Role of SALL4 in HER2+ Breast Cancer Progression: Regulating PI3K/AKT Pathway
Abstract
1. Introduction
2. Results
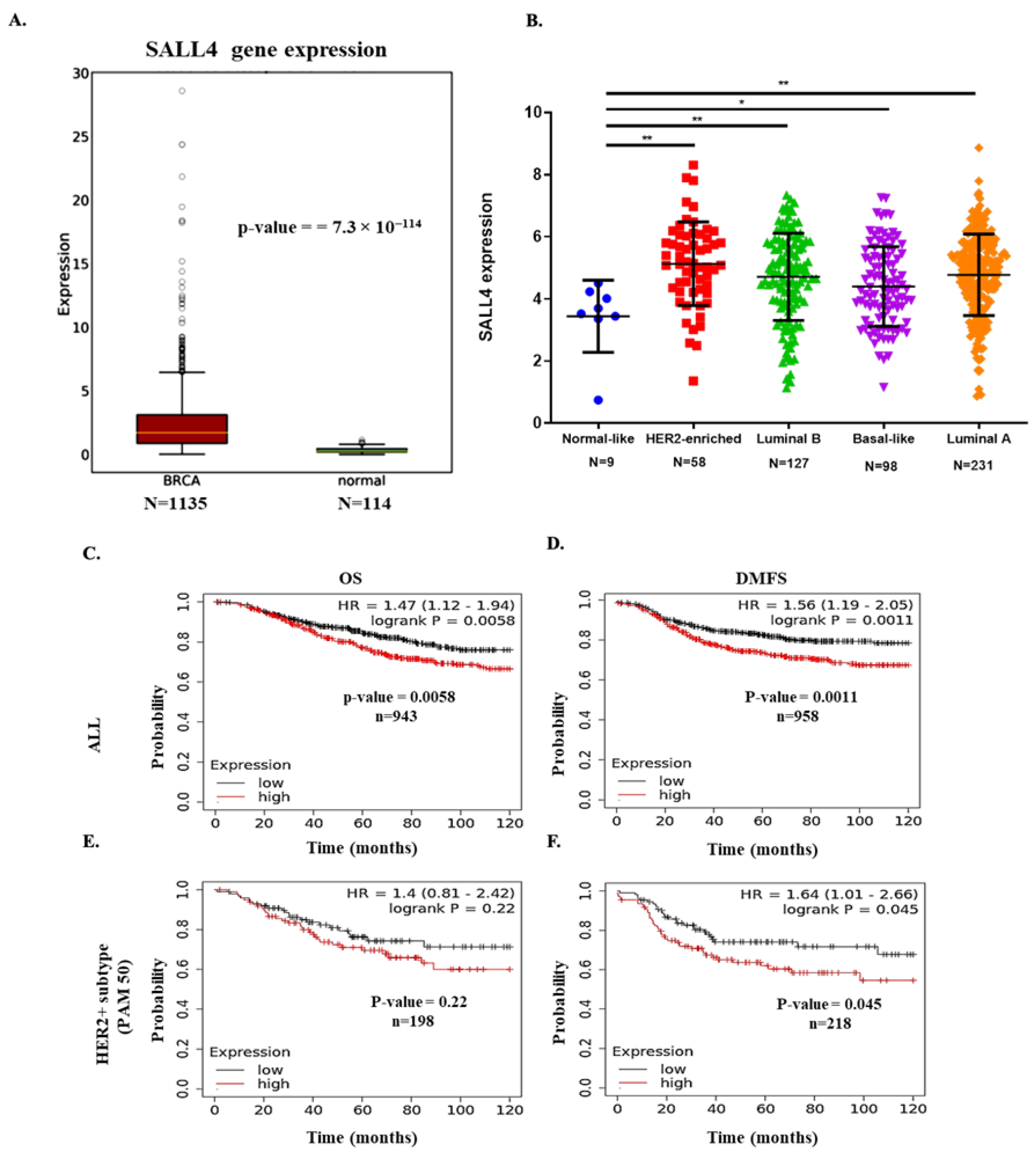
2.1. SALL4 Expression in BC Patient’s Samples
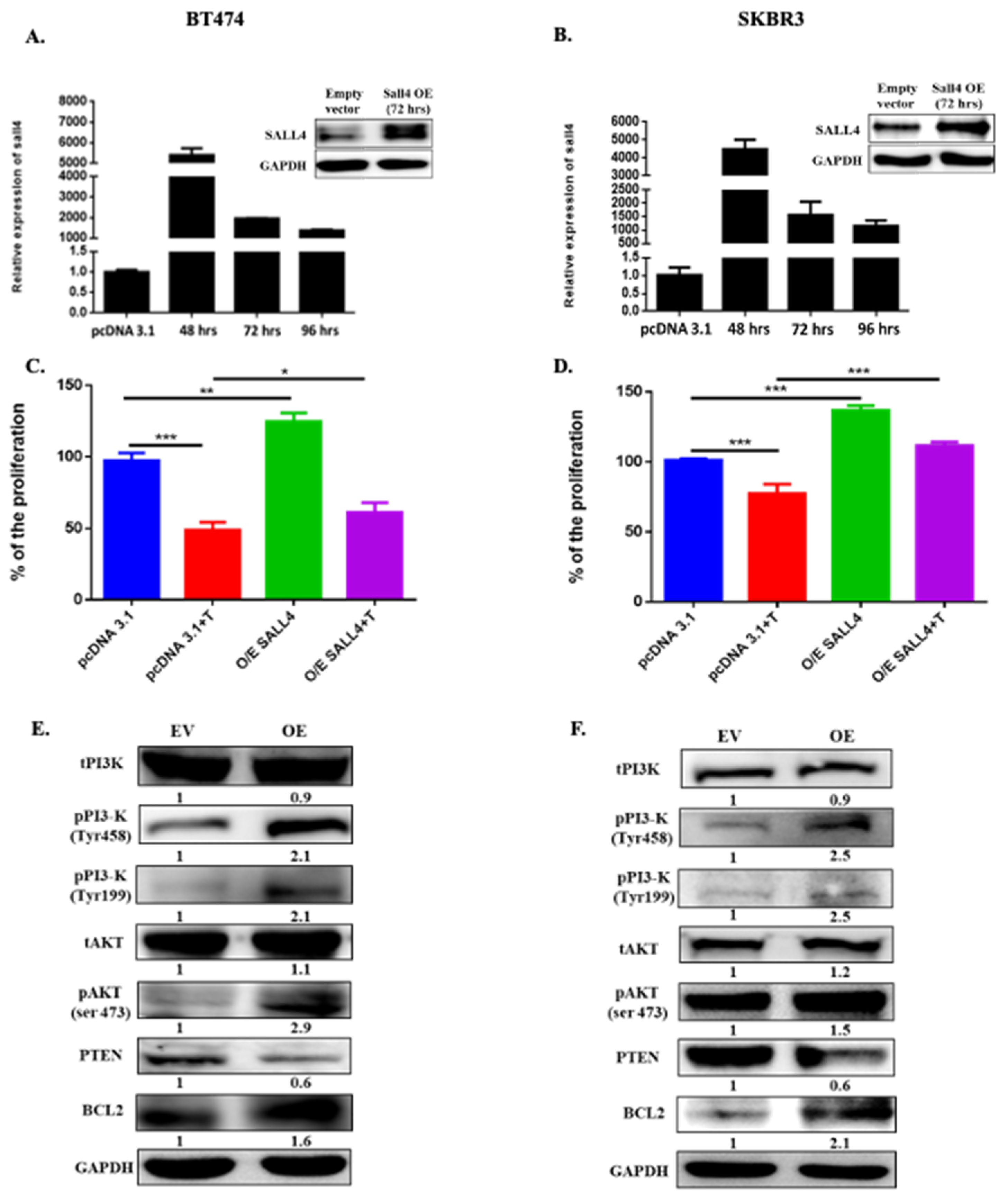
2.2. Ectopically SALL4 Expression Enhances Proliferation and Hinders Partially the Trastuzumab Effect
2.3. SALL4 Follows the PI3K/AKT Pathway for Cell Proliferation
2.4. RBBp4 as a Bridge between SALL4 and PI3K/AKT
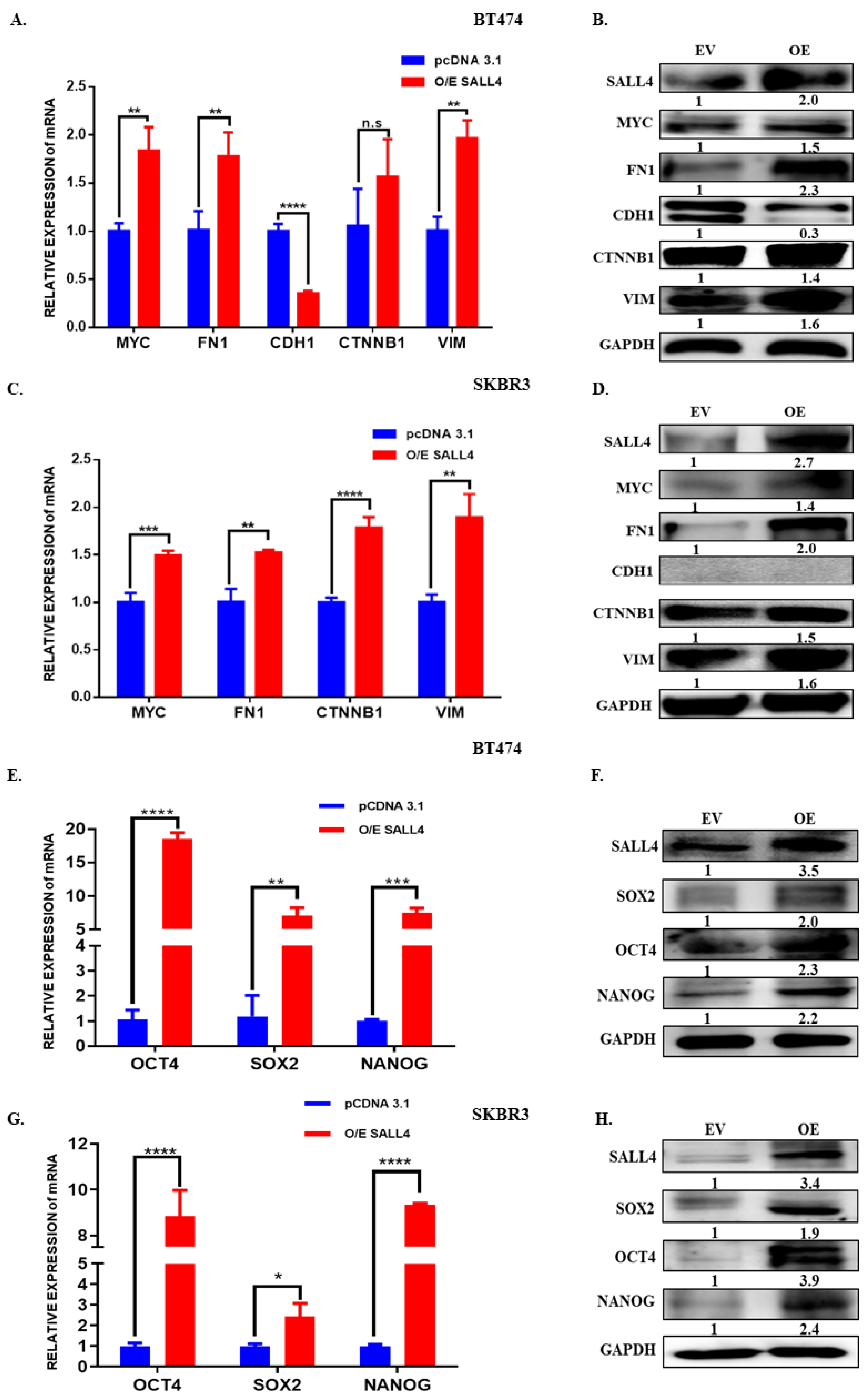
2.5. SALL4 Promotes EMT and Stemness
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Plasmids and siRNA Transfection
4.3. RNA Extraction and Quantitative Real-Time PCR
4.4. Cell Proliferation Assay
4.5. Clinical Samples and RNA Isolation
4.6. Western Blot and Co-Immunoprecipitation Analysis
4.7. In Silico Survival Analysis
4.8. Data Collection and Gene Expression Analysis from OncoDB and TCGA Database
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Callahan, R.; Hurvitz, S. Human Epidermal Growth Factor Receptor-2-Positive Breast Cancer: Current Management of Early, Advanced, and Recurrent Disease. Curr. Opin. Obstet. Gynecol. 2011, 23, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Pupa, S.M.; Ligorio, F.; Cancila, V.; Franceschini, A.; Tripodo, C.; Vernieri, C.; Castagnoli, L. HER2 Signaling and Breast Cancer Stem Cells: The Bridge behind HER2-Positive Breast Cancer Aggressiveness and Therapy Refractoriness. Cancers 2021, 13, 4778. [Google Scholar] [CrossRef] [PubMed]
- Yao, E.; Zhou, W.; Lee-Hoeflich, S.T.; Truong, T.; Haverty, P.M.; Eastham-Anderson, J.; Lewin-Koh, N.; Gunter, B.; Belvin, M.; Murray, L.J.; et al. Suppression of HER2/HER3-Mediated Growth of Breast Cancer Cells with Combinations of GDC-0941 PI3K Inhibitor, Trastuzumab, and Pertuzumab. Clin. Cancer Res. 2009, 15, 4147–4156. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Manning, B.D. The PI3K–AKT Network at the Interface of Oncogenic Signalling and Cancer Metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef]
- Park, J.-Y.; Kang, S.-E.; Ahn, K.S.; Um, J.-Y.; Yang, W.M.; Yun, M.; Lee, S.-G. Inhibition of the PI3K-AKT-MTOR Pathway Suppresses the Adipocyte-Mediated Proliferation and Migration of Breast Cancer Cells. J. Cancer 2020, 11, 2552–2559. [Google Scholar] [CrossRef]
- Nagata, Y.; Lan, K.-H.; Zhou, X.; Tan, M.; Esteva, F.J.; Sahin, A.A.; Klos, K.S.; Li, P.; Monia, B.P.; Nguyen, N.T.; et al. PTEN Activation Contributes to Tumor Inhibition by Trastuzumab, and Loss of PTEN Predicts Trastuzumab Resistance in Patients. Cancer Cell 2004, 6, 117–127. [Google Scholar] [CrossRef]
- Tekesin, K.; Emin Gunes, M.; Bayrak, S.; Akar, E.; Ozturk, T.; Altinay, S.; Tural, D. PTEN Loss Is a Predictive Marker for HER2-Positive Metastatic Breast Cancer Patients Treated with Trastuzumab-Based Therapies. J. BUON 2019, 24, 1920–1926. [Google Scholar]
- Yue, X.; Xiao, L.; Yang, Y.; Liu, W.; Zhang, K.; Shi, G.; Zhou, H.; Geng, J.; Ning, X.; Wu, J.; et al. High Cytoplasmic Expression of SALL4 Predicts a Malignant Phenotype and Poor Prognosis of Breast Invasive Ductal Carcinoma. Neoplasma 2015, 62, 980–988. [Google Scholar] [CrossRef]
- Sweetman, D.; Münsterberg, A. The Vertebrate Spalt Genes in Development and Disease. Dev. Biol. 2006, 293, 285–293. [Google Scholar] [CrossRef]
- Nicolè, L.; Sanavia, T.; Veronese, N.; Cappellesso, R.; Luchini, C.; Dabrilli, P.; Fassina, A. Oncofetal Gene SALL4 and Prognosis in Cancer: A Systematic Review with Meta-Analysis. Oncotarget 2017, 8, 22968–22979. [Google Scholar] [CrossRef] [PubMed]
- Moein, S.; Tenen, D.G.; Amabile, G.; Chai, L. SALL4: An Intriguing Therapeutic Target in Cancer Treatment. Cells 2022, 11, 2601. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Xu, L.; Bi, W.; Ou, W.-B. SALL4 Oncogenic Function in Cancers: Mechanisms and Therapeutic Relevance. Int. J. Mol. Sci. 2022, 23, 2053. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Li, L.; Wang, D.; Yang, S.; Chen, X.; Zhou, S.; Zhong, S.; Zhao, J.; Tang, J. Higher Expression of SALL4 Predicts Poor Cancer Prognosis: A Meta-Analysis. Cancer Biomark. 2017, 19, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Dirican, E.; Akkiprik, M. Functional and Clinical Significance of SALL4 in Breast Cancer. Tumor Biol. 2016, 37, 11701–11709. [Google Scholar] [CrossRef] [PubMed]
- Yanagihara, N.; Kobayashi, D.; Kuribayashi, K.; Tanaka, M.; Hasegawa, T.; Watanabe, N. Significance of SALL4 as a Drug-Resistant Factor in Lung Cancer. Int. J. Oncol. 2015, 46, 1527–1534. [Google Scholar] [CrossRef]
- Chen, Y.-Y.; Li, Z.-Z.; Ye, Y.-Y.; Xu, F.; Niu, R.-J.; Zhang, H.-C.; Zhang, Y.-J.; Liu, Y.-B.; Han, B.-S. Knockdown of SALL4 Inhibits the Proliferation and Reverses the Resistance of MCF-7/ADR Cells to Doxorubicin Hydrochloride. BMC Mol. Biol. 2016, 17, 6. [Google Scholar] [CrossRef]
- Jiang, G.; Liu, C.-T. Knockdown of SALL4 Overcomes Cisplatin-Resistance through AKT/MTOR Signaling in Lung Cancer Cells. Int. J. Clin. Exp. Pathol. 2018, 11, 634–641. [Google Scholar]
- Yang, C.; Zhang, X.; Gao, C.; Du, K.; Liu, Y. NRBP1 Negatively Regulates SALL4 to Reduce the Invasion and Migration, Promote Apoptosis and Increase the Sensitivity to Chemotherapy Drugs of Breast Cancer Cells. Oncol. Lett. 2022, 23, 139. [Google Scholar] [CrossRef]
- Li, A.; Jiao, Y.; Yong, K.J.; Wang, F.; Gao, C.; Yan, B.; Srivastava, S.; Lim, G.S.D.; Tang, P.; Yang, H.; et al. SALL4 Is a New Target in Endometrial Cancer. Oncogene 2015, 34, 63–72. [Google Scholar] [CrossRef]
- Lu, J.; Jeong, H.; Kong, N.; Yang, Y.; Carroll, J.; Luo, H.R.; Silberstein, L.E.; YupoMa; Chai, L. Stem Cell Factor SALL4 Represses the Transcriptions of PTEN and SALL1 through an Epigenetic Repressor Complex. PLoS ONE 2009, 4, e5577. [Google Scholar] [CrossRef]
- Lai, A.Y.; Wade, P.A. Cancer Biology and NuRD: A Multifaceted Chromatin Remodelling Complex. Nat. Rev. Cancer 2011, 11, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.H.; Jobichen, C.; Chia, C.S.B.; Chan, T.H.M.; Tang, J.P.; Chung, T.X.Y.; Li, J.; Poulsen, A.; Hung, A.W.; Koh-Stenta, X.; et al. Targeting Cancer Addiction for SALL4 by Shifting Its Transcriptome with a Pharmacologic Peptide. Proc. Natl. Acad. Sci. USA 2018, 115, E7119–E7128. [Google Scholar] [CrossRef] [PubMed]
- Godoy-Ortiz, A.; Sanchez-Muñoz, A.; Chica Parrado, M.R.; Álvarez, M.; Ribelles, N.; Rueda Dominguez, A.; Alba, E. Deciphering HER2 Breast Cancer Disease: Biological and Clinical Implications. Front. Oncol. 2019, 9, 1124. [Google Scholar] [CrossRef] [PubMed]
- Bhagwat, A.S.; Vakoc, C.R. Targeting Transcription Factors in Cancer. Trends Cancer 2015, 1, 53–65. [Google Scholar] [CrossRef]
- Lambert, M.; Jambon, S.; Depauw, S.; David-Cordonnier, M.-H. Targeting Transcription Factors for Cancer Treatment. Molecules 2018, 23, 1479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Jiang, F.; Wang, X.; Li, G. Knockdown of SALL4 Inhibits Proliferation, Migration, and Invasion in Osteosarcoma Cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 763–771. [Google Scholar] [CrossRef]
- He, J.; Zhou, M.; Chen, X.; Yue, D.; Yang, L.; Qin, G.; Zhang, Z.; Gao, Q.; Wang, D.; Zhang, C.; et al. Inhibition of SALL4 Reduces Tumorigenicity Involving Epithelial-Mesenchymal Transition via Wnt/β-Catenin Pathway in Esophageal Squamous Cell Carcinoma. J. Exp. Clin. Cancer Res. 2016, 35, 98. [Google Scholar] [CrossRef]
- Liu, K.F.; Shan, Y.X. Effects of SiRNA-Mediated Silencing of Sal-like 4 Expression on Proliferation and Apoptosis of Prostate Cancer C4-2 Cells. Genet. Mol. Res. 2016, 15, 1–9. [Google Scholar] [CrossRef]
- Liu, C.; Yao, F.; Mao, X.; Li, W.; Chen, H. Effect of SALL4 on the Proliferation, Invasion and Apoptosis of Breast Cancer Cells. Technol. Cancer Res. Treat. 2020, 19, 153303382098007. [Google Scholar] [CrossRef]
- Kobayashi, D.; Kuribayshi, K.; Tanaka, M.; Watanabe, N. Watanabe SALL4 Is Essential for Cancer Cell Proliferation and Is Overexpressed at Early Clinical Stages in Breast Cancer. Int. J. Oncol. 2011, 38, 933–939. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chandarlapaty, S.; Sakr, R.A.; Giri, D.; Patil, S.; Heguy, A.; Morrow, M.; Modi, S.; Norton, L.; Rosen, N.; Hudis, C.; et al. Frequent Mutational Activation of the PI3K-AKT Pathway in Trastuzumab-Resistant Breast Cancer. Clin. Cancer Res. 2012, 18, 6784–6791. [Google Scholar] [CrossRef]
- Liu, C.; Wu, H.; Li, Y.; Shen, L.; Yu, R.; Yin, H.; Sun, T.; Sun, C.; Zhou, Y.; Du, Z. SALL4 Suppresses PTEN Expression to Promote Glioma Cell Proliferation via PI3K/AKT Signaling Pathway. J. Neurooncol. 2017, 135, 263–272. [Google Scholar] [CrossRef]
- Denslow, S.A.; Wade, P.A. The Human Mi-2/NuRD Complex and Gene Regulation. Oncogene 2007, 26, 5433–5438. [Google Scholar] [CrossRef] [PubMed]
- Kitange, G.J.; Mladek, A.C.; Schroeder, M.A.; Pokorny, J.C.; Carlson, B.L.; Zhang, Y.; Nair, A.A.; Lee, J.-H.; Yan, H.; Decker, P.A.; et al. Retinoblastoma Binding Protein 4 Modulates Temozolomide Sensitivity in Glioblastoma by Regulating DNA Repair Proteins. Cell Rep. 2016, 14, 2587–2598. [Google Scholar] [CrossRef] [PubMed]
- Khateb, A.; Deshpande, A.; Feng, Y.; Finlay, D.; Lee, J.S.; Lazar, I.; Fabre, B.; Li, Y.; Fujita, Y.; Zhang, T.; et al. The Ubiquitin Ligase RNF5 Determines Acute Myeloid Leukemia Growth and Susceptibility to Histone Deacetylase Inhibitors. Nat. Commun. 2021, 12, 5397. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Kohashi, K.; Yoshizumi, T.; Okumura, Y.; Tanaka, Y.; Shimokawa, M.; Iwasaki, T.; Aishima, S.; Maehara, Y.; Oda, Y. Coexpression of SALL4 with HDAC1 and/or HDAC2 Is Associated with Underexpression of PTEN and Poor Prognosis in Patients with Hepatocellular Carcinoma. Hum. Pathol. 2017, 64, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Tsang, J.Y.S.; Su, X.; Li, P.; Sun, W.; Wong, I.L.K.; Choy, K.; Yang, Q.; Tse, G.M.K.; Chan, T.H.; et al. SALL4 Promotes Tumor Progression in Breast Cancer by Targeting EMT. Mol. Carcinog. 2020, 59, 1209–1226. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, P.; Shao, M.; Zang, X.; Zhang, J.; Mao, F.; Qian, H.; Xu, W. SALL4 Activates TGF-β/SMAD Signaling Pathway to Induce EMT and Promote Gastric Cancer Metastasis. Cancer Manag. Res. 2018, 10, 4459–4470. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Yang, X.; Fang, C.; Xu, H.; Xi, X. SALL4 as an Epithelial-Mesenchymal Transition and Drug Resistance Inducer through the Regulation of c-Myc in Endometrial Cancer. PLoS ONE 2015, 10, e0138515. [Google Scholar] [CrossRef]
- Forghanifard, M.M.; Moghbeli, M.; Raeisossadati, R.; Tavassoli, A.; Mallak, A.J.; Boroumand-Noughabi, S.; Abbaszadegan, M.R. Role of SALL4 in the Progression and Metastasis of Colorectal Cancer. J. Biomed. Sci. 2013, 20, 6. [Google Scholar] [CrossRef] [PubMed]
- Itou, J.; Tanaka, S.; Li, W.; Iida, A.; Sehara-Fujisawa, A.; Sato, F.; Toi, M. The Sal-like 4-Integrin A6β1 Network Promotes Cell Migration for Metastasis via Activation of Focal Adhesion Dynamics in Basal-like Breast Cancer Cells. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2017, 1864, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Itou, J.; Matsumoto, Y.; Yoshikawa, K.; Toi, M. Sal-like 4 (SALL4) Suppresses CDH1 Expression and Maintains Cell Dispersion in Basal-like Breast Cancer. FEBS Lett. 2013, 587, 3115–3121. [Google Scholar] [CrossRef]
- bin Cho, K.; Cho, M.K.; Lee, W.Y.; Kang, K.W. Overexpression of C-Myc Induces Epithelial Mesenchymal Transition in Mammary Epithelial Cells. Cancer Lett. 2010, 293, 230–239. [Google Scholar] [CrossRef]
- Jeong, H.-W.; Cui, W.; Yang, Y.; Lu, J.; He, J.; Li, A.; Song, D.; Guo, Y.; Liu, B.H.; Chai, L. SALL4, a Stem Cell Factor, Affects the Side Population by Regulation of the ATP-Binding Cassette Drug Transport Genes. PLoS ONE 2011, 6, e18372. [Google Scholar] [CrossRef]
- Tang, G.; Cho, M.; Wang, X. OncoDB: An Interactive Online Database for Analysis of Gene Expression and Viral Infection in Cancer. Nucleic Acids Res. 2022, 50, D1334–D1339. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pattanayak, B.; Lameirinhas, A.; Torres-Ruiz, S.; Burgués, O.; Rovira, A.; Martínez, M.T.; Tapia, M.; Zazo, S.; Albanell, J.; Rojo, F.; et al. Role of SALL4 in HER2+ Breast Cancer Progression: Regulating PI3K/AKT Pathway. Int. J. Mol. Sci. 2022, 23, 13292. https://doi.org/10.3390/ijms232113292
Pattanayak B, Lameirinhas A, Torres-Ruiz S, Burgués O, Rovira A, Martínez MT, Tapia M, Zazo S, Albanell J, Rojo F, et al. Role of SALL4 in HER2+ Breast Cancer Progression: Regulating PI3K/AKT Pathway. International Journal of Molecular Sciences. 2022; 23(21):13292. https://doi.org/10.3390/ijms232113292
Chicago/Turabian StylePattanayak, Birlipta, Ana Lameirinhas, Sandra Torres-Ruiz, Octavio Burgués, Ana Rovira, María Teresa Martínez, Marta Tapia, Sandra Zazo, Joan Albanell, Federico Rojo, and et al. 2022. "Role of SALL4 in HER2+ Breast Cancer Progression: Regulating PI3K/AKT Pathway" International Journal of Molecular Sciences 23, no. 21: 13292. https://doi.org/10.3390/ijms232113292
APA StylePattanayak, B., Lameirinhas, A., Torres-Ruiz, S., Burgués, O., Rovira, A., Martínez, M. T., Tapia, M., Zazo, S., Albanell, J., Rojo, F., Bermejo, B., & Eroles, P. (2022). Role of SALL4 in HER2+ Breast Cancer Progression: Regulating PI3K/AKT Pathway. International Journal of Molecular Sciences, 23(21), 13292. https://doi.org/10.3390/ijms232113292





