang, Y.; Knutson, T.P.; Xu, Y.; Williams, J.W.; Schwertfeger, K.L. Characterizing macrophage diversity in metastasis-bearing lungs reveals a lipid-associated macrophage subset. Cancer Res. 2021, 81, 5284–5295. [Google Scholar] [CrossRef]
- Zilionis, R.; Engblom, C.; Pfirschke, C.; Savova, V.; Zemmour, D.; Saatcioglu, H.D.; Krishnan, I.; Maroni, G.; Meyerovitz, C.V.; Kerwin, C.M. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 2019, 50, 1317–1334. [Google Scholar] [CrossRef]
- Wu, S.Z.; Al-Eryani, G.; Roden, D.L.; Junankar, S.; Harvey, K.; Andersson, A.; Thennavan, A.; Wang, C.; Torpy, J.R.; Bartonicek, N. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 2021, 53, 1334–1347. [Google Scholar] [CrossRef]
- Bi, K.; He, M.X.; Bakouny, Z.; Kanodia, A.; Napolitano, S.; Wu, J.; Grimaldi, G.; Braun, D.A.; Cuoco, M.S.; Mayorga, A. Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell 2021, 39, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.H.; Sun, L.; Mochizuki, A.Y.; Reynoso, J.G.; Orpilla, J.; Chow, F.; Kienzler, J.C.; Everson, R.G.; Nathanson, D.A.; Bensinger, S.J.; et al. Neoadjuvant PD-1 blockade induces T cell and cDC1 activation but fails to overcome the immunosuppressive tumor associated macrophages in recurrent glioblastoma. Nat. Commun. 2021, 12, 6938. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S.M. MicroRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Mcgeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Vannini, I.; Fanini, F.; Fabbri, M. Emerging roles of microRNAs in cancer. Curr. Opin. Genet. Dev. 2018, 48, 128–133. [Google Scholar] [CrossRef]
- Saw, P.E.; Xu, X.; Chen, J.; Song, E.W. Non-coding RNAs: The new central dogma of cancer biology. Sci. China Life Sci. 2021, 64, 22–50. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Curtale, G.; Rubino, M.; Locati, M. MicroRNAs as molecular switches in macrophage activation. Front. Immunol. 2019, 10, 799. [Google Scholar] [CrossRef]
- Cortez-Retamozo, V.; Etzrodt, M.; Newton, A.; Rauch, P.J.; Chudnovskiy, A.; Berger, C.; Ryan, R.J.; Iwamoto, Y.; Marinelli, B.; Gorbatov, R. Origins of tumor-associated macrophages and neutrophils. Proc. Natl. Acad. Sci. USA 2012, 109, 2491–2496. [Google Scholar] [CrossRef]
- Geissmann, F.; Manz, M.G.; Jung, S.; Sieweke, M.H.; Merad, M.; Ley, K. Development of monocytes, macrophages, and dendritic cells. Science 2010, 327, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Ran, D.; Shia, W.; Lo, M.; Fan, J.; Knorr, D.A.; Ferrell, P.I.; Ye, Z.; Yan, M.; Cheng, L.; Kaufman, D.S. RUNX1a enhances hematopoietic lineage commitment from human embryonic stem cells and inducible pluripotent stem cells. Blood J. Am. Soc. Hematol. 2013, 121, 2882–2890. [Google Scholar] [CrossRef] [PubMed]
- Ghani, S.; Riemke, P.; Schönheit, J.; Lenze, D.; Stumm, J.; Hoogenkamp, M.; Lagendijk, A.; Heinz, S.; Bonifer, C.; Bakkers, J. Macrophage development from HSCs requires PU. 1-Coordinated microRNA expression. Blood J. Am. Soc. Hematol. 2011, 118, 2275–2284. [Google Scholar] [CrossRef] [PubMed]
- Van Overmeire, E.; Stijlemans, B.; Heymann, F.; Keirsse, J.; Morias, Y.; Elkrim, Y.; Brys, L.; Abels, C.; Lahmar, Q.; Ergen, C. M-CSF and GM-CSF receptor signaling differentially regulate monocyte maturation and macrophage polarization in the tumor microenvironment. Cancer Res. 2016, 76, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Haneklaus, M.; Gerlic, M.; O’Neill, L.A.; Masters, S.L. MiR-223: Infection, inflammation and cancer. J. Intern. Med. 2013, 274, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Zhao, J.; Wang, L.; Gao, C.; Liang, S.; An, D.; Bai, J.; Chen, Y.; Han, H.; Qin, H. MiR-148a-3p mediates Notch signaling to promote the differentiation and M1 activation of macrophages. Front. Immunol. 2017, 8, 1327. [Google Scholar] [CrossRef]
- Batool, A.; Wang, Y.Q.; Hao, X.X.; Chen, S.R.; Liu, Y.X. A miR-125b/CSF1-CX3CL1/tumor-associated macrophage recruitment axis controls testicular germ cell tumor growth. Cell Death Dis. 2018, 9, 962. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, K.; Xu, Y.; Gao, Y.; Li, C.; Wang, R.; Chen, L. The role of microRNA-26a in human cancer progression and clinical application. Tumor Biol. 2016, 37, 7095–7108. [Google Scholar] [CrossRef]
- Chai, Z.; Zhu, X.; Ao, J.; Wang, W.; Gao, D.; Kong, J.; Zhang, N.; Zhang, Y.; Ye, B.; Ma, D. MicroRNA-26a suppresses recruitment of macrophages by down-regulating macrophage colony-stimulating factor expression through the PI3K/Akt pathway in hepatocellular carcinoma. J. Hematol. Oncol. 2015, 8, 56. [Google Scholar] [CrossRef]
- Frank, A.; Ebersberger, S.; Fink, A.F.; Lampe, S.; Weigert, A.; Schmid, T.; Ebersberger, I.; Syed, S.N.; Brüne, B. Apoptotic tumor cell-derived microRNA-375 uses CD36 to alter the tumor-associated macrophage phenotype. Nat. Commun. 2019, 10, 1135. [Google Scholar] [CrossRef]
- Murray, P.J. Macrophage polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef]
- Zhao, S.; Mi, Y.; Guan, B.; Zheng, B.; Wei, P.; Gu, Y.; Zhang, Z.; Cai, S.; Xu, Y.; Li, X.; et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J. Hematol. Oncol. 2020, 13, 156. [Google Scholar] [CrossRef]
- Shinohara, H.; Kuranaga, Y.; Kumazaki, M.; Sugito, N.; Yoshikawa, Y.; Takai, T.; Taniguchi, K.; Ito, Y.; Akao, Y. Regulated polarization of Tumor-Associated macrophages by miR-145 via colorectal Cancer-Derived extracellular vesicles. J. Immunol. 2017, 199, 1505–1515. [Google Scholar] [CrossRef]
- Yuan, R.; Li, S.; Geng, H.; Wang, X.; Guan, Q.; Li, X.; Ren, C.; Yuan, X. Reversing the polarization of tumor-associated macrophages inhibits tumor metastasis. Int. Immunopharmacol. 2017, 49, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Wang, S.; Sun, M.; Zhang, C.; Wei, C.; Yang, C.; Dou, R.; Liu, Q.; Xiong, B. MiR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J. Hematol. Oncol. 2019, 12, 20. [Google Scholar] [CrossRef]
- Lin, Z.Y.; Huang, Y.Q.; Zhang, Y.Q.; Han, Z.D.; He, H.C.; Ling, X.H.; Fu, X.; Dai, Q.S.; Cai, C.; Chen, J.H. MicroRNA-224 inhibits progression of human prostate cancer by downregulating TRIB1. Int. J. Cancer 2014, 135, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Zang, X.; Zhang, X.; Zhao, X.; Hu, H.; Qiao, M.; Deng, Y.; Chen, D. Targeted delivery of miRNA 155 to tumor associated macrophages for tumor immunotherapy. Mol. Pharm. 2019, 16, 1714–1722. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Sun, Y.; Wang, X.; Park, J.; Zhang, Y.; Li, M.; Yin, J.; Liu, Q.; Wei, M. Progress on the relationship between miR-125 family and tumorigenesis. Exp. Cell Res. 2015, 339, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Wei, J.; Wang, F.; Kong, L.Y.; Ling, X.Y.; Nduom, E.; Gabrusiewicz, K.; Doucette, T.; Yang, Y.; Yaghi, N.K.; et al. Effect of miR-142-3p on the M2 macrophage and therapeutic efficacy against murine glioblastoma. J. Natl. Cancer Inst. 2014, 106, dju162. [Google Scholar] [CrossRef]
- Wang, L.; Hu, Y.Y.; Zhao, J.L.; Huang, F.; Liang, S.Q.; Dong, L.; Chen, Y.; Yu, H.C.; Bai, J.; Yang, J.M.; et al. Targeted delivery of miR-99b reprograms tumor-associated macrophage phenotype leading to tumor regression. J. Immunother. Cancer 2020, 8, e000517. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Xu, Q.I.; Yao, W.; Wu, Q.; Yuan, J.; Yan, W.; Xu, T.; Ji, X.; Ni, C. Long non-coding RNA-ATB promotes EMT during silica-induced pulmonary fibrosis by competitively binding miR-200c. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2018, 1864, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.M.; Christenson, J.L.; O’Neill, K.I.; Hafeez, S.A.; Ihle, C.L.; Spoelstra, N.S.; Slansky, J.E.; Richer, J.K. MicroRNA-200c restoration reveals a cytokine profile to enhance M1 macrophage polarization in breast cancer. NPJ Breast Cancer 2021, 7, 64. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Cui, H.; Xie, N.; Tan, Z.; Yang, S.; Icyuz, M.; Thannickal, V.J.; Abraham, E.; Liu, G. MiR-125a-5p regulates differential activation of macrophages and inflammation. J. Biol. Chem. 2013, 288, 35428–35436. [Google Scholar] [CrossRef] [PubMed]
- Gerloff, D.; Lützkendorf, J.; Moritz, R.K.; Wersig, T.; Mäder, K.; Müller, L.P.; Sunderkötter, C. Melanoma-derived exosomal miR-125b-5p educates tumor associated macrophages (TAMs) by targeting lysosomal acid lipase a (LIPA). Cancers 2020, 12, 464. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, D. Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Pascut, D.; Pratama, M.Y.; Vo, N.; Masadah, R.; Tiribelli, C. The crosstalk between tumor cells and the microenvironment in hepatocellular carcinoma: The role of exosomal microRNAs and their clinical implications. Cancers 2020, 12, 823. [Google Scholar] [CrossRef]
- Li, B.; Cao, Y.; Sun, M.; Feng, H. Expression, regulation, and function of exosome-derived miRNAs in cancer progression and therapy. FASEB J. 2021, 35, e21916. [Google Scholar] [CrossRef]
- Aziz, F. The emerging role of miR-223 as novel potential diagnostic and therapeutic target for inflammatory disorders. Cell. Immunol. 2016, 303, 1–6. [Google Scholar] [CrossRef]
- Yang, M.; Chen, J.; Su, F.; Yu, B.; Su, F.; Lin, L.; Liu, Y.; Huang, J.; Song, E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer 2011, 10, 117. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, B.; Xiao, S.; Li, Y.; Chen, Q. MiR-125a/b inhibits tumor-associated macrophages mediated in cancer stem cells of hepatocellular carcinoma by targeting CD90. J. Cell. Biochem. 2019, 120, 3046–3055. [Google Scholar] [CrossRef]
- Lan, J.; Sun, L.; Xu, F.; Liu, L.; Hu, F.; Song, D.; Hou, Z.; Wu, W.; Luo, X.; Wang, J.; et al. M2 Macrophage-Derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019, 79, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Sun, J.; Chen, J.; Xu, C.; Zhang, L. Downregulation of miR-95 in gastric cancer promotes EMT via regulation of Slug, thereby promoting migration and invasion. Oncol. Rep. 2019, 41, 1395–1403. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Meng, W.; Yue, P.; Li, X. M2 macrophage-derived extracellular vesicles promote gastric cancer progression via a microRNA-130b-3p/MLL3/GRHL2 signaling cascade. J. Exp. Clin. Cancer Res. 2020, 39, 134. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Cai, S.; Shu, Y.; Deng, X.; Zhang, Y.; He, N.; Wan, L.; Chen, X.; Qu, Y.; Yu, S. Exosomal miR-487a derived from m2 macrophage promotes the progression of gastric cancer. Cell Cycle 2021, 20, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yao, S.; Hu, Y.; Feng, Y.; Li, M.; Bian, Z.; Zhang, J.; Qin, Y.; Qi, X.; Zhou, L.; et al. The immune-microenvironment confers chemoresistance of colorectal cancer through Macrophage-Derived IL6. Clin. Cancer Res. 2017, 23, 7375–7387. [Google Scholar] [CrossRef]
- Zheng, P.; Chen, L.; Yuan, X.; Luo, Q.; Liu, Y.; Xie, G.; Ma, Y.; Shen, L. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J. Exp. Clin. Cancer Res. 2017, 36, 53. [Google Scholar] [CrossRef]
- Zhu, X.; Shen, H.; Yin, X.; Yang, M.; Wei, H.; Chen, Q.; Feng, F.; Liu, Y.; Xu, W.; Li, Y. Macrophages derived exosomes deliver miR-223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J. Exp. Clin. Cancer Res. 2019, 38, 81. [Google Scholar] [CrossRef]
- Binenbaum, Y.; Fridman, E.; Yaari, Z.; Milman, N.; Schroeder, A.; Ben, D.G.; Shlomi, T.; Gil, Z. Transfer of miRNA in Macrophage-Derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. 2018, 78, 5287–5299. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, J.; Chen, Z.; Wang, H.; Xue, H.; Yang, C.; Guo, Q.; Qi, Y.; Guo, X.; Qian, M.; et al. Transfer of MicroRNA via Macrophage-Derived extracellular vesicles promotes Proneural-to-Mesenchymal transition in glioma stem cells. Cancer Immunol. Res. 2020, 8, 966–981. [Google Scholar] [CrossRef]
- Aucher, A.; Rudnicka, D.; Davis, D.M. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J. Immunol. 2013, 191, 6250–6260. [Google Scholar] [CrossRef]
- Hu, Y.; Li, D.; Wu, A.; Qiu, X.; Di, W.; Huang, L.; Qiu, L. TWEAK-stimulated macrophages inhibit metastasis of epithelial ovarian cancer via exosomal shuttling of microRNA. Cancer Lett. 2017, 393, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Moradi-Chaleshtori, M.; Shojaei, S.; Mohammadi-Yeganeh, S.; Hashemi, S.M. Transfer of miRNA in tumor-derived exosomes suppresses breast tumor cell invasion and migration by inducing M1 polarization in macrophages. Life Sci. 2021, 282, 119800. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Wu, Q.; Wu, X.; Zhu, Q.; Wang, X.; Jiang, L.; Chen, X.; Wang, X. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget 2016, 7, 43076–43087. [Google Scholar] [CrossRef] [PubMed]
- Xun, J.; Du, L.; Gao, R.; Shen, L.; Wang, D.; Kang, L.; Chen, C.; Zhang, Z.; Zhang, Y.; Yue, S.; et al. Cancer-derived exosomal miR-138-5p modulates polarization of tumor-associated macrophages through inhibition of KDM6B. Theranostics 2021, 11, 6847–6859. [Google Scholar] [CrossRef] [PubMed]
- Takano, Y.; Masuda, T.; Iinuma, H.; Yamaguchi, R.; Sato, K.; Tobo, T.; Hirata, H.; Kuroda, Y.; Nambara, S.; Hayashi, N.; et al. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget 2017, 8, 78598–78613. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ying, X.; Wang, X.; Wu, X.; Zhu, Q.; Wang, X. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol. Rep. 2017, 38, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.; Hung, J.; Chang, W.; Jian, S.; Lin, Y.; Pan, Y.; Wu, C.; Kuo, P. Hypoxic lung-cancer-derived extracellular vesicle microRNA-103a increases the oncogenic effects of macrophages by targeting PTEN. Mol. Ther. 2018, 26, 568–581. [Google Scholar] [CrossRef]
- Park, J.E.; Dutta, B.; Tse, S.W.; Gupta, N.; Tan, C.F.; Low, J.K.; Yeoh, K.W.; Kon, O.L.; Tam, J.P.; Sze, S.K. Hypoxia-induced tumor exosomes promote M2-like macrophage polarization of infiltrating myeloid cells and microRNA-mediated metabolic shift. Oncogene 2019, 38, 5158–5173. [Google Scholar] [CrossRef]
- Jang, J.; Lee, J.; Jeon, Y.; Kim, C. Exosome derived from epigallocatechin gallate treated breast cancer cells suppresses tumor growth by inhibiting tumor-associated macrophage infiltration and M2 polarization. BMC Cancer 2013, 13, 421. [Google Scholar] [CrossRef]
- Su, M.; Aldawsari, H.; Amiji, M. Pancreatic cancer cell exosome-mediated macrophage reprogramming and the role of microRNAs 155 and 125b2 transfection using nanoparticle delivery systems. Sci. Rep. 2016, 6, 30110. [Google Scholar] [CrossRef]
- Meng, X.; Yin, J.; Yu, X.; Guo, Y. MicroRNA-205-5p promotes unstable atherosclerotic plaque formation in vivo. Cardiovasc. Drug Ther. 2020, 34, 25–39. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Zhang, N.; Lin, Z. MicroRNA-125a-5p modulates macrophage polarization by targeting E26 transformation-specific variant 6 gene during orthodontic tooth movement. Arch. Oral Biol. 2021, 124, 105060. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; He, X.; Xu, Y.; Zhang, W.; Fu, F. MiR-200b is upregulated in plasma-derived exosomes and functions as an oncogene by promoting macrophage M2 polarization in ovarian cancer. J. Ovarian Res. 2021, 14, 74. [Google Scholar] [CrossRef] [PubMed]
- Noorolyai, S.; Duijf, P.H.; Silvestris, N.; Kolahian, S.; Hashemzadeh, S.; Fallahvazirabad, A.; Baradaran, B. The impact of microRNAs on myeloid-derived suppressor cells in cancer. Hum. Immunol. 2021, 82, 668–678. [Google Scholar]
- Gajeton, J.; Krukovets, I.; Muppala, S.; Verbovetskiy, D.; Zhang, J.; Stenina-Adognravi, O. Hyperglycemia-Induced miR-467 drives tumor inflammation and growth in breast cancer. Cancers 2021, 13, 1346. [Google Scholar] [CrossRef]
- Mirzaei, S.; Saebfar, H.; Gholami, M.H.; Hashemi, F.; Zarrabi, A.; Zabolian, A.; Entezari, M.; Hushmandi, K.; Samarghandian, S.; Aref, A.R. MicroRNAs regulating SOX2 in cancer progression and therapy response. Expert Rev. Mol. Med. 2021, 23, e13. [Google Scholar] [CrossRef]
- Lee, J.Y.; Ryu, D.; Lim, S.W.; Ryu, K.J.; Choi, M.E.; Yoon, S.E.; Kim, K.; Park, C.; Kim, S.J. Exosomal miR-1305 in the oncogenic activity of hypoxic multiple myeloma cells: A biomarker for predicting prognosis. J. Cancer 2021, 12, 2825. [Google Scholar] [CrossRef]
- Gong, X.; Liu, Y.; Zheng, C.; Tian, P.; Peng, M.; Pan, Y.; Li, X. Establishment of a 4-miRNA prognostic model for risk stratification of patients with pancreatic adenocarcinoma. Front. Oncol. 2022, 12, 827259. [Google Scholar] [CrossRef]
- Cai, J.; Qiao, B.; Gao, N.; Lin, N.; He, W. Oral squamous cell carcinoma-derived exosomes promote M2 subtype macrophage polarization mediated by exosome-enclosed miR-29a-3p. Am. J. Physiol.-Cell Physiol. 2019, 316, C731–C740. [Google Scholar] [CrossRef]
- Jia, X.; Hu, X.; Han, S.; Miao, X.; Liu, H.; Li, X.; Lin, Z.; Wang, Z.; Gong, W. Increased M1 macrophages in young miR-15a/16−/− mice with tumour grafts or dextran sulphate sodium-induced colitis. Scand. J. Immunol. 2018, 88, e12703. [Google Scholar] [CrossRef]
- Xia, X.; Zhang, K.; Luo, G.; Cen, G.; Cao, J.; Huang, K.; Qiu, Z. Downregulation of miR-301a-3p sensitizes pancreatic cancer cells to gemcitabine treatment via PTEN. Am. J. Transl. Res. 2017, 9, 1886–1895. [Google Scholar]
- Ghafouri-Fard, S.; Abak, A.; Avval, S.T.; Shoorei, H.; Taheri, M.; Samadian, M. The impact of non-coding RNAs on macrophage polarization. Biomed. Pharmacother. 2021, 142, 112112. [Google Scholar] [CrossRef]
- Cooks, T.; Pateras, I.S.; Jenkins, L.M.; Patel, K.M.; Robles, A.I.; Morris, J.; Forshew, T.; Appella, E.; Gorgoulis, V.G.; Harris, C.C. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 2018, 9, 771. [Google Scholar] [CrossRef] [PubMed]
- Ke, M.; Zhang, Z.; Cong, L.; Zhao, S.; Li, Y.; Wang, X.; Lv, Y.; Zhu, Y.; Dong, J. MicroRNA-148b-colony-stimulating factor-1 signaling-induced tumor-associated macrophage infiltration promotes hepatocellular carcinoma metastasis. Biomed. Pharmacother. 2019, 120, 109523. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yan, M.; Sun, F.; Song, J.; Hu, X.; Yu, S.; Tang, L.; Deng, S. MiR-498 inhibits autophagy and M2-like polarization of tumor-associated macrophages in esophageal cancer via MDM2/ATF3. Epigenomics 2021, 13, 1013–1030. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Luo, R.; Wang, J.; Luan, X.; Wu, D.; Chen, H.; Hou, Q.; Mao, G.; Li, X. Tumor cell-derived exosomal miR-770 inhibits M2 macrophage polarization via targeting MAP3K1 to inhibit the invasion of non-small cell lung cancer cells. Front. Cell Dev. Biol. 2021, 9, 1409. [Google Scholar] [CrossRef] [PubMed]
- Moradi-Chaleshtori, M.; Bandehpour, M.; Heidari, N.; Mohammadi-Yeganeh, S.; Hashemi, S.M. Exosome-mediated miR-33 transfer induces M1 polarization in mouse macrophages and exerts antitumor effect in 4T1 breast cancer cell line. Int. Immunopharmacol. 2021, 90, 107198. [Google Scholar] [CrossRef]
- Moradi-Chaleshtori, M.; Bandehpour, M.; Soudi, S.; Mohammadi-Yeganeh, S.; Hashemi, S.M. In vitro and in vivo evaluation of anti-tumoral effect of M1 phenotype induction in macrophages by miR-130 and miR-33 containing exosomes. Cancer Immunol. Immunother. 2021, 70, 1323–1339. [Google Scholar] [CrossRef]
- Wang, N.; Tan, H.; Feng, Y.; Zhang, C.; Chen, F.; Feng, Y. MicroRNA-23a in human cancer: Its roles, mechanisms and therapeutic relevance. Cancers 2018, 11, 7. [Google Scholar] [CrossRef]
- Li, Y.; Chen, D.; Jin, L.U.; Liu, J.; Li, Y.; Su, Z.; Qi, Z.; Shi, M.; Jiang, Z.; Yang, S. Oncogenic microRNA-142-3p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Oncol. Lett. 2016, 11, 1235–1241. [Google Scholar] [CrossRef]
- Banerjee, S.; Xie, N.; Cui, H.; Tan, Z.; Yang, S.; Icyuz, M.; Abraham, E.; Liu, G. MicroRNA let-7c regulates macrophage polarization. J. Immunol. 2013, 190, 6542–6549. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Li, X.; Chang, H.; Zhou, W.; Jin, B. MiRNA-99b inhibits the invasion ability of glioma cells by targeting mTOR. Chin. Pharmacol. Bull. 2018, 473–479. Available online: https://pesquisa.bvsalud.org/portal/resource/pt/wpr-705069 (accessed on 24 October 2022).
- Liu, Y.; Li, X.; Zhang, Y.; Wang, H.; Rong, X.; Peng, J.; He, L.; Peng, Y. An miR-340-5p-macrophage feedback loop modulates the progression and tumor microenvironment of glioblastoma multiforme. Oncogene 2019, 38, 7399–7415. [Google Scholar] [CrossRef]
- Garofalo, M.; Croce, C.M. MicroRNAs as therapeutic targets in chemoresistance. Drug Resist. Updates 2013, 16, 47–59. [Google Scholar] [CrossRef]
- Jiang, Q.; Yuan, Y.; Gong, Y.; Luo, X.; Su, X.; Hu, X.; Zhu, W. Therapeutic delivery of microRNA-143 by cationic lipoplexes for non-small cell lung cancer treatment in vivo. J. Cancer Res. Clin. Oncol. 2019, 145, 2951–2967. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Z.; Chen, C.; Liu, Y.; Si, Q.; Chuang, T.H.; Li, N.; Gomez-Cabrero, A.; Reisfeld, R.A.; Xiang, R.; et al. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene 2014, 33, 3014–3023. [Google Scholar] [CrossRef]
- Zhang, J.; Shan, W.F.; Jin, T.T.; Wu, G.Q.; Xiong, X.X.; Jin, H.Y.; Zhu, S.M. Propofol exerts anti-hepatocellular carcinoma by microvesicle-mediated transfer of miR-142-3p from macrophage to cancer cells. J. Transl. Med. 2014, 12, 279. [Google Scholar] [CrossRef]
- Chuang, H.; Su, Y.; Liu, H.; Chen, C.; Chiu, S.; Cho, D.; Lin, S.; Chen, Y.; Lin, C. Preclinical evidence of STAT3 inhibitor pacritinib overcoming temozolomide resistance via downregulating miR-21-enriched exosomes from M2 glioblastoma-associated macrophages. J. Clin. Med. 2019, 8, 959. [Google Scholar] [CrossRef]
- Van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Querfeld, C.; Foss, F.M.; Kim, Y.H.; Pinter-Brown, L.; William, B.M.; Porcu, P.; Pacheco, T.; Haverkos, B.M.; Desimone, J.; Guitart, J. Phase 1 trial of cobomarsen, an inhibitor of Mir-155, in cutaneous T cell lymphoma. Blood 2018, 132, 2903. [Google Scholar] [CrossRef]
- Wang, T.; Ren, Y.; Liu, R.; Ma, J.; Shi, Y.; Zhang, L.; Bu, R. MiR-195-5p suppresses the proliferation, migration, and invasion of oral squamous cell carcinoma by targeting TRIM14. Biomed. Res. Int. 2017, 2017, 7378148. [Google Scholar] [CrossRef] [PubMed]
- Pakravan, G.; Foroughmand, A.M.; Peymani, M.; Ghaedi, K.; Hashemi, M.; Hajjari, M.; Nasr-Esfahani, M.H. Downregulation of miR-130a, antagonized doxorubicin-induced cardiotoxicity via increasing the PPARγ expression in mESCs-derived cardiac cells. Cell Death Dis. 2018, 9, 758. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R.; Yang, C.H.; Pfeffer, L.M. The role of miR-21 in cancer. Drug Dev. Res. 2015, 76, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, S.; Möller, C.; Jirström, K.; Lee, A.; Busch, S.; Lamb, R.; Landberg, G. Downregulation of miR-92a is associated with aggressive breast cancer features and increased tumour macrophage infiltration. PLoS ONE 2012, 7, e36051. [Google Scholar] [CrossRef]
- Anandappa, G.; Lampis, A.; Cunningham, D.; Khan, K.H.; Kouvelakis, K.; Vlachogiannis, G.; Hedayat, S.; Tunariu, N.; Rao, S.; Watkins, D. MiR-31-3p expression and benefit from Anti-EGFR inhibitors in metastatic colorectal cancer patients enrolled in the prospective phase II PROSPECT-C TrialmiR-31-3p: A novel biomarker for Anti-EGFR treatment. Clin. Cancer Res. 2019, 25, 3830–3838. [Google Scholar] [CrossRef]
- Søkilde, R.; Persson, H.; Ehinger, A.; Pirona, A.C.; Fernö, M.; Hegardt, C.; Larsson, C.; Loman, N.; Malmberg, M.; Rydén, L. Refinement of breast cancer molecular classification by miRNA expression profiles. BMC Genom. 2019, 20, 503. [Google Scholar] [CrossRef]
- Dastmalchi, N.; Safaralizadeh, R.; Baradaran, B.; Hosseinpourfeizi, M.; Baghbanzadeh, A. An update review of deregulated tumor suppressive microRNAs and their contribution in various molecular subtypes of breast cancer. Gene 2020, 729, 144301. [Google Scholar] [CrossRef]
- Schotte, D.; Pieters, R.; Den Boer, M.L. MicroRNAs in acute leukemia: From biological players to clinical contributors. Leukemia 2012, 26, 1–12. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, D.; Du, W.; Gu, D.; Yang, R. MicroRNA and leukemia: Tiny molecule, great function. Crit. Rev. Oncol./Hematol. 2010, 74, 149–155. [Google Scholar] [CrossRef]
| MiRNAs Related to Macrophages | Function | Interactions between miRNAs and Macrophages | Ref. |
|---|---|---|---|
| Main Tumor-promoters | |||
| miR-21a | Tumor-promoting | Inducing M2 polarization of macrophages | [116] |
| miR-205 | Tumor-promoting | Recruiting macrophages, inducing inflammation in TME | [131] |
| miR-125a-5p | Tumor-promoting | Inducing M2 polarization of macrophages | [132] |
| miR-200b | Tumor-promoting | Triggering M2 polarization of macrophages | [133] |
| miR-200c | Tumor-promoting/suppressor | Inducing M1 or M2 polarization | [134] |
| miR-467 | Tumor-promoting | Enhancing TAM infiltration in TME | [135] |
| miR-223 | Tumor-promoting | Inducing IL-6 secretion by macrophages | [108] |
| miR-138-5p | Tumor-promoting | Inducing M2 polarization | [124] |
| miR-145 | Tumor-promoting | Inducing M2 polarization | [136] |
| miR-1305 | Tumor-promoting | Inducing M2 polarization | [137] |
| miR-375 | Tumor-promoting | Facilitating macrophage recruitment, M2 polarization | [90] |
| miR-934 | Tumor-promoting | Inducing M2 polarization | [138] |
| miR-29a-3p | Tumor-promoting | Inducing M2 polarization | [139] |
| miR-222-3p | Tumor-promoting | Inducing M2 polarization | [123] |
| miR-940 | Tumor-promoting | Inducing M2 polarization | [126] |
| miR-16 | Tumor-suppressor | Inducing M1 polarization | [140] |
| miR-103a | Tumor-promoting | Inducing M2 polarization | [127] |
| miR-301a-3p | Tumor-promoting | Enrichment of M2 macrophages | [141] |
| miR-132 | Tumor-promoting | Inducing M2b polarization | [142] |
| miR-1246 | Tumor-promoting | Reprograming to M2 polarization | [143] |
| miR-148b | Tumor-promoting | Inducing TAM infiltration in tumors | [144] |
| Main Tumor-suppressors | |||
| miR-498 | Tumor-suppressor | Preventing M2 polarization of macrophages | [145] |
| miR-770 | Tumor-suppressor | Preventing M2 polarization of macrophages | [146] |
| miR-125b | Tumor-suppressor | Reducing number of Macrophages in TME, repolarization of M1 macrophages | [87,98] |
| miR-33 | Tumor-suppressor | Inducing M1 polarization of macrophages | [147] |
| miR-130 | Tumor-suppressor | Triggering M1 polarization of macrophages | [148] |
| miR-23a | Tumor-suppressor | Promoting M1 polarization and inhibits M2 polarization | [149] |
| miR-155 | Tumor-suppressor | Reprogramming to an M1-like phenotype | [97] |
| miR-142-3p | Tumor-suppressor | Preventing M2 polarization of macrophages, triggering apoptosis in M2 macrophages | [150] |
| let-7c | Tumor-suppressor | Preventing M1 polarization and inducing M2 polarization of macrophages | [151] |
| miR-26a | Tumor-suppressor | suppress the recruitment of macrophages | [89] |
| miR-195-5p | Tumor-suppressor | Inhibits IL-4-mediated M2 polarization of macrophages | [95] |
| miR-224 | Tumor-suppressor | suppressed M2 macrophage polarization in the TME | [96] |
| miR-99b | Tumor-suppressor | induce M1 polarization of macrophages and enhance antitumor immunity by enhancing the ability of macrophages in phagocytosis and antigen presentation | [152] |
| miR-340-5p | Tumor-suppressor | Downregulation of miR-340-5p promoted TAM recruitment and M2 polarization | [153] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Chen, B.; Zhang, Z.; Huang, Y.; Li, J.; Wei, Q.; Cao, D.; Ai, J. Crosstalk between Tumor-Associated Macrophages and MicroRNAs: A Key Role in Tumor Microenvironment. Int. J. Mol. Sci. 2022, 23, 13258. https://doi.org/10.3390/ijms232113258
Zhou X, Chen B, Zhang Z, Huang Y, Li J, Wei Q, Cao D, Ai J. Crosstalk between Tumor-Associated Macrophages and MicroRNAs: A Key Role in Tumor Microenvironment. International Journal of Molecular Sciences. 2022; 23(21):13258. https://doi.org/10.3390/ijms232113258
Chicago/Turabian StyleZhou, Xianghong, Bo Chen, Zilong Zhang, Yin Huang, Jinze Li, Qiang Wei, Dehong Cao, and Jianzhong Ai. 2022. "Crosstalk between Tumor-Associated Macrophages and MicroRNAs: A Key Role in Tumor Microenvironment" International Journal of Molecular Sciences 23, no. 21: 13258. https://doi.org/10.3390/ijms232113258
APA StyleZhou, X., Chen, B., Zhang, Z., Huang, Y., Li, J., Wei, Q., Cao, D., & Ai, J. (2022). Crosstalk between Tumor-Associated Macrophages and MicroRNAs: A Key Role in Tumor Microenvironment. International Journal of Molecular Sciences, 23(21), 13258. https://doi.org/10.3390/ijms232113258






