Transcriptomic and Metabolomic Analysis of Korean Pine Cell Lines with Different Somatic Embryogenic Potential
Abstract
:1. Introduction
2. Results
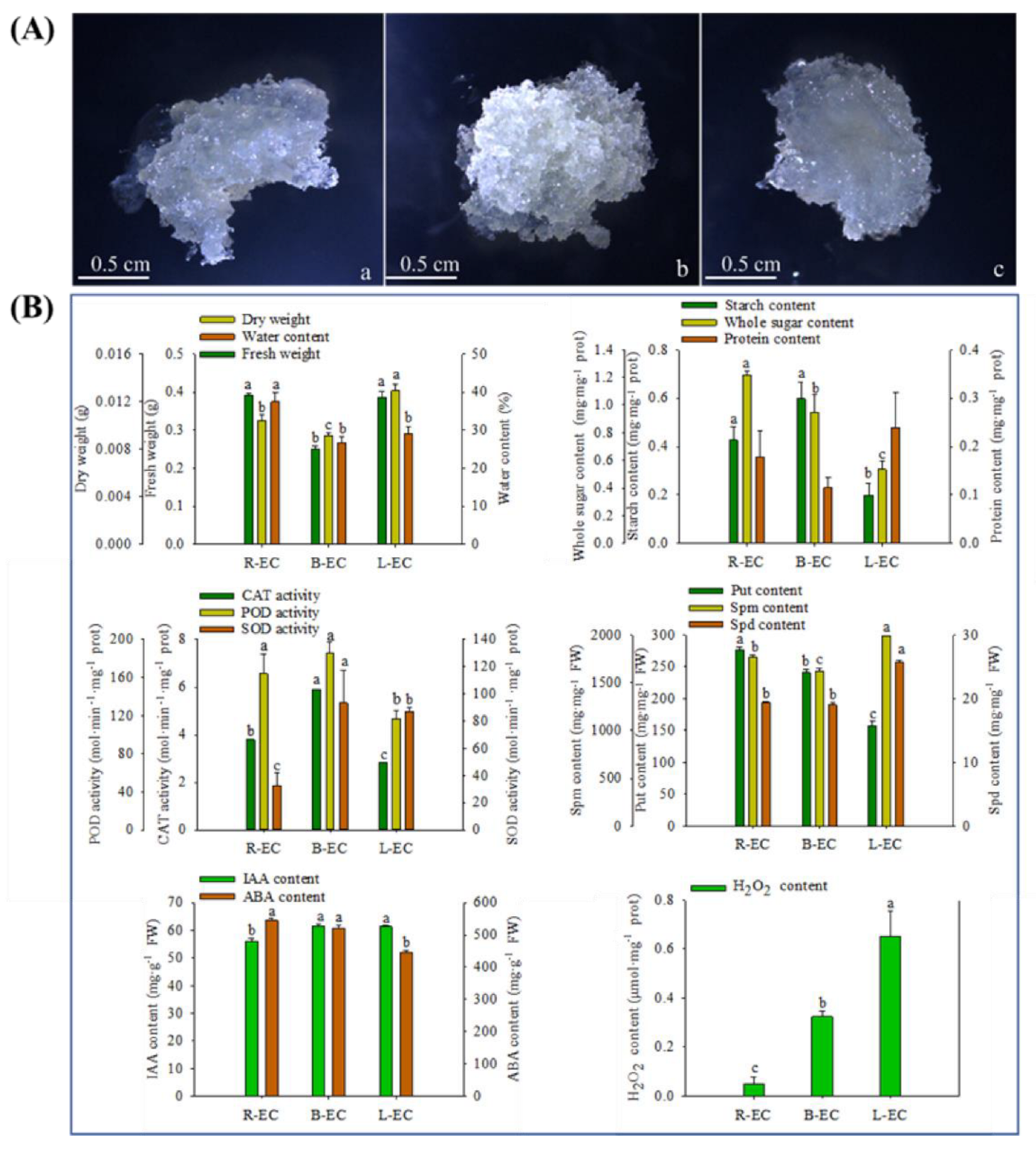
2.1. Morphological and Physiological Responses between Different Cell Lines
2.2. Transcriptome Analysis among R-EC, L-EC, and B-EC
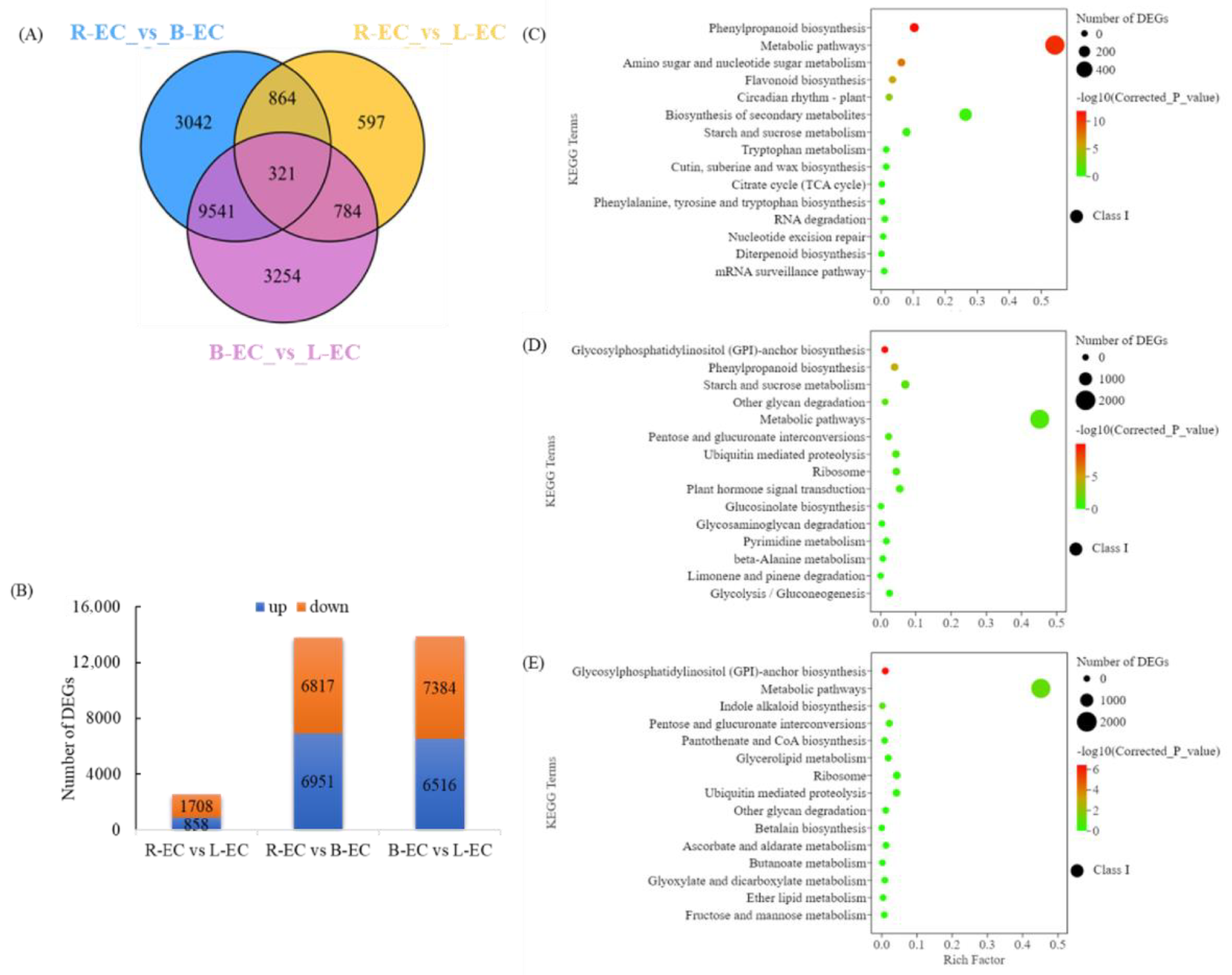
2.2.1. Analysis of Differentially Expressed Genes DEGs in R endash EC, L endash EC, and B endash EC
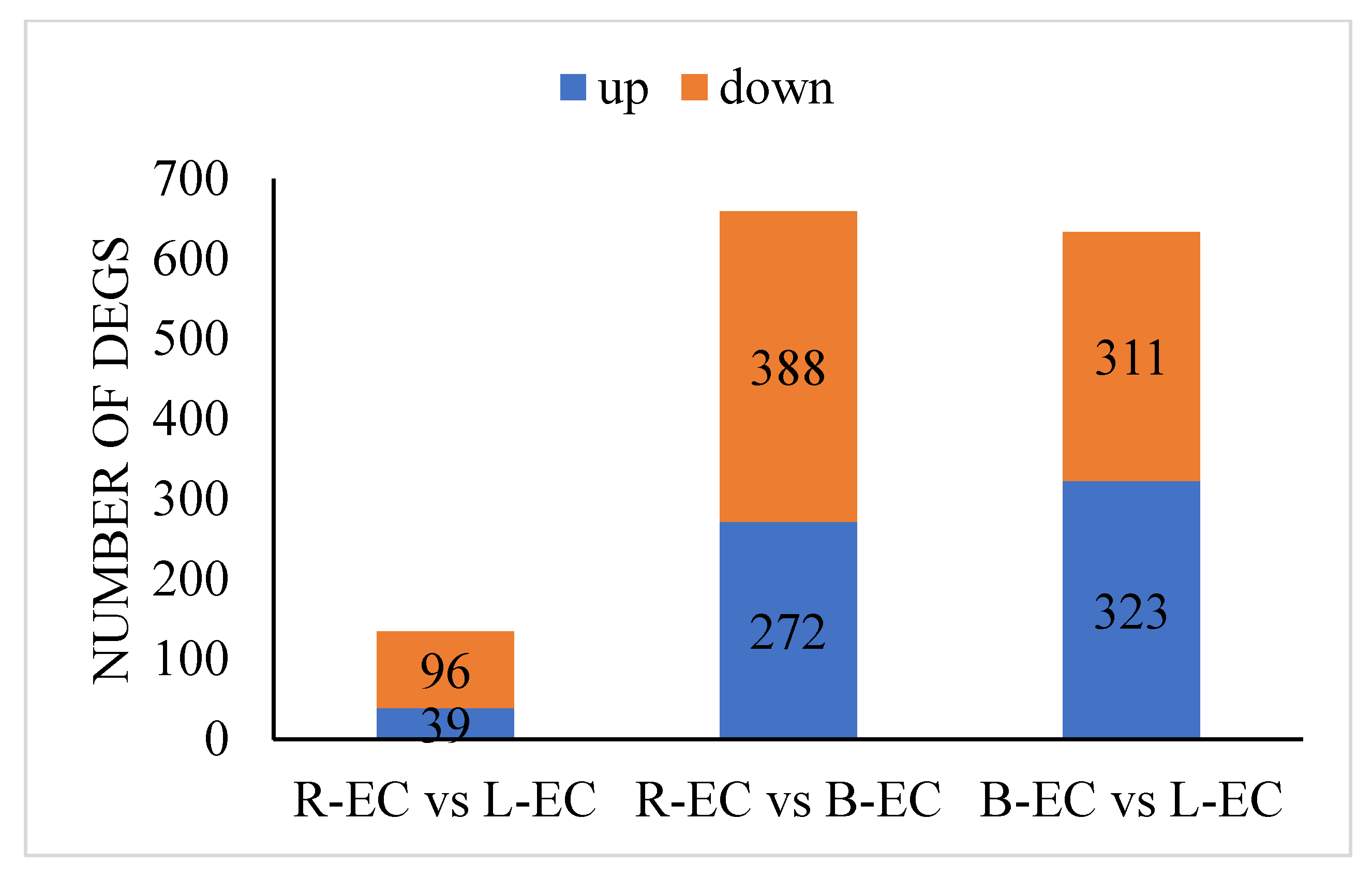
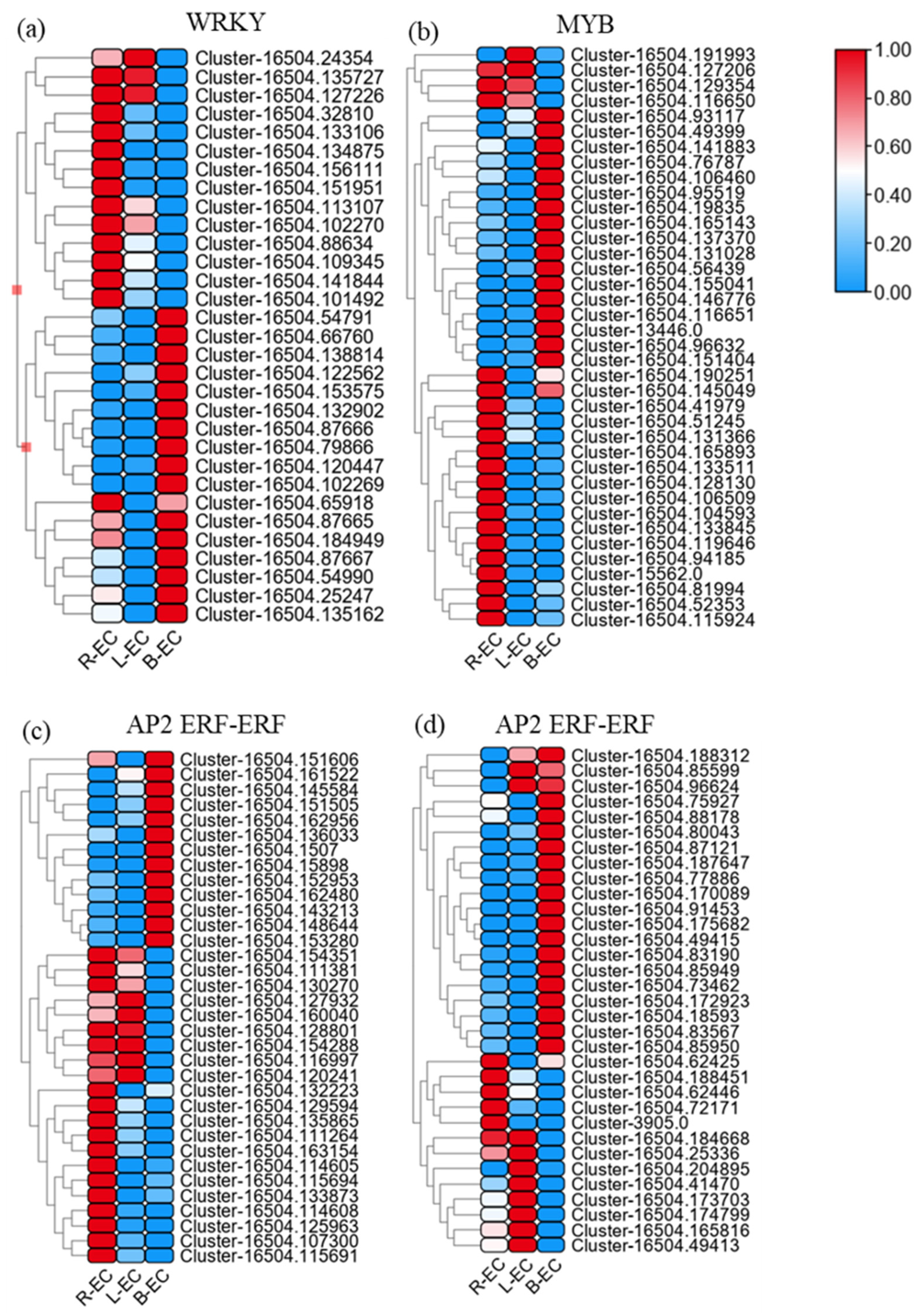
2.2.2. Transcription Factor Expression among R-EC, L-EC, and B-EC
2.3. Metabolomic Analysis of R-EC, L-EC, and B-EC
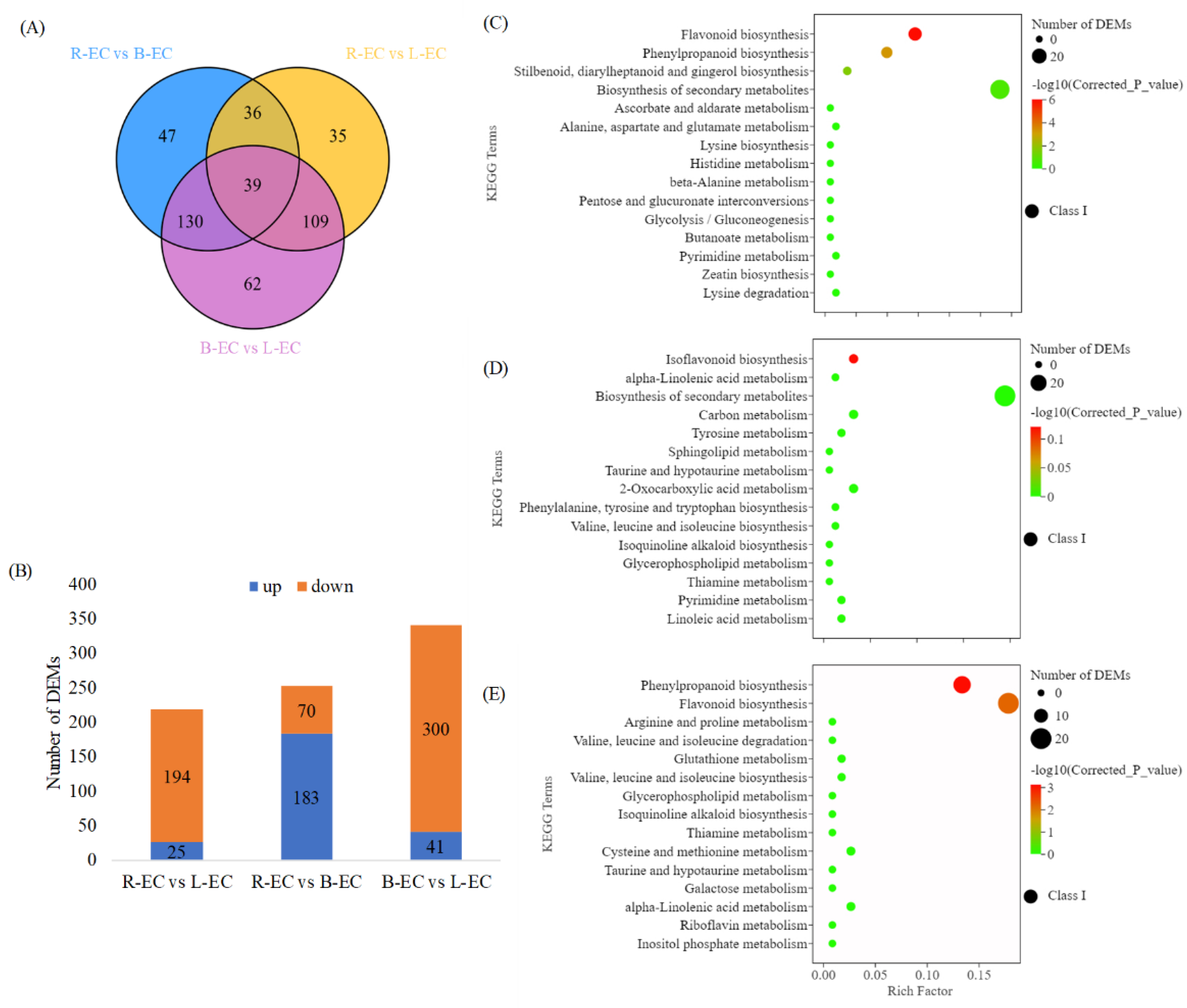
2.3.1. Differential Metabolite Analysis in R-EC, L-EC, and B-EC
2.3.2. Functional Analysis of DEGs and DEMs among R-EC, L-EC, and B-EC
Differences in Hormone Signaling Function between Different Cell Lines
Differences in Starch and Sugar Metabolism between Different Cell Lines
Differences in Phenylpropane Metabolic Pathways between Different Cell Lines
Differences in Flavonoid Metabolic Pathways among Different Cell Lines
3. Discussion
3.1. Influence of Transcription Factors during Somatic Embryogenesis
3.2. Effects of Plant Hormones on Somatic Embryogenesis Potential
3.3. The Role of Starch and Sugar Metabolism in Somatic Embryogenesis
3.4. The Role of Phenylpropane Metabolism in Somatic Embryogenesis
3.5. The Role of Flavonoid Metabolism in Somatic Embryogenesis
4. Materials and Methods
4.1. Initiation of Korean Pine Embryogenic Callus
4.2. Experimental Methods
4.2.1. Morphological Observation
4.2.2. Physiological Measurements
4.2.3. Transcriptomic Analysis
4.2.4. Metabolomics Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Arrillaga, I.; Morcillo, M.; Zanón, I.; Lario, F.; Segura, J.; Sales, E. New approaches to optimize somatic embryogenesis in maritime pine. Front. Plant Sci. 2019, 10, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pullman, G.S. Embryogenic tissue initiation in Loblolly pine (Pinus Taeda L.). In Step Wise Protocols for Somatic Embryogenesis of Important Woody Plants, 2nd ed.; Shri Mohan, J., Gupta, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 13–31. [Google Scholar]
- Peng, C.; Gao, F.; Wang, H.; Shen, H.; Yang, L. Optimization of maturation process for somatic embryo production and cryopreservation of embryogenic tissue in Pinus koraiensis. Plant Cell Tissue Organ Cult. 2021, 144, 185–194. [Google Scholar] [CrossRef]
- Peng, C.; Gao, F.; Wang, H.; Tretyakova, I.N.; Nosov, A.M.; Shen, H.; Yang, L. Morphological and Physiological Indicators for Screening Cell Lines with High Potential for Somatic Embryo Maturation at an Early Stage of Somatic Embryogenesis in Pinus Koraiensis. Plants 2022, 11, 1867. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Gao, F.; Wang, H.; Shen, H.; Yang, L. Physiological and Biochemical Traits in Korean Pine Somatic Embryogenesis. Forests 2020, 11, 577. [Google Scholar] [CrossRef]
- Guo, H.; Guo, H.; Zhang, L.; Fan, Y.; Wu, J.; Tang, Z.; Zhang, Y.; Fan, Y.; Zeng, F. Dynamic Transcriptome Analysis Reveals Uncharacterized Complex Regulatory Pathway Underlying Genotype-Recalcitrant Somatic Embryogenesis Transdifferentiation in Cotton. Genes 2020, 11, 519. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, A.L.; Elbl, P.; Navarro, B.V.; de Oliveira, L.F.; Salvato, F.; Balbuena, T.S.; Floh, E.I. Quantitative proteomic analysis of Araucaria angustifolia (Bertol.) Kuntze cell lines with contrasting embryogenic potential. J. Proteom. 2016, 130, 180–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klimaszewska, K.; Hargreaves, C.; Lelu-Walter, M.A.; Trontin, J.F. Advances in Conifer Somatic Embryogenesis Since Year 2000. Methods Mol. Biol. 2016, 1359, 131–166. [Google Scholar]
- Klimaszewska, K.; Noceda, C.; Pelletier, G.; Label, P.; Rodriguez, R.; Lelu-Walter, M.-A. Biological characterization of young and aged embryogenic cultures of Pinus pinaster (Ait.). Vitr. Cell. Dev. Biol.-Plant 2009, 45, 20–33. [Google Scholar] [CrossRef]
- Li, W.-F.; Zhang, S.-G.; Han, S.-Y.; Wu, T.; Zhang, J.-H.; Qi, L.-W. Regulation of LaMYB33 by miR159 during maintenance of embryogenic potential and somatic embryo maturation in Larix kaempferi (Lamb.) Carr. Plant Cell Tissue Organ Cult. 2013, 113, 131–136. [Google Scholar] [CrossRef]
- Lelu-Walter, M.A.; Paques, L.E. Simplified and improved somatic embryogenesis of hybrid larches (Larix × eurolepis and Larix × marschlinsii). Perspectives for breeding. Ann. For. Sci. 2009, 66, 1. [Google Scholar] [CrossRef]
- Plačková, L.; Hrdlička, J.; Smýkalová, I.; Cvečková, M.; Novák, O.; Griga, M.; Doležal, K. Cytokinin profiling of long-term in vitro pea (Pisum sativum L.) shoot cultures. Plant Growth Regul. 2015, 77, 125–132. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, N.; Zhao, S. Functional characterization of a WRKY family gene involved in somatic embryogenesis in Panax ginseng. Protoplasma 2020, 257, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Altamura, M.M.; Della Rovere, F.; Fattorini, L.; D’Angeli, S.; Falasca, G. Recent advances on genetic and physiological bases of in vitro somatic embryo formation. Vitr. Embryog. High. Plants 2016, 1359, 47–85. [Google Scholar]
- Piyatrakul, P.; Putranto, R.-A.; Martin, F.; Rio, M.; Dessailly, F.; Leclercq, J.; Dufayard, J.-F.; Lardet, L.; Montoro, P. Some ethylene biosynthesis and AP2/ERF genes reveal a specific pattern of expression during somatic embryogenesis in Hevea brasiliensis. BMC Plant Biol. 2012, 12, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olivares-García, C.A.; Mata-Rosas, M.; Peña-Montes, C.; Quiroz-Figueroa, F.; Segura-Cabrera, A.; Shannon, L.M.; Loyola-Vargas, V.M.; Monribot-Villanueva, J.L.; Elizalde-Contreras, J.M.; Ibarra-Laclette, E. Phenylpropanoids are connected to cell wall fortification and stress tolerance in avocado somatic embryogenesis. Int. J. Mol. Sci. 2020, 21, 5679. [Google Scholar] [CrossRef]
- Jamaluddin, N.D.; Noor, N.M.; Goh, H.-H. Genome-wide transcriptome profiling of Carica papaya L. embryogenic callus. Physiol. Mol. Biol. Plants 2017, 23, 357–368. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Zhang, J.; Hou, Y.; Yao, Y.; Ji, Q. RNA-Seq screening of differentially-expressed genes during somatic embryogenesis in Fragaria× ananassa Duch.‘Benihopp’. J. Hortic. Sci. Biotechnol. 2015, 90, 671–681. [Google Scholar] [CrossRef]
- Zhai, L.; Xu, L.; Wang, Y.; Zhu, X.; Feng, H.; Li, C.; Luo, X.; Everlyne, M.M.; Liu, L. Transcriptional identification and characterization of differentially expressed genes associated with embryogenesis in radish (Raphanus sativus L.). Sci. Rep. 2016, 6, 21652. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Xu, X.; Liu, Z.; Zhang, Z.; XuHan, X.; Lin, Y.; Lai, Z. Global scale transcriptome analysis reveals differentially expressed genes involve in early somatic embryogenesis in Dimocarpus longan Lour. BMC Genom. 2020, 21, 4. [Google Scholar] [CrossRef] [Green Version]
- Navarro, B.V.; Elbl, P.; De Souza, A.P.; Jardim, V.; de Oliveira, L.F.; Macedo, A.F.; Dos Santos, A.L.; Buckeridge, M.S.; Floh, E.I. Carbohydrate-mediated responses during zygotic and early somatic embryogenesis in the endangered conifer, Araucaria angustifolia. PLoS ONE 2017, 12, e0180051. [Google Scholar] [CrossRef] [Green Version]
- Dossou, S.S.K.; Xu, F.; Cui, X.; Sheng, C.; Zhou, R.; You, J.; Tozo, K.; Wang, L. Comparative metabolomics analysis of different sesame (Sesamum indicum L.) tissues reveals a tissue-specific accumulation of metabolites. BMC Plant Biol. 2021, 21, 352. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, S.; Wang, J. Transcriptome analysis of callus from Picea balfouriana. BMC Genom. 2014, 15, 553. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Guo, H.; Zhang, L.; Tang, Z.; Yu, X.; Wu, J.; Zeng, F. Metabolome and transcriptome association analysis reveals dynamic regulation of purine metabolism and flavonoid synthesis in transdifferentiation during somatic embryogenesis in cotton. Int. J. Mol. Sci. 2019, 20, 2070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, S.E.; Zheng, Q.; Zheng, Y. Transcriptome analysis indicates that GmAGAMOUS-Like 15 may enhance somatic embryogenesis by promoting a dedifferentiated state. Plant Signal. Behav. 2016, 11, e1197463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, H.; Tian, S.; Dong, H.; Guo, C. Pleiotropic effects of TaMYB3R1 on plant development and response to osmotic stress in transgenic Arabidopsis. Gene 2015, 558, 227–234. [Google Scholar] [CrossRef]
- Tsuwamoto, R.; Yokoi, S.; Takahata, Y. Arabidopsis EMBRYOMAKER encoding an AP2 domain transcription factor plays a key role in developmental change from vegetative to embryonic phase. Plant Mol. Biol. 2010, 73, 481–492. [Google Scholar] [CrossRef]
- El Ouakfaoui, S.; Schnell, J.; Abdeen, A.; Colville, A.; Labbé, H.; Han, S.; Baum, B.; Laberge, S.; Miki, B. Control of somatic embryogenesis and embryo development by AP2 transcription factors. Plant. Mol. Biol. 2010, 74, 313–326. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Wang, Q.; Schmitz, G.; Müller, D.; Theres, K. The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J. 2012, 71, 61–70. [Google Scholar] [CrossRef]
- Tretyakova, I.N.; Kudoyarova, G.R.; Park, M.E.; Kazachenko, A.S.; Shuklina, A.S.; Akhiyarova, G.R.; Korobova, A.V.; Veselov, S.U. Content and immunohistochemical localization of hormones during in vitro somatic embryogenesis in long-term proliferating Larix sibirica cultures. Plant Cell Tissue Organ Cult. 2019, 136, 511–522. [Google Scholar] [CrossRef]
- Andreae, W.; Good, N.E. The Formation of Indoleacetylaspartic Acid in Pea Seedlings. Plant Physiol. 1955, 30, 380. [Google Scholar] [CrossRef]
- Goldental-Cohen, S.; Israeli, A.; Ori, N.; Yasuor, H. Auxin response dynamics during wild-type and entire flower development in tomato. Plant Cell Physiol. 2017, 58, 1661–1672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, F.; Hu, L.; Yuan, D.; Xu, J.; Gao, W.; He, L.; Yang, X.; Zhang, X. Comparative transcriptome analysis between somatic embryos (SE s) and zygotic embryos in cotton: Evidence for stress response functions in SE development. Plant Biotechnol. J. 2014, 12, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Etienne, H.; Sotta, B.; Montoro, P.; Miginiac, E.; Carron, M.-P. Relations between exogenous growth regulators and endogenous indole-3-acetic acid and abscisic acid in the expression of somatic embryogenesis in Hevea brasiliensis (Müll. Arg.). Plant Sci. 1993, 88, 91–96. [Google Scholar] [CrossRef]
- Ivanova, A.; Velcheva, M.; Denchev, P.; Atanassov, A.; Van Onckelen, H.A. Endogenous hormone levels during direct somatic embryogenesis in Medicago falcata. Physiol. Plant. 1994, 92, 85–89. [Google Scholar] [CrossRef]
- Ranwala, A.P.; Miller, W.B. Purification and characterization of an endoamylase from tulip (Tulipa gesneriana) bulbs. Physiol. Plant. 2000, 109, 388–395. [Google Scholar] [CrossRef]
- Maadon, S.N.; Rohani, E.R.; Ismail, I.; Baharum, S.N.; Normah, M.N. Somatic embryogenesis and metabolic differences between embryogenic and non-embryogenic structures in mangosteen. Plant Cell Tissue Organ Cult. 2016, 127, 443–459. [Google Scholar] [CrossRef]
- Chakhchar, A.; Lamaoui, M.; Wahbi, S.; Ferradous, A.; El Mousadik, A.; Ibnsouda-Koraichi, S.; Filali-Maltouf, A.; El Modafar, C. Leaf water status, osmoregulation and secondary metabolism as a model for depicting drought tolerance in Argania spinosa. Acta Physiol. Plant. 2015, 37, 80. [Google Scholar] [CrossRef]
- Zhang, C.-H.; Ma, T.; Luo, W.-C.; Xu, J.-M.; Liu, J.-Q.; Wan, D.-S. Identification of 4CL genes in desert poplars and their changes in expression in response to salt stress. Genes 2015, 6, 901–917. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.P.; Matthews, M.L.; Williams, C.M.; Shi, R.; Yang, C.; Tunlaya-Anukit, S.; Chen, H.-C.; Li, Q.; Liu, J.; Lin, C.-Y. Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat. Commun. 2018, 9, 1579. [Google Scholar] [CrossRef] [Green Version]
- Moura, J.C.M.S.; Bonine, C.A.V.; de Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef]
- Dauwe, R.; Morreel, K.; Goeminne, G.; Gielen, B.; Rohde, A.; Van Beeumen, J.; Ralph, J.; Boudet, A.M.; Kopka, J.; Rochange, S.F.; et al. Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J. 2007, 52, 263–285. [Google Scholar] [CrossRef] [PubMed]
- Santos, E.L.; Maia, B.; Ferriani, A.P.; Teixeira, S.D. Flavonoids: Classification, biosynthesis and chemical ecology. Flavonoids-Biosynth. Hum. Health 2017, 13, 78–94. [Google Scholar]
- Koirala, N.; Thuan, N.H.; Ghimire, G.P.; Van Thang, D.; Sohng, J.K. Methylation of flavonoids: Chemical structures, bioactivities, progress and perspectives for biotechnological production. Enzym. Microb. Technol. 2016, 86, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Moriguchi, T.; Kita, M.; Tomono, Y.; Endo-Inagaki, T.; Omura, M. One type of chalcone synthase gene expressed during embryogenesis regulates the flavonoid accumulation in citrus cell cultures. Plant Cell Physiol. 1999, 40, 651–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.A.; Abbasi, B.H.; Ali, H.; Ali, M.; Adil, M.; Hussain, I. Temporal variations in metabolite profiles at different growth phases during somatic embryogenesis of Silybum marianum L. Plant Cell Tissue Organ Cult. 2015, 120, 127–139. [Google Scholar] [CrossRef]
- Kocsy, G.; Tari, I.; Vanková, R.; Zechmann, B.; Gulyás, Z.; Poór, P.; Galiba, G. Redox control of plant growth and development. Plant Sci. 2013, 211, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, T.; Hayashi, N.; Sasamoto, H. Atomic force microscopy and laser confocal scanning microscopy analysis of callose fibers developed from protoplasts of embryogenic cells of a conifer. Planta 2005, 223, 40–45. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Deng, C.; Xia, Y.; Kong, L.; Zhang, H.; Zhang, S.; Wang, J. Identification of novel miRNAs and miRNA expression profiling in embryogenic tissues of Picea balfouriana treated by 6-benzylaminopurine. PLoS ONE 2017, 12, e0176112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, X.; Zhang, T.-t.; Zhang, J. Effect of subculture times on genetic fidelity, endogenous hormone level and pharmaceutical potential of Tetrastigma hemsleyanum callus. Plant Cell Tissue Organ Cult. 2015, 122, 67–77. [Google Scholar] [CrossRef]
- Dutra, N.T.; Silveira, V.; de Azevedo, I.G.; Gomes-Neto, L.R.; Façanha, A.R.; Steiner, N.; Guerra, M.P.; Floh, E.I.S.; Santa-Catarina, C. Polyamines affect the cellular growth and structure of pro-embryogenic masses in Araucaria angustifolia embryogenic cultures through the modulation of proton pump activities and endogenous levels of polyamines. Physiol. Plant. 2013, 148, 121–132. [Google Scholar] [CrossRef]
- Yuan, H.; Zeng, X.; Shi, J.; Xu, Q.; Wang, Y.; Jabu, D.; Sang, Z.; Nyima, T. Time-course comparative metabolite profiling under osmotic stress in tolerant and sensitive Tibetan hulless barley. BioMed Res. Int. 2018, 2018, 9425409. [Google Scholar] [CrossRef] [PubMed]





| Sample | Raw Reads | Clean Reads | Clean Bases (G) | Error Rate (%) | Q20 (%) | GC Content (%) |
|---|---|---|---|---|---|---|
| R-EC1 | 57,338,114 | 55,752,360 | 8.36 | 0.03 | 97.12 | 44.34 |
| R-EC2 | 55,311,330 | 54,021,080 | 8.1 | 0.03 | 97.07 | 44.19 |
| R-EC3 | 61,795,852 | 60,159,682 | 9.02 | 0.03 | 97.18 | 44.17 |
| L-EC1 | 60,854,538 | 59,090,214 | 8.86 | 0.03 | 97.19 | 44.27 |
| L-EC2 | 70,881,322 | 68,699,534 | 10.3 | 0.03 | 97.19 | 44.16 |
| L-EC3 | 56,300,036 | 54,869,884 | 8.23 | 0.03 | 97.06 | 44.22 |
| B-EC1 | 65,271,562 | 63,299,676 | 9.49 | 0.03 | 97.11 | 44.28 |
| B-EC2 | 61,163,512 | 59,224,850 | 8.88 | 0.03 | 97.17 | 44.27 |
| B-EC3 | 73,620,870 | 70,887,378 | 10.63 | 0.03 | 97.14 | 44.27 |
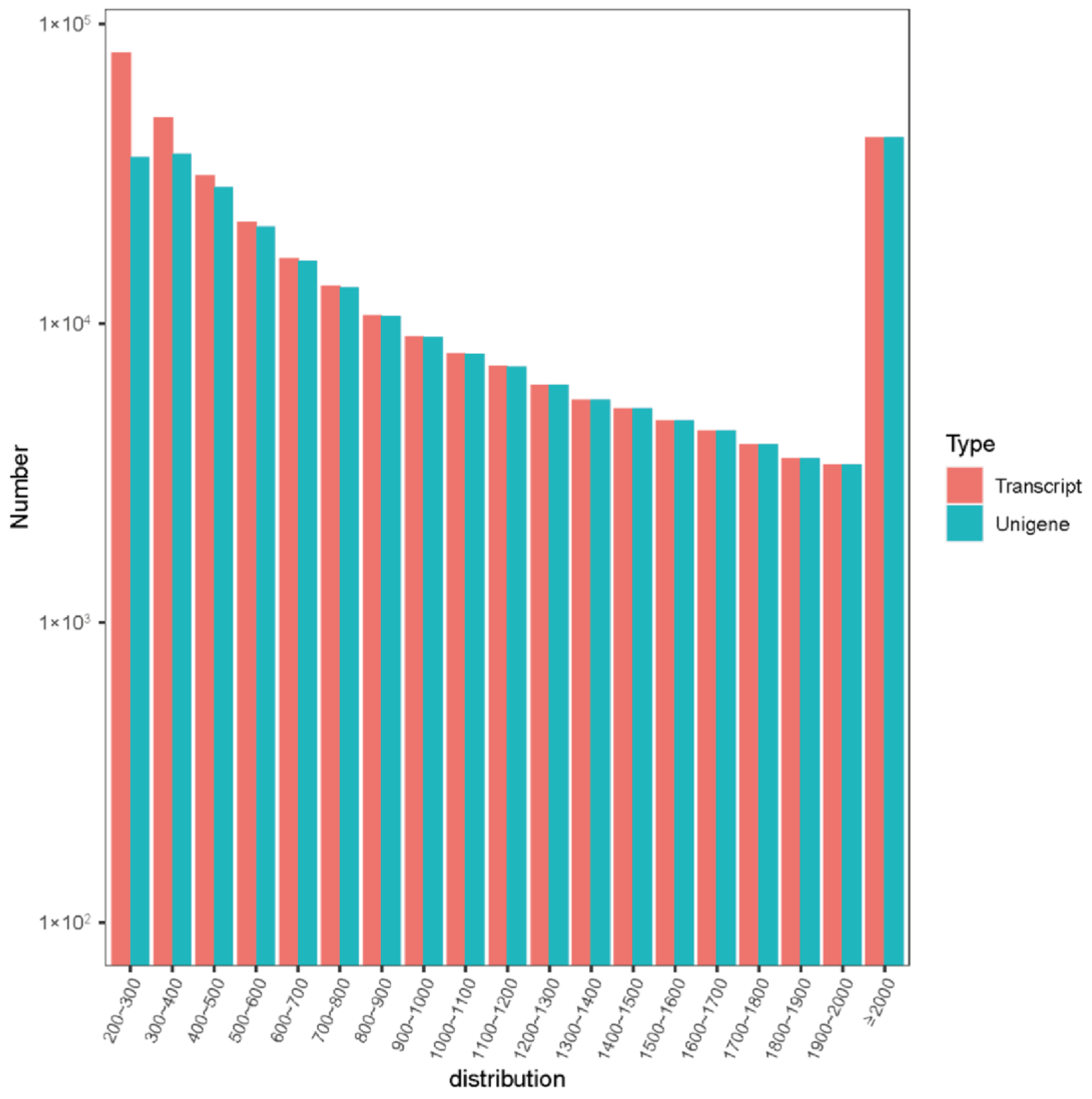
| Type | Number | Mean Length | N50 | N90 | Total Bases |
|---|---|---|---|---|---|
| Transcript | 325,415 | 959 | 1718 | 366 | 312,096,367 |
| Unigene | 264,676 | 1115 | 1842 | 459 | 295,135,923 |
| Sample | WRKY Family | MYB Family | AP2/ERF-ERF Family | |||
|---|---|---|---|---|---|---|
| up | down | up | down | up | down | |
| R-EC_vs._L-EC | 0 | 4 | 1 | 13 | 2 | 14 |
| R-EC_vs._B-EC | 4 | 15 | 6 | 15 | 27 | 26 |
| B-EC_vs._L-EC | 6 | 13 | 13 | 27 | 20 | 26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, C.; Gao, F.; Tretyakova, I.N.; Nosov, A.M.; Shen, H.; Yang, L. Transcriptomic and Metabolomic Analysis of Korean Pine Cell Lines with Different Somatic Embryogenic Potential. Int. J. Mol. Sci. 2022, 23, 13301. https://doi.org/10.3390/ijms232113301
Peng C, Gao F, Tretyakova IN, Nosov AM, Shen H, Yang L. Transcriptomic and Metabolomic Analysis of Korean Pine Cell Lines with Different Somatic Embryogenic Potential. International Journal of Molecular Sciences. 2022; 23(21):13301. https://doi.org/10.3390/ijms232113301
Chicago/Turabian StylePeng, Chunxue, Fang Gao, Iraida Nikolaevna Tretyakova, Alexander Mikhaylovich Nosov, Hailong Shen, and Ling Yang. 2022. "Transcriptomic and Metabolomic Analysis of Korean Pine Cell Lines with Different Somatic Embryogenic Potential" International Journal of Molecular Sciences 23, no. 21: 13301. https://doi.org/10.3390/ijms232113301






