Genome-Wide Analysis of SAUR Gene Family Identifies a Candidate Associated with Fruit Size in Loquat (Eriobotrya japonica Lindl.)
Abstract
:1. Introduction
2. Results
2.1. Identification and Annotation of the SAUR Family
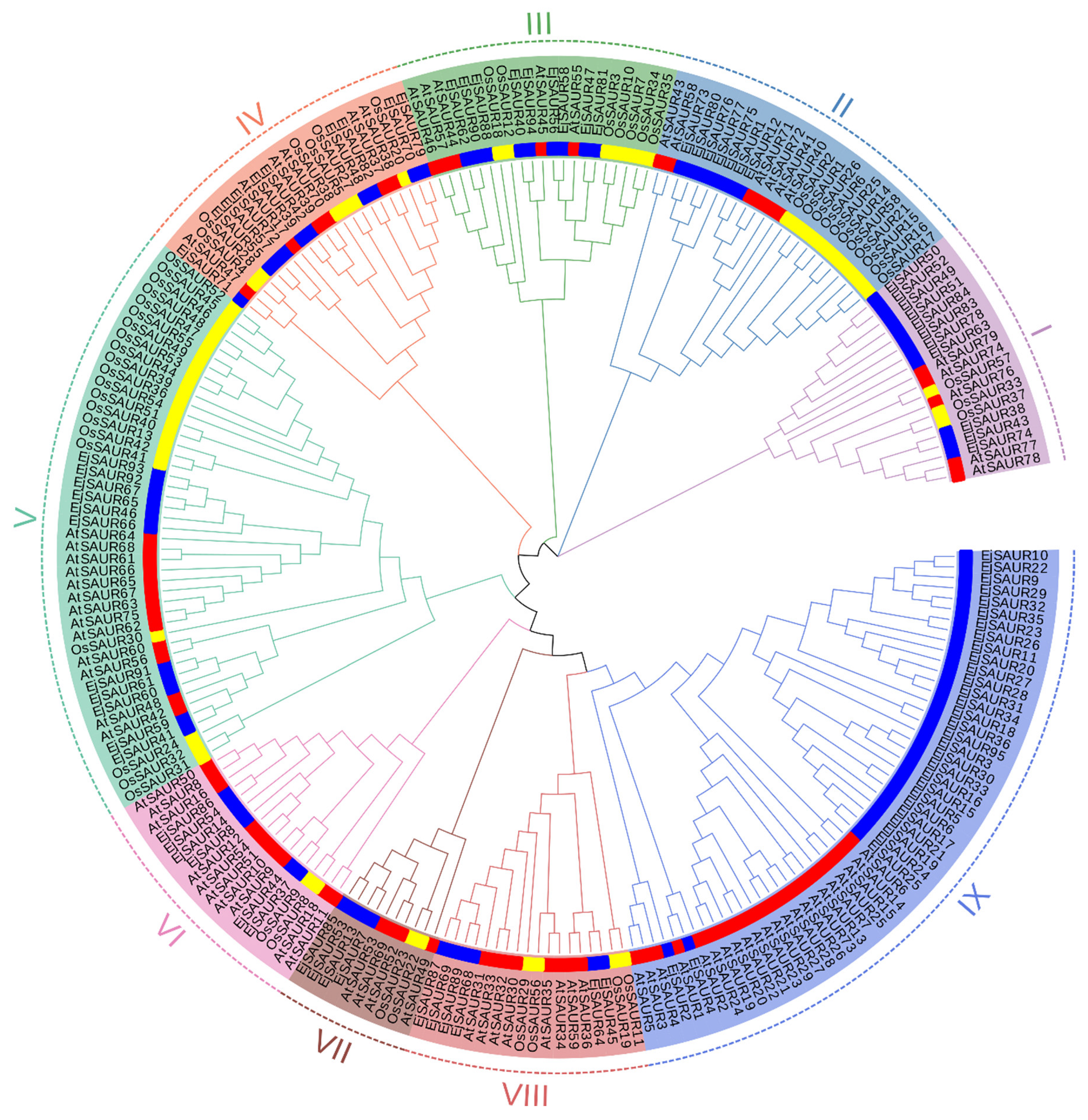
2.2. Phylogenetic Relationships, Gene Structure, and Conserved Motifs
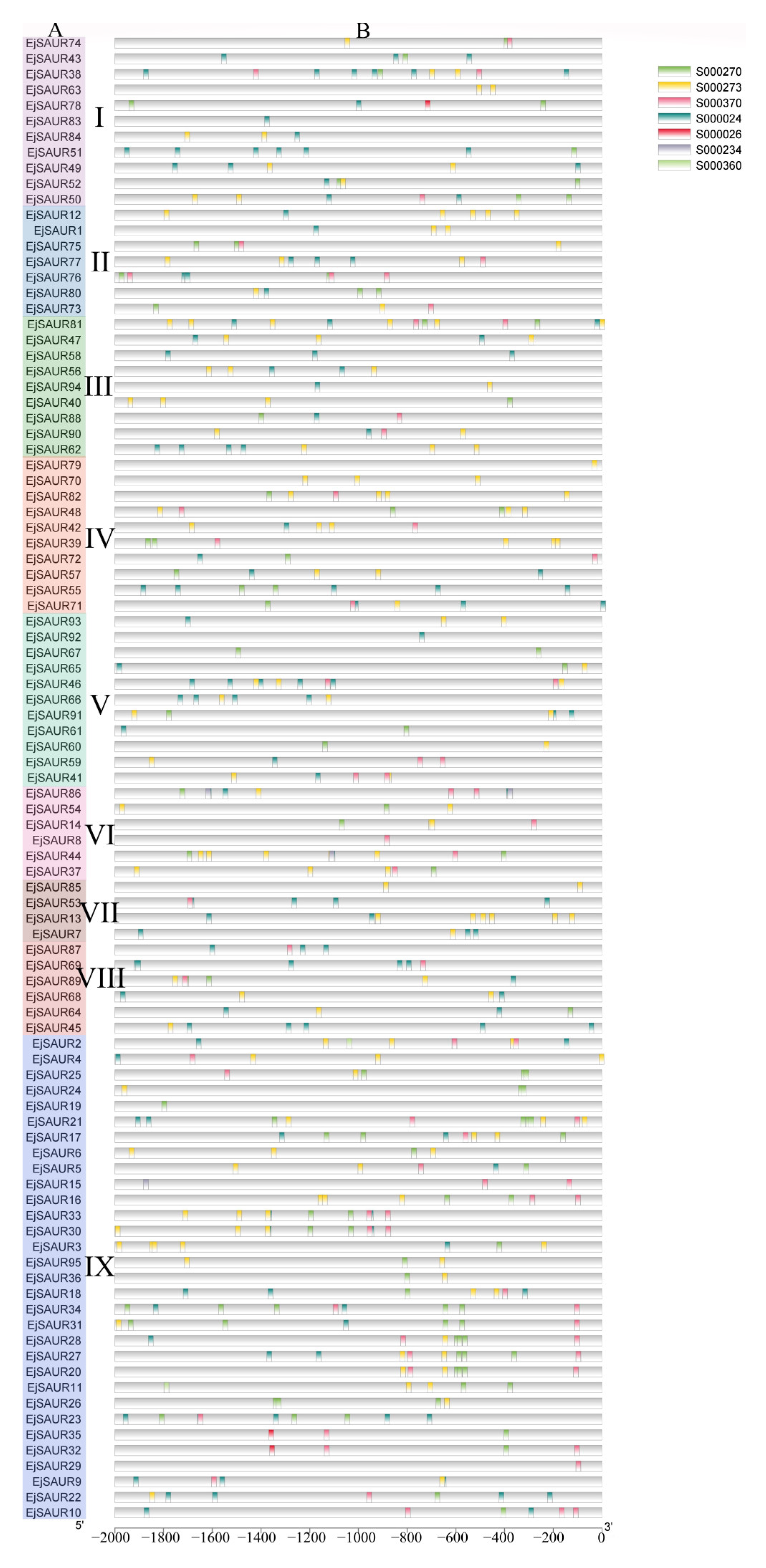
2.3. Cis Elements in the Promoters of EjSAURs
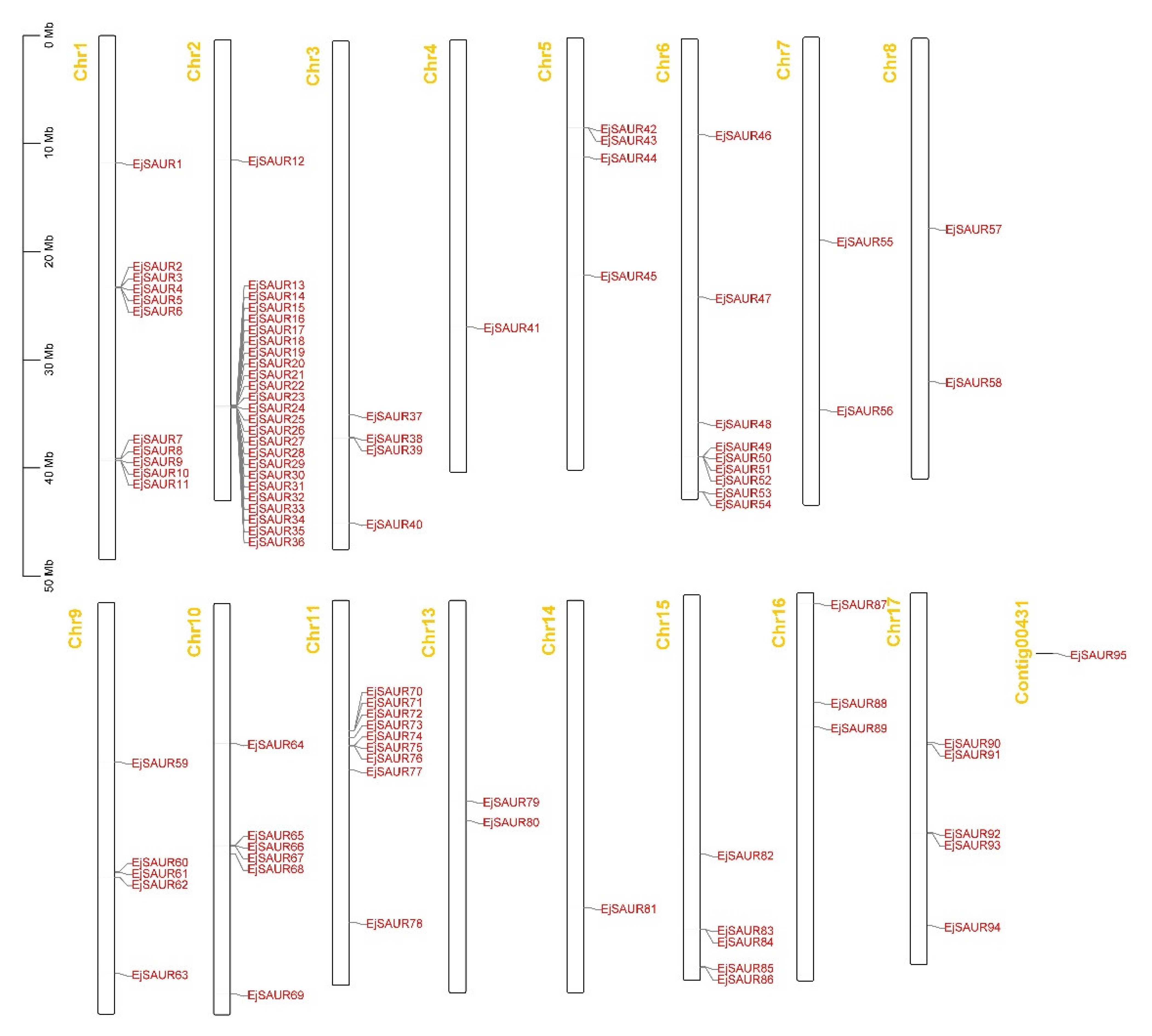
2.4. Chromosomal Locations, Gene Duplication, and Synteny Analysis
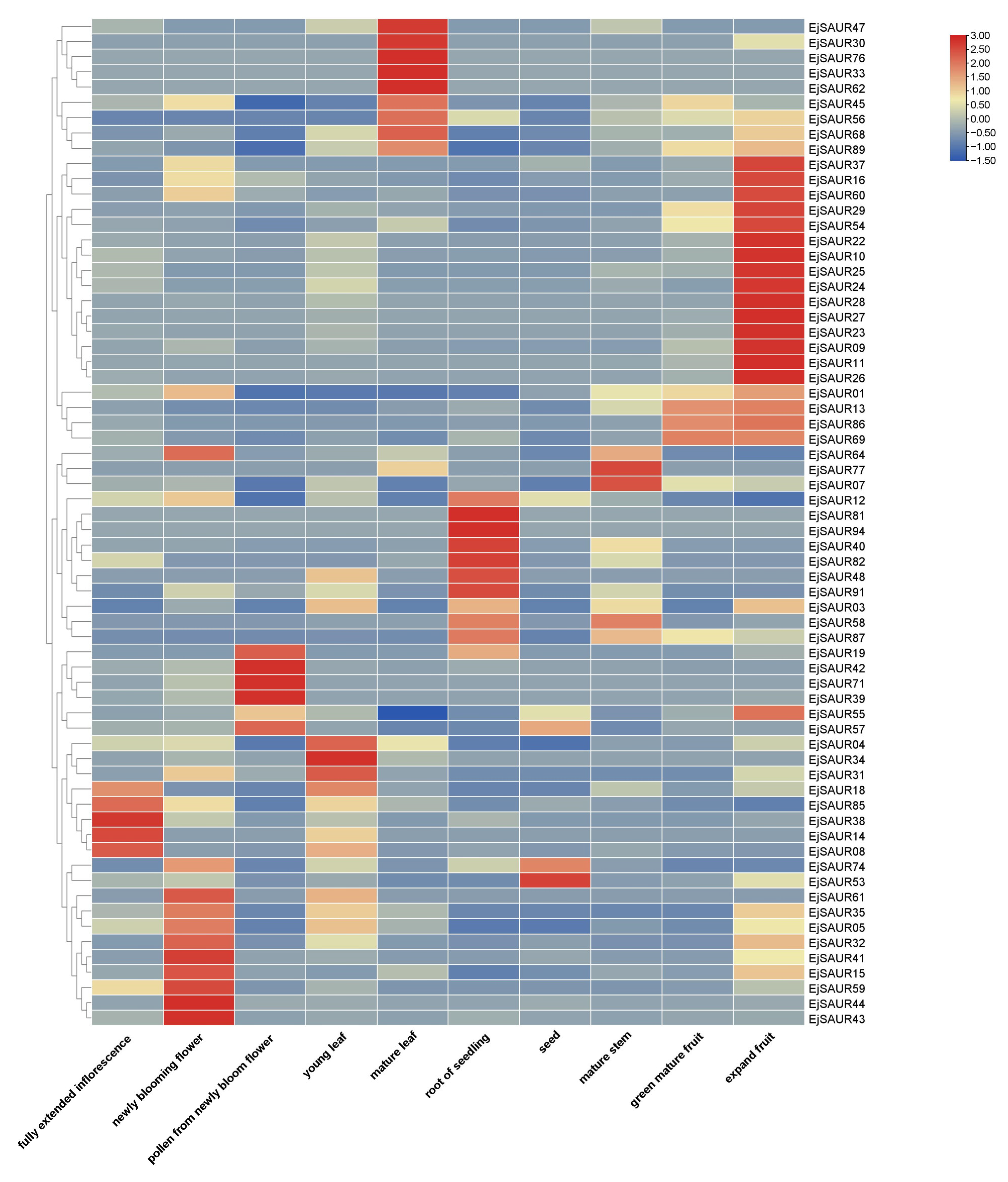
2.5. Expression Profiles of EjSAURs in Different Tissues
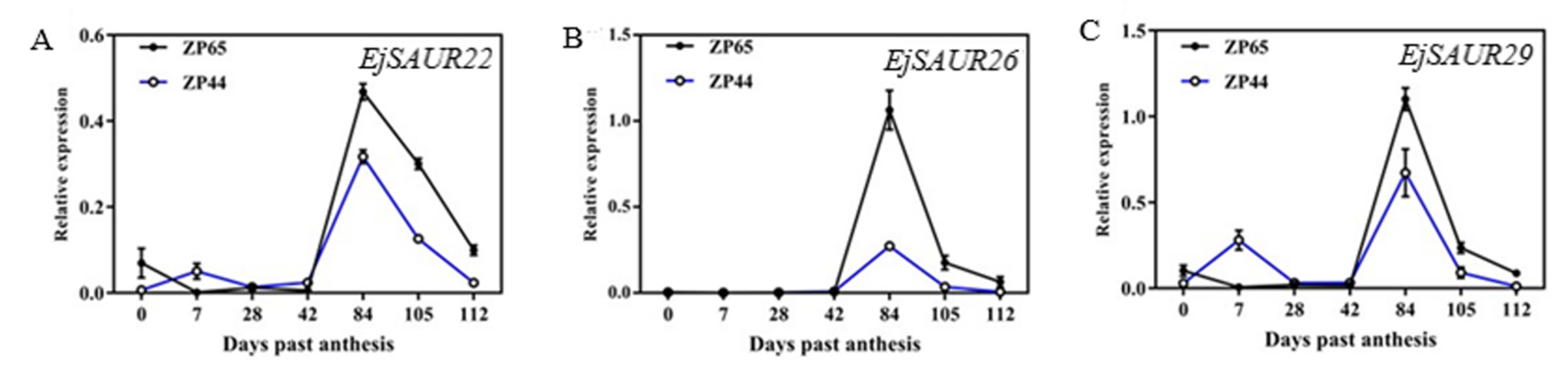
2.6. Observation of Fruit Development and Expression Patterns of Three EjSAURs
2.7. EjSAUR22, EjSAUR26, and EjSAUR29 Responses to IAA treatment
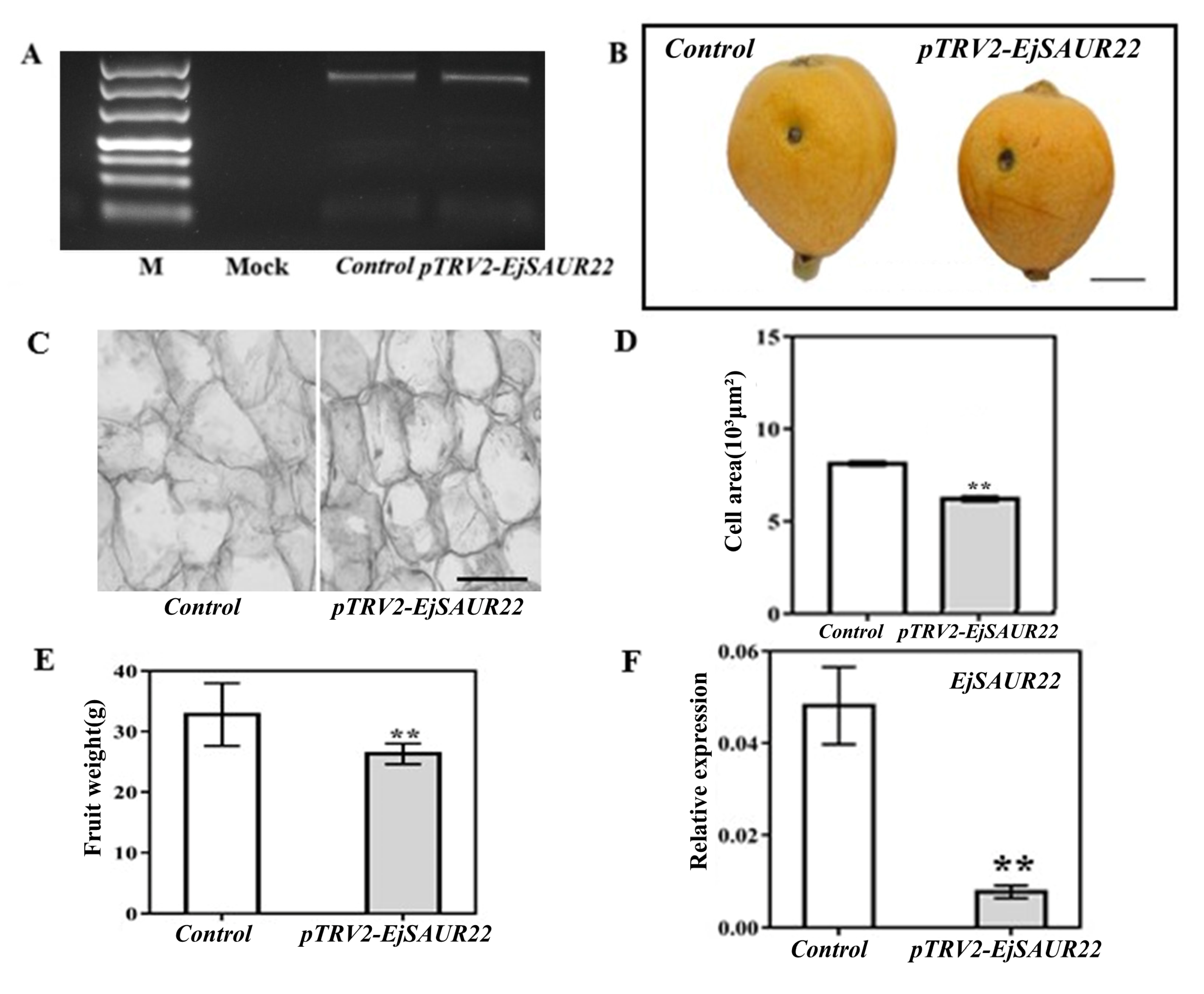
2.8. VIGS Support EjSAUR22′s Role in Cell Expansion and Fruit Size
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of the SAUR Gene Family
4.3. Phylogenetic Analysis
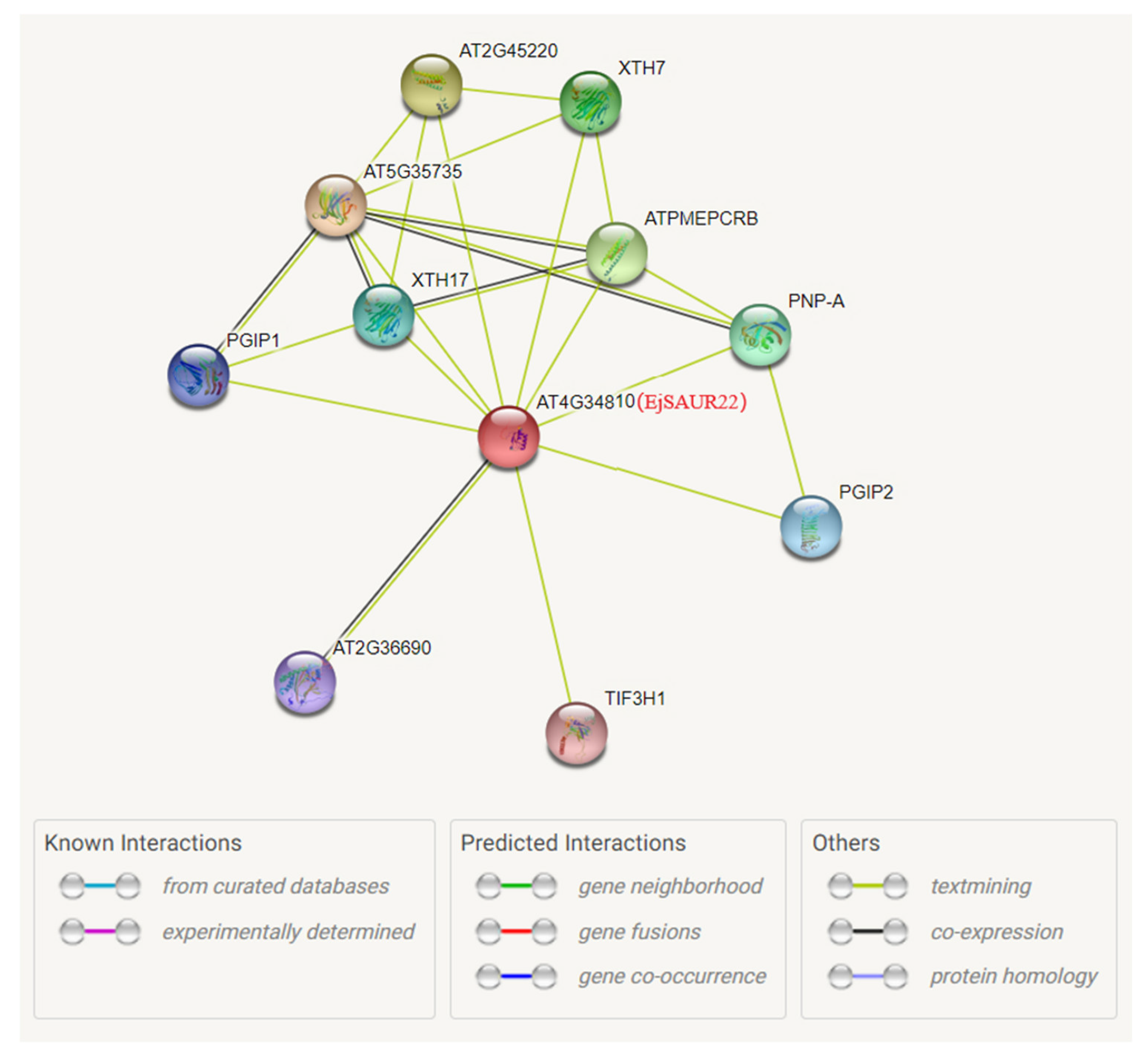
4.4. Chromosomal Locations, Gene Duplication, and Protein–-Protein Interaction Network
4.5. Analysis of Gene Structure, Conserved Motifs, and cis-Elements of EjSAURs
4.6. RNA Extraction and qRT-PCR
4.7. Virus Induced Gene Silencing
4.8. RNA-Seq Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hoffmann, M.; Hentrich, M.; Pollmann, S. Auxin-Oxylipin Crosstalk: Relationship of Antagonists. J. Integr. Plant Biol. 2011, 53, 429–445. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Hayashi, K.; Kinoshita, T. Auxin Activates the Plasma Membrane H+-ATPase by Phosphorylation during Hypocotyl Elongation in Arabidopsis. Plant Physiol. 2012, 159, 632–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Hao, X.; Cao, J. Small Auxin Upregulated RNA (SAUR) Gene Family in Maize: Identification, Evolution, and Its Phylogenetic Comparison with Arabidopsis, Rice, and Sorghum. J. Integr. Plant Biol. 2014, 56, 133–150. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-Responsive Gene Expression: Genes, Promoters and Regulatory Factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J.; Hagen, G. Auxin Response Factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.P.; Nourizadeh, S.D.; Peer, W.A.; Xu, J.; Bandyopadhyay, A.; Murphy, A.S.; Goldsbrough, P.B. Arabidopsis AtGSTF2 Is Regulated by Ethylene and Auxin, and Encodes a Glutathione S-Transferase That Interacts with Flavonoids. Plant J. 2003, 36, 433–442. [Google Scholar] [CrossRef]
- Franco, A.R.; Gee, M.A.; Guilfoyle, T.J. Induction and Superinduction of Auxin-Responsive MRNAs with Auxin and Protein Synthesis Inhibitors. J. Biol. Chem. 1990, 265, 15845–15849. [Google Scholar] [CrossRef]
- McClure, B.A.; Guilfoyle, T. Characterization of a Class of Small Auxin-Inducible Soybean Polyadenylated RNAs. Plant Mol. Biol. 1987, 9, 611–623. [Google Scholar] [CrossRef]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Genome-Wide Analysis, Evolutionary Expansion, and Expression of Early Auxin-Responsive SAUR Gene Family in Rice (Oryza sativa). Genomics 2006, 88, 360–371. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Bai, Y.; Shen, C.; Wu, Y.; Zhang, S.; Jiang, D.; Guilfoyle, T.J.; Chen, M.; Qi, Y. Auxin-Related Gene Families in Abiotic Stress Response in Sorghum Bicolor. Funct. Integr. Genom. 2010, 10, 533–546. [Google Scholar] [CrossRef]
- Xie, R.; Dong, C.; Ma, Y.; Deng, L.; He, S.; Yi, S.; Lv, Q.; Zheng, Y. Comprehensive Analysis of SAUR Gene Family in Citrus and Its Transcriptional Correlation with Fruitlet Drop from Abscission Zone A. Funct. Integr. Genom. 2015, 15, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Huang, X.; Bao, Y.; Wang, B.; Zeng, H.; Cheng, W.; Tang, M.; Li, Y.; Ren, J.; Sun, Y. Genome-Wide Identification of SAUR Genes in Watermelon (Citrullus lanatus). Physiol. Mol. Biol. Plants 2017, 23, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Lu, S.; Xie, M.; Wu, M.; Ding, S.; Khaliq, A.; Ma, Z.; Mao, J.; Chen, B. Identification and Expression Analysis of the Small Auxin-up RNA (SAUR) Gene Family in Apple by Inducing of Auxin. Gene 2020, 750, 144725. [Google Scholar] [CrossRef] [PubMed]
- Stortenbeker, N.; Bemer, M. The SAUR Gene Family: The Plant’s Toolbox for Adaptation of Growth and Development. J. Exp. Bot. 2019, 70, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Park, J.-E.; Kim, Y.-S.; Yoon, H.-K.; Park, C.-M. Functional Characterization of a Small Auxin-up RNA Gene in Apical Hook Development in Arabidopsis. Plant Sci. 2007, 172, 150–157. [Google Scholar] [CrossRef]
- Chae, K.; Isaacs, C.G.; Reeves, P.H.; Maloney, G.S.; Muday, G.K.; Nagpal, P.; Reed, J.W. Arabidopsis SMALL AUXIN UP RNA63 Promotes Hypocotyl and Stamen Filament Elongation. Plant J. 2012, 71, 684–697. [Google Scholar] [CrossRef]
- Hou, K.; Wu, W.; Gan, S.S. SAUR36, a Small Auxin up RNA Gene, Is Involved in the Promotion of Leaf Senescence in Arabidopsis. Plant Physiol. 2013, 161, 1002–1009. [Google Scholar] [CrossRef] [Green Version]
- Wen, Z.; Mei, Y.; Zhou, J.; Cui, Y.; Wang, D.; Wang, N.N. SAUR49 Can Positively Regulate Leaf Senescence by Suppressing SSPP in Arabidopsis. Plant Cell Physiol. 2020, 61, 644–658. [Google Scholar] [CrossRef]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inzé, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 Subfamily of SMALL AUXIN UP RNA Genes Promote Cell Expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef] [Green Version]
- Kant, S.; Bi, Y.M.; Zhu, T.; Rothstein, S.J. SAUR39, a Small Auxin-up RNA Gene, Acts as a Negative Regulator of Auxin Synthesis and Transport in Rice. Plant Physiol. 2009, 151, 691–701. [Google Scholar] [CrossRef]
- Li, M.; Chen, R.; Gu, H.; Cheng, D.; Guo, X.; Shi, C.; Li, L.; Xu, G.; Gu, S.; Wu, Z.; et al. Grape Small Auxin Upregulated RNA (SAUR) 041 Is a Candidate Regulator of Berry Size in Grape. Int. J. Mol. Sci. 2021, 22, 1818. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Zhao, C.; Li, S.; Guo, Y.; Xu, H.; Hu, G.; Liu, Z.; Chen, X.; Chen, J.; Lin, S.; et al. Integration of Genomics, Transcriptomics and Metabolomics Identifies Candidate Loci Underlying Fruit Weight in Loquat. Hortic. Res. 2022, 9, uhac037. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Zhu, Y.; Zhang, L.; Yang, X.; Gao, Y.; Lin, S. The Cellular Physiology of Loquat (Eriobotrya japonica Lindl.) Fruit with a Focus on How Cell Division and Cell Expansion Processes Contribute to Pome Morphogenesis. Sci. Hortic. 2017, 224, 142–149. [Google Scholar] [CrossRef]
- Su, W.; Jing, Y.; Lin, S.; Yue, Z.; Yang, X.; Xu, J.; Wu, J.; Zhang, Z.; Xia, R.; Zhu, J.; et al. Polyploidy Underlies Co-Option and Diversification of Biosynthetic Triterpene Pathways in the Apple Tribe. Proc. Natl. Acad. Sci. USA 2021, 118, e2101767118. [Google Scholar] [CrossRef]
- Pattison, R.J.; Csukasi, F.; Catalá, C. Mechanisms Regulating Auxin Action during Fruit Development. Physiol. Plant 2014, 151, 62–72. [Google Scholar] [CrossRef]
- Sundberg, E.; Østergaard, L. Distinct and Dynamic Auxin Activities during Reproductive Development. Cold Spring Harbor. Perspect. Biol. 2009, 1, a001628. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; An, H.; Xu, F.; Zhang, X. Chromosome-Level Genome Assembly and Annotation of the Loquat (Eriobotrya japonica) Genome. Gigascience 2020, 9, giaa015. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y. A Draft Genome, Resequencing, and Metabolomes Reveal the Genetic Background and Molecular Basis of the Nutritional and Medicinal Properties of Loquat (Eriobotrya japonica (Thunb.) Lindl). Hortic. Res. 2021, 8, 231. [Google Scholar] [CrossRef]
- Navarro, E.; Mallén, A.; Hueso, M. Dynamic Variations of 3′UTR Length Reprogram the MRNA Regulatory Landscape. Biomedicines 2021, 9, 1560. [Google Scholar] [CrossRef]
- Aguilar-Hernández, V.; Guzmán, P. Spliceosomal Introns in the 5′ Untranslated Region of Plant BTL RING-H2 Ubiquitin Ligases Are Evolutionary Conserved and Required for Gene Expression. BMC Plant Biol. 2013, 13, 179. [Google Scholar] [CrossRef]
- Kamo, K.; Kim, A.Y.; Park, S.H.; Joung, Y.H. The 5′UTR-Intron of the Gladiolus Polyubiquitin Promoter GUBQ1 Enhances Translation Efficiency in Gladiolus and Arabidopsis. BMC Plant Biol. 2012, 12, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, W.; Yan, H.; Luo, S.; Pan, F.; Wang, Y.; Xiang, Y. Genome-Wide Analysis of Poplar SAUR Gene Family and Expression Profiles under Cold, Polyethylene Glycol and Indole-3-Acetic Acid Treatments. Plant Physiol. Biochem. 2018, 128, 50–65. [Google Scholar] [CrossRef]
- Muranty, H.; Troggio, M.; Sadok, I.B.; Rifaï, M.A.; Auwerkerken, A.; Banchi, E.; Velasco, R.; Stevanato, P.; Van De Weg, W.E.; Di Guardo, M. Accuracy and Responses of Genomic Selection on Key Traits in Apple Breeding. Hortic. Res. 2015, 2, 15060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurens, F.; Aranzana, M.J.; Arus, P.; Bassi, D.; Bink, M.; Bonany, J.; Caprera, A.; Corelli-Grappadelli, L.; Costes, E.; Durel, C.E. An Integrated Approach for Increasing Breeding Efficiency in Apple and Peach in Europe. Hortic. Res. 2018, 5, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Cao, K.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Zhao, P.; Guo, J.; Ding, T.; Guan, L. Genomic Analyses of an Extensive Collection of Wild and Cultivated Accessions Provide New Insights into Peach Breeding History. Genome biology 2019, 20, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting Subcellular Localization of Proteins for Gram-Negative Bacteria by Support Vector Machines Based on n-Peptide Compositions. Protein. Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Yu, Z.; Yao, X.; Chen, J.; Chen, X.; Zhou, H.; Lou, Y.; Ming, F.; Jin, Y. Genome-Wide Identification and Characterization of Small Auxin-up RNA (SAUR) Gene Family in Plants: Evolution and Expression Profiles during Normal Growth and Stress Response. BMC Plant Biol. 2021, 21, 4. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Shao, Z.; Wang, M.; Gan, X.; Yang, X.; Lin, S. EjBZR1 Represses Fruit Enlargement by Binding to the EjCYP90 Promoter in Loquat. Hortic. Res. 2021, 8, 152. [Google Scholar] [CrossRef] [PubMed]
- You, F.M.; Huo, N.; Gu, Y.Q.; Luo, M.C.; Ma, Y.; Hane, D.; Lazo, G.R.; Dvorak, J.; Anderson, O.D. BatchPrimer3: A High Throughput Web Application for PCR and Sequencing Primer Design. BMC Bioinform. 2008, 9, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, W.; Yuan, Y.; Zhang, L.; Jiang, Y.; Gan, X.; Bai, Y.; Peng, J.; Wu, J.; Liu, Y.; Lin, S. Selection of the Optimal Reference Genes for Expression Analyses in Different Materials of Eriobotrya Japonica. Plant Methods 2019, 15, 7. [Google Scholar] [CrossRef]





| Gene Name | Gene ID | Location | Peptide Length | MW/Da | PI | Predicted Subcellular Location |
|---|---|---|---|---|---|---|
| EjSAUR1 | EVM0023123 | Chr1:11774882–11776212(−) | 139 | 16,031.33 | 9.38 | Nuclear(1.968)/Mitochondrial(1.116) |
| EjSAUR2 | EVM0027975 | Chr1:23253858–23254133(−) | 91 | 10,303.12 | 9.37 | Mitochondrial(2.134) |
| EjSAUR3 | EVM0026311 | Chr1:23277661–23278316(−) | 102 | 11,240.81 | 6.39 | Mitochondrial(1.514)/Nuclear(1.100) |
| EjSAUR4 | EVM0036225 | Chr1:23310700–23311301(−) | 94 | 10,565.18 | 8.57 | Extracellular(1.554)/PlasmaMembrane(1.101) |
| EjSAUR5 | EVM0001621 | Chr1:23313466–23314268(+) | 93 | 10,399.74 | 6.96 | Nuclear(2.403) |
| EjSAUR6 | EVM0027340 | Chr1:23320718–23321002(+) | 94 | 10,519.81 | 6.89 | Nuclear(2.747) |
| EjSAUR7 | EVM0024197 | Chr1:39105016–39105753(+) | 151 | 16,938.55 | 9.03 | Nuclear(2.201) |
| EjSAUR8 | EVM0008023 | Chr1:39155495–39156275(−) | 105 | 12,020.02 | 8.51 | Mitochondrial(2.036)/Cytoplasmic(1.594) |
| EjSAUR9 | EVM0019503 | Chr1:39309289–39309934(+) | 101 | 11,396.06 | 6.56 | Mitochondrial(1.637)/Nuclear(1.271) |
| EjSAUR10 | EVM0040814 | Chr1:39327746–39328520(+) | 101 | 11,312.01 | 6.71 | Mitochondrial(1.705) |
| EjSAUR11 | EVM0039098 | Chr1:39335634–39336324(+) | 101 | 11,356.99 | 6.26 | Mitochondrial(1.737) |
| EjSAUR12 | EVM0025395 | Chr2:11090245–11091063(−) | 139 | 15,820.21 | 9.21 | Nuclear(2.199) |
| EjSAUR13 | EVM0010716 | Chr2:33801351–33802064(+) | 146 | 16,414 | 9.21 | Nuclear(2.310) |
| EjSAUR14 | EVM0039525 | Chr2:33843069–33843505(−) | 104 | 11,890.8 | 7.75 | Mitochondrial(1.932)/Cytoplasmic(1.595) |
| EjSAUR15 | EVM0005420 | Chr2:33869889–33870586(+) | 99 | 11,181.98 | 7.88 | Mitochondrial(1.083)/Extracellular(1.060) |
| EjSAUR16 | EVM0022268 | Chr2:33874632–33875391(+) | 100 | 11,280.98 | 6.06 | PlasmaMembrane(1.800) |
| EjSAUR17 | EVM0042792 | Chr2:33884220–33884807(−) | 127 | 14,144.29 | 6.71 | PlasmaMembrane(1.550)/Extracellular(1.459) |
| EjSAUR18 | EVM0004006 | Chr2:33885766–33886265(+) | 98 | 11,175.67 | 7.9 | Mitochondrial(1.992) |
| EjSAUR19 | EVM0002248 | Chr2:33893889–33894164(−) | 91 | 10,015.55 | 6.03 | Extracellular(0.995)/Nuclear(0.962)/Chloroplast(0.957) |
| EjSAUR20 | EVM0003298 | Chr2:33906669–33906974(−) | 101 | 11,214.91 | 5.23 | Mitochondrial(1.203)/Extracellular(1.104) |
| EjSAUR21 | EVM0028994 | Chr2:33907908–33908180(+) | 90 | 10,060.39 | 4.92 | Cytoplasmic(1.643)/Nuclear(1.285) |
| EjSAUR22 | EVM0027703 | Chr2:33924785–33925382(+) | 101 | 11,287.08 | 8.64 | Mitochondrial(1.456)/Extracellular(1.106)/Nuclear(1.053) |
| EjSAUR23 | EVM0022346 | Chr2:33927004–33927649(−) | 101 | 11,289.86 | 6.9 | Mitochondrial(1.148)/Nuclear(1.073) |
| EjSAUR24 | EVM0035336 | Chr2:33934148–33934842(+) | 91 | 9982.47 | 6.03 | Nuclear(1.420)/Extracellular(1.227) |
| EjSAUR25 | EVM0035171 | Chr2:33936811–33937086(−) | 91 | 10,019.61 | 8.73 | Nuclear(1.371) |
| EjSAUR26 | EVM0031995 | Chr2:33938646–33939222(+) | 101 | 11,267.96 | 6.82 | PlasmaMembrane(1.157)/Mitochondrial(1.088) |
| EjSAUR27 | EVM0031846 | Chr2:33946504–33947087(+) | 100 | 11,129.82 | 5.28 | Mitochondrial(1.658) |
| EjSAUR28 | EVM0002370 | Chr2:33967678–33968177(+) | 101 | 11,321.04 | 6.57 | Mitochondrial(2.149) |
| EjSAUR29 | EVM0015711 | Chr2:33972636–33973301(+) | 101 | 11,104.83 | 5.71 | Extracellular(1.824) |
| EjSAUR30 | EVM0038688 | Chr2:34011539–34011841(+) | 100 | 11,492.37 | 9.3 | PlasmaMembrane(1.571) |
| EjSAUR31 | EVM0025517 | Chr2:34018506–34019087(+) | 101 | 11,293.97 | 5.79 | Mitochondrial(2.138) |
| EjSAUR32 | EVM0044407 | Chr2:34021147–34021686(+) | 100 | 10,969.69 | 5.24 | Mitochondrial(1.235)/Extracellular(1.134) |
| EjSAUR33 | EVM0039690 | Chr2:34032156–34032458(+) | 100 | 11,512.44 | 9.46 | PlasmaMembrane(1.862) |
| EjSAUR34 | EVM0026210 | Chr2:34039113–34039637(+) | 101 | 11,302 | 6.72 | Mitochondrial(2.080) |
| EjSAUR35 | EVM0026900 | Chr2:34041830–34042147(+) | 105 | 11,625.53 | 7.77 | Extracellular(1.488)/Mitochondrial(1.154) |
| EjSAUR36 | EVM0001246 | Chr2:34086922–34087275(+) | 117 | 13,500.49 | 7.88 | Nuclear(1.889)/Mitochondrial(1.787) |
| EjSAUR37 | EVM0038271 | Chr3:34602523–34602843(−) | 106 | 12,197.03 | 9.3 | Mitochondrial(2.380) |
| EjSAUR38 | EVM0005442 | Chr3:36683708–36684389(+) | 120 | 13,571.39 | 5.33 | Nuclear(1.986) |
| EjSAUR39 | EVM0025157 | Chr3:36746045–36748186(−) | 183 | 20,384.85 | 6.24 | Nuclear(2.636) |
| EjSAUR40 | EVM0038485 | Chr3:44606644–44607309(−) | 128 | 14,229.45 | 6.13 | Chloroplast(1.219)/Nuclear(1.118) |
| EjSAUR41 | EVM0015619 | Chr4:26550990–26551811(−) | 142 | 16,013.1 | 5.66 | Nuclear(2.491) |
| EjSAUR42 | EVM0030373 | Chr5:8343504–8346032(+) | 183 | 20,272.91 | 6.38 | Nuclear(2.709) |
| EjSAUR43 | EVM0043234 | Chr5:8361849–8362434(−) | 121 | 13,671.57 | 5.11 | Nuclear(1.523)/Mitochondrial(1.084) |
| EjSAUR44 | EVM0017373 | Chr5:11041105–11041507(+) | 106 | 11,997.75 | 8.6 | Mitochondrial(1.686)/Chloroplast(1.317)/Cytoplasmic(1.111) |
| EjSAUR45 | EVM0039643 | Chr5:21933735–21935000(−) | 173 | 19,518.59 | 10.09 | Mitochondrial(2.333)/Nuclear(1.759) |
| EjSAUR46 | EVM0043534 | Chr6:8833467–8843020(+) | 144 | 16,158.07 | 9.08 | PlasmaMembrane(2.867) |
| EjSAUR47 | EVM0001296 | Chr6:23893000–23895587(+) | 170 | 19,202.14 | 9.32 | Nuclear(1.616)/Extracellular(1.355)/Mitochondrial(1.262) |
| EjSAUR48 | EVM0040221 | Chr6:35531114–35531632(−) | 172 | 19,075.91 | 8.83 | Nuclear(2.561) |
| EjSAUR49 | EVM0018324 | Chr6:38647524–38647835(+) | 103 | 11,754.81 | 9.76 | Nuclear(1.809)/Mitochondrial(1.334) |
| EjSAUR50 | EVM0003367 | Chr6:38650612–38650929(+) | 105 | 12,021.13 | 10 | Mitochondrial(2.579) |
| EjSAUR51 | EVM0009009 | Chr6:38652604–38652915(+) | 103 | 11,852.93 | 9.85 | Mitochondrial(2.766) |
| EjSAUR52 | EVM0026005 | Chr6:38661854–38662329(+) | 103 | 11,766.62 | 9.83 | Mitochondrial(2.337) |
| EjSAUR53 | EVM0012665 | Chr6:41908807–41909985(+) | 150 | 16,730.32 | 9.54 | Mitochondrial(1.295)/Extracellular(1.238) |
| EjSAUR54 | EVM0029547 | Chr6:41946158–41947012(−) | 105 | 11,990.81 | 6.9 | Mitochondrial(1.711)/Cytoplasmic(1.321) |
| EjSAUR55 | EVM0017455 | Chr7:18784282–18785389(−) | 226 | 25,481.7 | 10.14 | Mitochondrial(1.378)/PlasmaMembrane(1.198) |
| EjSAUR56 | EVM0025877 | Chr7:34435942–34437008(+) | 155 | 17,389.32 | 9.39 | Nuclear(1.716)/Mitochondrial(1.423) |
| EjSAUR57 | EVM0008209 | Chr8:17546313–17546996(-) | 169 | 19,146.49 | 9.8 | Mitochondrial(1.960) |
| EjSAUR58 | EVM0002293 | Chr8:31723362–31723838(+) | 153 | 17,274.22 | 9.56 | Mitochondrial(2.006) |
| EjSAUR59 | EVM0005588 | Chr9:14721550–14722184(+) | 152 | 17,317.65 | 7.65 | Chloroplast(1.322)/Nuclear(1.146) |
| EjSAUR60 | EVM0034387 | Chr9:24812318–24812749(+) | 133 | 15,062.18 | 7.84 | Nuclear(2.062) |
| EjSAUR61 | EVM0032278 | Chr9:24905574–24906094(+) | 166 | 19,047.25 | 9.57 | Nuclear(2.880) |
| EjSAUR62 | EVM0042993 | Chr9:25377558–25377971(+) | 137 | 15,658.85 | 6.07 | Extracellular(1.704)/Nuclear(1.638) |
| EjSAUR63 | EVM0021105 | Chr9:34270371–34270790(−) | 139 | 16,677.25 | 5.85 | Mitochondrial(1.644)/Extracellular(1.343) |
| EjSAUR64 | EVM0027800 | Chr10:12919311–12920724(+) | 171 | 19,170.14 | 10.1 | Mitochondrial(2.625) |
| EjSAUR65 | EVM0001701 | Chr10:22309553–22310006(−) | 138 | 15,798.33 | 7.71 | PlasmaMembrane(1.540)/Nuclear(1.285)/Extracellular(1.056) |
| EjSAUR66 | EVM0028728 | Chr10:22335128–22335606(−) | 135 | 15,510.61 | 9.63 | PlasmaMembrane(2.371) |
| EjSAUR67 | EVM0008977 | Chr10:22405749–22406198(+) | 106 | 12,495.67 | 8.55 | PlasmaMembrane(1.849) |
| EjSAUR68 | EVM0017929 | Chr10:23141987–23142813(+) | 124 | 14,583.63 | 8.54 | Nuclear(1.893) |
| EjSAUR69 | EVM0006441 | Chr10:36037617–36038018(+) | 133 | 15,649.71 | 6.67 | Nuclear(2.017) |
| EjSAUR70 | EVM0015952 | Chr11:12020642–12021202(+) | 186 | 21,109.31 | 9.1 | Nuclear(2.785) |
| EjSAUR71 | EVM0038556 | Chr11:12028926–12029372(−) | 148 | 16,773.47 | 8.55 | Mitochondrial(1.690) |
| EjSAUR72 | EVM0003137 | Chr11:12032052–12032480(+) | 142 | 16,444.39 | 9.24 | PlasmaMembrane(1.521)/Mitochondrial(1.278) |
| EjSAUR73 | EVM0023832 | Chr11:12666841–12667188(+) | 115 | 13,221.58 | 9.63 | Mitochondrial(2.517) |
| EjSAUR74 | EVM0000097 | Chr11:13415260–13415932(+) | 119 | 13,144.02 | 6.06 | Mitochondrial(1.451)/Chloroplast(1.368)/Nuclear(1.136) |
| EjSAUR75 | EVM0037519 | Chr11:13443972–13444438(−) | 118 | 13,507.83 | 9.45 | Mitochondrial(2.147) |
| EjSAUR76 | EVM0015267 | Chr11:13450237–13450593(−) | 118 | 13,512.04 | 9.52 | Mitochondrial(2.192) |
| EjSAUR77 | EVM0028342 | Chr11:15696531–15696872(+) | 113 | 13,066.42 | 9.4 | Mitochondrial(2.054) |
| EjSAUR78 | EVM0044617 | Chr11:29707812–29708126(+) | 104 | 12,044.19 | 9.52 | Mitochondrial(1.827)/Nuclear(1.592) |
| EjSAUR79 | EVM0017994 | Chr13:18527573–18528148(+) | 191 | 21,616.83 | 9.1 | Nuclear(2.960) |
| EjSAUR80 | EVM0008731 | Chr13:20372332–20372661(−) | 109 | 12,670.87 | 9.71 | Mitochondrial(2.503) |
| EjSAUR81 | EVM0036677 | Chr14:28350218–28350736(+) | 172 | 19,317.36 | 9.21 | Extracellular(1.994)/Nuclear(1.716) |
| EjSAUR82 | EVM0001339 | Chr15:23948878–23949456(−) | 173 | 19,287.14 | 9.23 | Nuclear(2.277) |
| EjSAUR83 | EVM0019162 | Chr15:30879025–30879300(−) | 91 | 10,594.44 | 9.62 | Mitochondrial(1.726)/Nuclear(1.549) |
| EjSAUR84 | EVM0006879 | Chr15:30881712–30882023(−) | 103 | 11,948.13 | 9.86 | Mitochondrial(1.704)/Nuclear(1.409) |
| EjSAUR85 | EVM0011555 | Chr15:34348265–34349570(+) | 150 | 16,912.54 | 9.69 | Mitochondrial(2.123) |
| EjSAUR86 | EVM0038736 | Chr15:34388795–34389525(−) | 105 | 12,107.98 | 7.83 | Mitochondrial(1.870)/Nuclear(1.606) |
| EjSAUR87 | EVM0006208 | Chr16:993990–995003(−) | 133 | 15,745.86 | 7.14 | Nuclear(1.753) |
| EjSAUR88 | EVM0027522 | Chr16:10108766–10109161(−) | 131 | 15,539.26 | 8.79 | Mitochondrial(1.662) |
| EjSAUR89 | EVM0021582 | Chr16:12402979–12403859(−) | 123 | 14,451.43 | 7.96 | Nuclear(2.113) |
| EjSAUR90 | EVM0039776 | Chr17:13803945–13804328(−) | 127 | 14,713.15 | 8.43 | Extracellular(2.177)/Nuclear(1.739) |
| EjSAUR91 | EVM0020457 | Chr17:14042817–14043748(−) | 149 | 16,909.35 | 9.25 | Nuclear(1.794) |
| EjSAUR92 | EVM0017269 | Chr17:22150290–22150730(−) | 146 | 16,326.86 | 9.24 | PlasmaMembrane(1.510)/Nuclear(1.203) |
| EjSAUR93 | EVM0039025 | Chr17:22220726–22221196(−) | 156 | 17,535.48 | 9.59 | Nuclear(2.088) |
| EjSAUR94 | EVM0010047 | Chr17:30752223–30752606(−) | 127 | 14,313.37 | 6.31 | Nuclear(1.450) |
| EjSAUR95 | EVM0035435 | Contig00431:47729–48082(+) | 117 | 13,500.49 | 7.88 | Nuclear(1.889)/Mitochondrial(1.787) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Z.; Li, W.; Gan, X.; Zhao, C.; Paudel, D.; Su, W.; Lv, J.; Lin, S.; Liu, Z.; Yang, X. Genome-Wide Analysis of SAUR Gene Family Identifies a Candidate Associated with Fruit Size in Loquat (Eriobotrya japonica Lindl.). Int. J. Mol. Sci. 2022, 23, 13271. https://doi.org/10.3390/ijms232113271
Peng Z, Li W, Gan X, Zhao C, Paudel D, Su W, Lv J, Lin S, Liu Z, Yang X. Genome-Wide Analysis of SAUR Gene Family Identifies a Candidate Associated with Fruit Size in Loquat (Eriobotrya japonica Lindl.). International Journal of Molecular Sciences. 2022; 23(21):13271. https://doi.org/10.3390/ijms232113271
Chicago/Turabian StylePeng, Ze, Wenxiang Li, Xiaoqing Gan, Chongbin Zhao, Dev Paudel, Wenbing Su, Juan Lv, Shunquan Lin, Zongli Liu, and Xianghui Yang. 2022. "Genome-Wide Analysis of SAUR Gene Family Identifies a Candidate Associated with Fruit Size in Loquat (Eriobotrya japonica Lindl.)" International Journal of Molecular Sciences 23, no. 21: 13271. https://doi.org/10.3390/ijms232113271
APA StylePeng, Z., Li, W., Gan, X., Zhao, C., Paudel, D., Su, W., Lv, J., Lin, S., Liu, Z., & Yang, X. (2022). Genome-Wide Analysis of SAUR Gene Family Identifies a Candidate Associated with Fruit Size in Loquat (Eriobotrya japonica Lindl.). International Journal of Molecular Sciences, 23(21), 13271. https://doi.org/10.3390/ijms232113271







