A Novel Antimicrobial Peptide Spampcin56–86 from Scylla paramamosain Exerting Rapid Bactericidal and Anti-Biofilm Activity In Vitro and Anti-Infection In Vivo
Abstract
:1. Introduction
2. Results
2.1. Sequence Analysis and Expression Profiles of Spampcin Gene
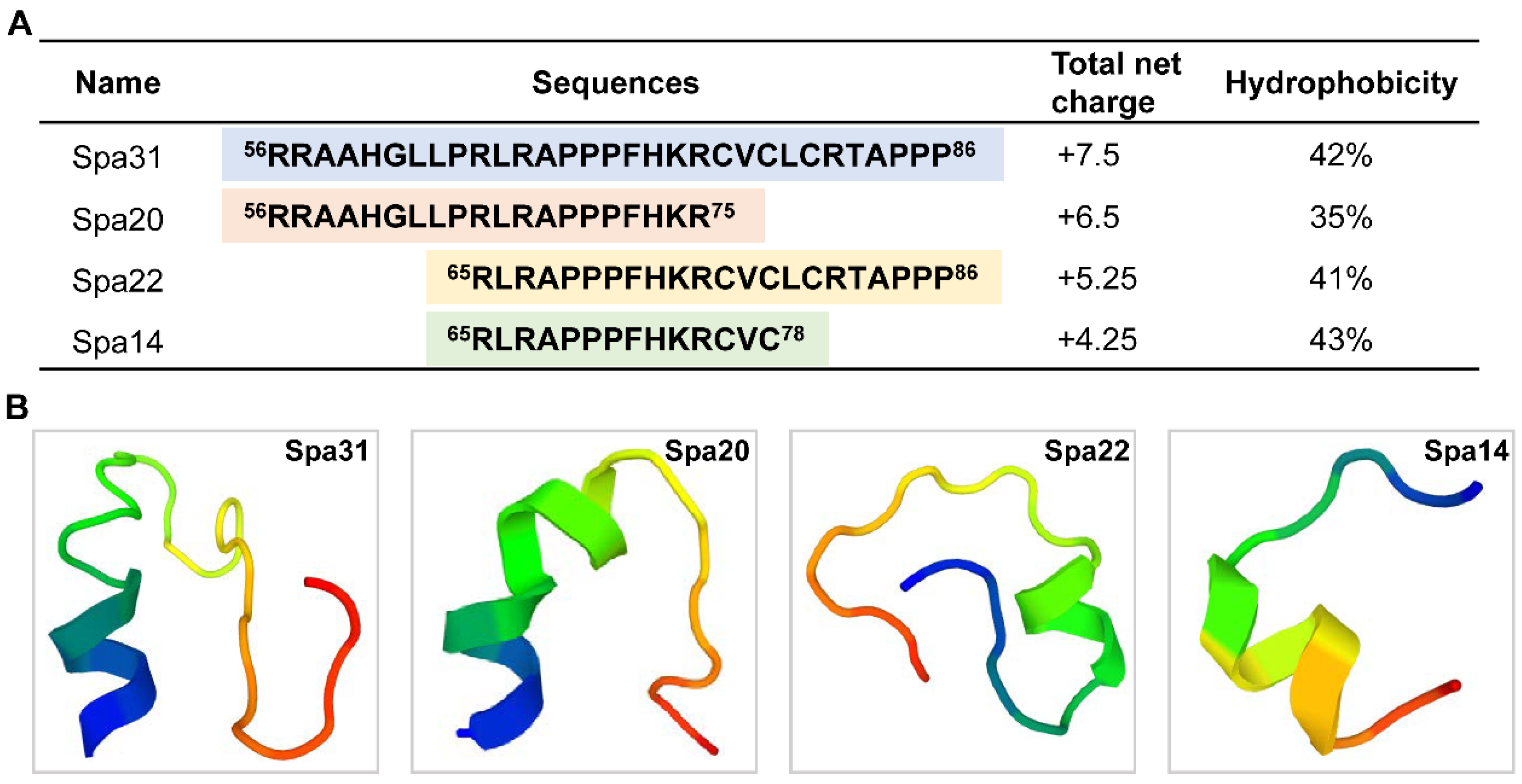
2.2. Truncated Peptides Derived from Spampcin Show Potent and Broad-Spectrum Antimicrobial Activity
2.3. Effective Bactericidal Activity of Spa31 against S. aureus and P. aeruginosa
2.4. Antibacterial Mechanism of Spa31 against P. aeruginosa
2.5. Anti-Biofilm Activity of Spa31 against P. aeruginosa
2.6. In Vitro Determination of Spa31 Properties and In Vivo Anti-P. aeruginosa Infection Analysis
3. Discussion
4. Materials and Methods
4.1. Microorganism Strains, Cell Lines, and Reagents
4.2. AMP Prediction and Truncated Peptide Synthesis
4.3. Antimicrobial Assay
4.4. Zone of Inhibition Assay
4.5. Time-Killing Kinetics
4.6. Scanning Electron Microscope (SEM) Observation
4.7. SYTO 9 and PI Staining Assay
4.8. NPN, LPS Inhibition, ATP Release, ROS Production Assays
4.8.1. NPN Staining Assay
4.8.2. LPS Inhibition Assay
4.8.3. ATP Release Assay
4.8.4. ROS Production Assay
4.9. Biofilm Inhibition Assays
4.9.1. Biofilm Formation Inhibition Assay
4.9.2. Mature Biofilm Inhibition Assay
4.10. Cytotoxicity and Hemolytic Activity Assay
4.10.1. Cytotoxicity Assay
4.10.2. Hemolytic Activity Assay
4.11. Evaluation of the In Vivo Infective Effect of Spa31 on Zebrafish D. rerio Infected with P. aeruginosa
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Bardan, A.; Nizet, V.; Gallo, R.L. Antimicrobial peptides and the skin. Expert Opin. Biol. 2004, 4, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Jenssen, H.; Hamill, P.; Hancock, R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, G.; Wu, H.; Huang, J.; Wang, W.; Ge, K.; Li, G.; Zhong, J.; Huang, Q. LAMP2: A major update of the database linking antimicrobial peptides. Database 2020, 2020, baaa061. [Google Scholar] [CrossRef]
- Hancock, R.E.; Haney, E.F.; Gill, E.E. The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, M.; Jenssen, H.; Kindrachuk, J.; Scott, W.R.; Elliott, M.; Hilpert, K.; Cheng, J.T.; Hancock, R.E.; Straus, S.K. Structural studies of a peptide with immune modulating and direct antimicrobial activity. Chem. Biol. 2010, 17, 970–980. [Google Scholar] [CrossRef] [Green Version]
- Bjorn, C.; Hakansson, J.; Myhrman, E.; Sjostrand, V.; Haug, T.; Lindgren, K.; Blencke, H.M.; Stensvag, K.; Mahlapuu, M. Anti-infectious and anti-inflammatory effects of peptide fragments sequentially derived from the antimicrobial peptide centrocin 1 isolated from the green sea urchin, Strongylocentrotus droebachiensis. AMB Express 2012, 2, 67. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Chen, F.; Wang, X.; Peng, H.; Zhang, H.; Wang, K.J. The synergistic effect of mud crab antimicrobial peptides Sphistin and Sph12-38 with antibiotics azithromycin and rifampicin enhances bactericidal activity against Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2020, 10, 572849. [Google Scholar] [CrossRef]
- Agerberth, B.; Lee, J.Y.; Bergman, T.; Carlquist, M.; Boman, H.G.; Mutt, V.; Jornvall, H. Amino acid sequence of PR-39. Isolation from pig intestine of a new member of the family of proline-arginine-rich antibacterial peptides. Eur. J. Biochem. 1991, 202, 849–854. [Google Scholar] [CrossRef]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef]
- Buelow, E.; Ploy, M.C.; Dagot, C. Role of pollution on the selection of antibiotic resistance and bacterial pathogens in the environment. Curr. Opin. Microbiol. 2021, 64, 117–124. [Google Scholar] [CrossRef]
- Chen, F.Y.; Liu, H.P.; Bo, J.; Ren, H.L.; Wang, K.J. Identification of genes differentially expressed in hemocytes of Scylla paramamosain in response to lipopolysaccharide. Fish Shellfish Immunol. 2010, 28, 167–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.F.; Qiao, K.; Huang, S.P.; Peng, H.; Huang, W.S.; Chen, F.Y.; Zhang, N.; Wang, G.Z.; Wang, K.J. The expression pattern of scygonadin during the ontogenesis of Scylla paramamosain predicting its potential role in reproductive immunity. Dev. Comp. Immunol. 2011, 35, 1078–1090. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.F.; Qiao, K.; Huang, S.P.; Peng, H.; Huang, W.S.; Chen, B.; Chen, F.Y.; Bo, J.; Wang, K.J. Quantitative gene expression and in situ localization of scygonadin potentially associated with reproductive immunity in tissues of male and female mud crabs, Scylla paramamosain. Fish Shellfish Immun. 2011, 31, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Qiao, K.; Xu, W.F.; Chen, H.Y.; Peng, H.; Zhang, Y.Q.; Huang, W.S.; Wang, S.P.; An, Z.; Shan, Z.G.; Chen, F.Y.; et al. A new antimicrobial peptide SCY2 identified in Scylla paramamosain exerting a potential role of reproductive immunity. Fish Shellfish Immunol. 2016, 51, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, F.; Chen, H.Y.; Peng, H.; Hao, H.; Wang, K.J. A novel antimicrobial peptide Scyreprocin from mud crab Scylla paramamosain showing potent antifungal and anti-biofilm activity. Front. Microbiol. 2020, 11, 1589. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, F.; Li, S.; Peng, H.; Wang, K.J. A novel antimicrobial peptide Sparanegtin identified in Scylla paramamosain showing antimicrobial activity and immunoprotective role in vitro and vivo. Int. J. Mol. Sci. 2021, 23, 15. [Google Scholar] [CrossRef]
- Chen, Y.C.; Yang, Y.; Zhang, C.; Chen, H.Y.; Chen, F.; Wang, K.J. A novel antimicrobial peptide Sparamosin26–54 from the mud crab Scylla paramamosain showing potent antifungal activity against Cryptococcus neoformans. Front. Microbiol. 2021, 12, 746006. [Google Scholar] [CrossRef]
- Chen, B.; Fan, D.Q.; Zhu, K.X.; Shan, Z.G.; Chen, F.Y.; Hou, L.; Cai, L.; Wang, K.J. Mechanism study on a new antimicrobial peptide Sphistin derived from the N-terminus of crab histone H2A identified in haemolymphs of Scylla paramamosain. Fish Shellfish Immunol. 2015, 47, 833–846. [Google Scholar] [CrossRef]
- Shan, Z.G.; Zhu, K.X.; Chen, F.Y.; Liu, J.; Chen, B.; Qiao, K.; Peng, H.; Wang, K.J. In vivo activity and the transcriptional regulatory mechanism of the antimicrobial peptide SpHyastatin in Scylla paramamosain. Fish Shellfish Immunol. 2016, 59, 155–165. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, Y.; Zhu, X.W.; Liu, Y.Z.; Chen, F.Y.; Wang, K.J. A truncated peptide Sp-NPFin from the neuropeptide FII SpNPFII of Scylla paramamosain exhibiting potent antimicrobial activity. Aquaculture 2021, 533, 736145. [Google Scholar] [CrossRef]
- Lata, S.; Mishra, N.K.; Raghava, G.P.S. AntiBP2: Improved version of antibacterial peptide prediction. BMC Bioinform. 2010, 11, S19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, S.; Karnik, S.; Nilawe, P.; Jayaraman, V.K.; Idicula-Thomas, S. ClassAMP: A Prediction tool for classification of antimicrobial peptides. IEEE ACM Trans. Comput. Biol. Bioinform. 2012, 9, 1535–1538. [Google Scholar] [CrossRef]
- Wang, G.S.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fjell, C.D.; Hancock, R.E.W.; Cherkasov, A. AMPer: A database and an automated discovery tool for antimicrobial peptides. Bioinformatics 2007, 23, 1148–1155. [Google Scholar] [CrossRef] [PubMed]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Z.; Vasil, A.I.; Hale, J.D.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic alpha-helical cationic antimicrobial peptides. Biopolymers 2008, 90, 369–383. [Google Scholar] [CrossRef]
- Jiang, Z.; Mant, C.T.; Vasil, M.; Hodges, R.S. Role of positively charged residues on the polar and non-polar faces of amphipathic alpha-helical antimicrobial peptides on specificity and selectivity for Gram-negative pathogens. Chem. Biol. Drug Des. 2018, 91, 75–92. [Google Scholar] [CrossRef]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J. Biol. Chem. 2005, 280, 12316–12329. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Chen, Y.; Song, Z.; Tan, Z.; Cheng, J. Recent advances in design of antimicrobial peptides and polypeptides toward clinical translation. Adv. Drug Deliv. Rev. 2021, 170, 261–280. [Google Scholar] [CrossRef]
- Mant, C.T.; Jiang, Z.; Gera, L.; Davis, T.; Nelson, K.L.; Bevers, S.; Hodges, R.S. De novo designed amphipathic alpha-helical antimicrobial peptides incorporating Dab and Dap residues on the polar face to treat the Gram-negative pathogen, Acinetobacter baumannii. J. Med. Chem. 2019, 62, 3354–3366. [Google Scholar] [CrossRef] [PubMed]
- Wiradharma, N.; Liu, S.Q.; Yang, Y.Y. Branched and 4-arm starlike alpha-helical peptide structures with enhanced antimicrobial potency and selectivity. Small 2012, 8, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wang, Y.; Wang, M.; Tian, Y.; Kang, W.; Liu, H.; Wang, H.; Dou, J.; Zhou, C. Effective antimicrobial activity of Cbf-14, derived from a cathelin-like domain, against penicillin-resistant bacteria. Biomaterials 2016, 87, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Wang, J.; Gao, H.; Wang, Z.; Dong, N.; Ma, Q.; Shan, A. Antimicrobial properties and membrane-active mechanism of a potential alpha-helical antimicrobial derived from cathelicidin PMAP-36. PLoS ONE 2014, 9, e86364. [Google Scholar]
- Kwon, J.Y.; Kim, M.K.; Mereuta, L.; Seo, C.H.; Luchian, T.; Park, Y. Mechanism of action of antimicrobial peptide P5 truncations against Pseudomonas aeruginosa and Staphylococcus aureus. AMB Express 2019, 9, 122. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.R.; You, D.G.; Kim, H.K.; Sohn, J.W.; Kim, M.J.; Park, J.K.; Lee, G.Y.; Yoo, Y.D. Romo1-Derived Antimicrobial Peptide Is a New Antimicrobial Agent against Multidrug-Resistant Bacteria in a Murine Model of Sepsis. Mbio 2020, 11, e03258-19. [Google Scholar] [CrossRef] [Green Version]
- Xhindoli, D.; Pacor, S.; Benincasa, M.; Scocchi, M.; Gennaro, R.; Tossi, A. The human cathelicidin LL-37—A pore-forming antibacterial peptide and host-cell modulator. Biochim. Biophys. Acta 2016, 1858, 546–566. [Google Scholar] [CrossRef]
- Lam, S.J.; O’Brien-Simpson, N.M.; Pantarat, N.; Sulistio, A.; Wong, E.H.; Chen, Y.Y.; Lenzo, J.C.; Holden, J.A.; Blencowe, A.; Reynolds, E.C.; et al. Combating multidrug-resistant Gram-negative bacteria with structurally nanoengineered antimicrobial peptide polymers. Nat. Microbiol. 2016, 1, 16162. [Google Scholar] [CrossRef]
- Li, Y.X.; Zhong, Z.; Zhang, W.P.; Qian, P.Y. Discovery of cationic nonribosomal peptides as Gram-negative antibiotics through global genome mining. Nat. Commun. 2018, 9, 3273. [Google Scholar] [CrossRef] [Green Version]
- Tytler, E.M.; Anantharamaiah, G.M.; Walker, D.E.; Mishra, V.K.; Palgunachari, M.N.; Segrest, J.P. Molecular basis for prokaryotic specificity of magainin-induced lysis. Biochemistry 1995, 34, 4393–4401. [Google Scholar] [CrossRef]
- Narayana, J.L.; Huang, H.N.; Wu, C.J.; Chen, J.Y. Epinecidin-1 antimicrobial activity: In vitro membrane lysis and In vivo efficacy against Helicobacter pylori infection in a mouse model. Biomaterials 2015, 61, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Khara, J.S.; Lim, F.K.; Wang, Y.; Ke, X.Y.; Voo, Z.X.; Yang, Y.Y.; Lakshminarayanan, R.; Ee, P.L.R. Designing alpha-helical peptides with enhanced synergism and selectivity against Mycobacterium smegmatis: Discerning the role of hydrophobicity and helicity. Acta Biomater. 2015, 28, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Chen, F.; Chen, Y.C.; Peng, H.; Wang, K.J. The long-term effect of a nine amino-acid antimicrobial peptide AS-hepc3(48–56) against Pseudomonas aeruginosa with no detectable resistance. Front. Cell. Infect. Microbiol. 2021, 11, 752637. [Google Scholar] [CrossRef] [PubMed]
- Ezraty, B.; Gennaris, A.; Barras, F.; Collet, J.F. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017, 15, 385–396. [Google Scholar] [CrossRef]
- Rowe-Magnus, D.A.; Kao, A.Y.; Prieto, A.C.; Pu, M.; Kao, C. Cathelicidin peptides restrict bacterial growth via membrane perturbation and induction of reactive oxygen species. Mbio 2019, 10, 19. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Tailhades, J.; O’Brien-Simpson, N.M.; Separovic, F.; Otvos, L., Jr.; Hossain, M.A.; Wade, J.D. Proline-rich antimicrobial peptides: Potential therapeutics against antibiotic-resistant bacteria. Amino Acids 2014, 46, 2287–2294. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Overhage, J.; Campisano, A.; Bains, M.; Torfs, E.C.; Rehm, B.H.; Hancock, R.E. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 2008, 76, 4176–4182. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Tan, H.; Cheng, T.; Shen, H.; Shao, J.; Guo, Y.; Shi, S.; Zhang, X. Human beta-defensin 3 inhibits antibiotic-resistant Staphylococcus biofilm formation. J. Surg. Res. 2013, 183, 204–213. [Google Scholar] [CrossRef]
- Sutton, J.M.; Pritts, T.A. Human beta-defensin 3: A novel inhibitor of Staphylococcus-produced biofilm production. Commentary on “Human beta-defensin 3 inhibits antibiotic-resistant Staphylococcus biofilm formation”. J. Surg. Res. 2014, 186, 99–100. [Google Scholar] [CrossRef] [Green Version]
- Okuda, K.; Zendo, T.; Sugimoto, S.; Iwase, T.; Tajima, A.; Yamada, S.; Sonomoto, K.; Mizunoe, Y. Effects of Bacteriocins on methicillin-resistant Staphylococcus aureus biofilm. Antimicrob. Agents Chemother. 2013, 57, 5572–5579. [Google Scholar] [CrossRef] [PubMed]
- Luca, V.; Stringaro, A.; Colone, M.; Pini, A.; Mangoni, M.L. Esculentin(1–21), an amphibian skin membrane-active peptide with potent activity on both planktonic and biofilm cells of the bacterial pathogen Pseudomonas aeruginosa. Cell. Mol. Life Sci. 2013, 70, 2773–2786. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Chen, C.X.; Chen, J.X.; Zhou, S.S.; Zhao, Y.R.; Xu, M.L.; Xu, H. Dual mode of anti-biofilm action of G3 against Streptococcus mutans. ACS Appl. Mater. Inter. 2020, 12, 27866–27875. [Google Scholar] [CrossRef] [PubMed]
- Choyam, S.; Jain, P.M.; Kammara, R. Characterization of a potent new-generation antimicrobial peptide of Bacillus. Front. Microbiol. 2021, 12, 710741. [Google Scholar] [CrossRef] [PubMed]
- Vishweshwaraiah, Y.L.; Acharya, A.; Hegde, V.; Prakash, B. Rational design of hyperstable antibacterial peptides for food preservation. NPJ Sci. Food 2021, 5, 26. [Google Scholar] [CrossRef] [PubMed]
- Xian, W.; Hennefarth, M.R.; Lee, M.W.; Do, T.; Lee, E.Y.; Alexandrova, A.N.; Wong, G.C.L. Histidine-mediated Ion specific effects enable salt tolerance of a pore-forming marine antimicrobial peptide. Angew. Chem. Int. Ed. 2022, 61, e202108501. [Google Scholar] [CrossRef]
- Bowdish, D.M.E.; Davidson, D.J.; Lau, Y.E.; Lee, K.; Scott, M.G.; Hancock, R.E.W. Impact of LL-37 on anti-infective immunity. J. Leukoc. Biol. 2005, 77, 451–459. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.H.; Cho, Y.; Lehrer, R.I. Effects of pH and salinity on the antimicrobial properties of clavanins. Infect. Immun. 1997, 65, 2898–2903. [Google Scholar] [CrossRef] [Green Version]
- Goldman, M.J.; Anderson, G.M.; Stolzenberg, E.D.; Kari, U.P.; Zasloff, M.; Wilson, J.M. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell 1997, 88, 553–560. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Saravanan, R.; Kwak, S.K.; Leong, S.S.J. Biomolecular engineering of a human beta defensin model for increased salt resistance. Chem. Eng. Sci. 2013, 95, 128–137. [Google Scholar] [CrossRef]
- Lyu, Y.; Yang, Y.; Lyu, X.; Dong, N.; Shan, A. Antimicrobial activity, improved cell selectivity and mode of action of short PMAP-36-derived peptides against bacteria and Candida. Sci. Rep. 2016, 6, 27258. [Google Scholar] [CrossRef] [PubMed]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorli, L.; Luque, S.; Gomez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Ye, F.; Zhang, H.; Zhou, Y.; Li, J.; Wu, Q.; Xu, X.; Wu, Q.; Wei, B.; et al. A small-molecule inhibitor of the anthranilyl-CoA synthetase PqsA for the treatment of multidrug-resistant Pseudomonas aeruginosa. Microbiol. Spectr. 2022, 10, e0276421. [Google Scholar] [CrossRef]
- Miyajima, Y.; Hiramatsu, K.; Mizukami, E.; Morinaga, R.; Ishii, H.; Shirai, R.; Kishi, K.; Tokimatsu, I.; Saikawa, T.; Kadota, J. In vitro and in vivo potency of polymyxin B against IMP-type metallo-beta-lactamase-producing Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 2008, 32, 437–440. [Google Scholar] [CrossRef]
- Pan, C.Y.; Wu, J.L.; Hui, C.F.; Lin, C.H.; Chen, J.Y. Insights into the antibacterial and immunomodulatory functions of the antimicrobial peptide, epinecidin-1, against Vibrio vulnificus infection in zebrafish. Fish Shellfish Immunol. 2011, 31, 1019–1025. [Google Scholar] [CrossRef]
- Chen, C.; Wang, A.; Zhang, F.; Zhang, M.; Yang, H.; Li, J.; Su, P.; Chen, Y.; Yu, H.; Wang, Y. The protective effect of fish-derived cathelicidins on bacterial infections in zebrafish, Danio rerio. Fish Shellfish Immunol. 2019, 92, 519–527. [Google Scholar] [CrossRef]
- Bulet, P.; Dimarcq, J.L.; Hetru, C.; Lagueux, M.; Charlet, M.; Hegy, G.; Van Dorsselaer, A.; Hoffmann, J.A. A novel inducible antibacterial peptide of Drosophila carries an O-glycosylated substitution. J. Biol. Chem. 1993, 268, 14893–14897. [Google Scholar] [CrossRef]
- Zhou, K.; Qin, Y.; Song, Y.; Zhao, K.; Pan, W.; Nan, X.; Wang, Y.; Wang, Q.; Li, W. A novel Ig domain-containing C-type lectin triggers the intestine-hemocyte axis to regulate antibacterial immunity in crab. J. Immunol. 2022, 208, 2343–2362. [Google Scholar] [CrossRef]
- Poonam, K.; Rutusmita, M.; Neha, A.; Apurva, C.; Rashmi, G.; Partha, R.; Ramasare, P. Antifungal and anti-biofilm activity of essential oil active components against Cryptococcus neoformans and Cryptococcus laurentii. Front. Microbiol. 2017, 8, 2161. [Google Scholar]
- Berditsch, M.; Jger, T.; Strempel, N.; Schwartz, T.; Ulrich, A.S. Synergistic effect of membrane-active peptides polymyxin B and gramicidin S on multidrug-resistant strains and biofilms of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2015, 59, 5288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]







| Name | Support Vector Machine | Random Forest | Artificial Neural Network | Discriminant Analysis |
|---|---|---|---|---|
| Spampcin | 1.000 | 0.943 | AMP | 1.000 |
| Spa31 | 0.978 | 0.873 | AMP | 0.930 |
| Spa20 | 0.921 | 0.771 | AMP | 0.869 |
| Spa22 | 0.912 | 0.794 | AMP | 0.974 |
| Spa14 | 0.782 | 0.601 | AMP | 0.568 |
| Microorganism | CGMCC NO. | MIC a (μM) | MBC b/MFC c (μM) | |||
|---|---|---|---|---|---|---|
| Spa31 | Spa31 | Spa20 | Spa22 | Spa14 | ||
| Gram-positive bacteria | ||||||
| Staphylococcus aureus | 1.2465 | 1.5–3 | 3–6 | 12–24 | 6–12 | >96 |
| Listeria monocytogenes | 1.10753 | 1.5–3 | 1.5–3 | 24–48 | 3–6 | 12–24 |
| Enterococcus faecalis | 1.2135 | 1.5–3 | 1.5–3 | >96 | 3–6 | >96 |
| Enterococcus faecium | 1.131 | 0–1.5 | 1.5–3 | >96 | 3–6 | >96 |
| Staphylococcus epidermidis | 1.4260 | 3–6 | 3–6 | 6–12 | 6–12 | >96 |
| Gram-negative bacteria | ||||||
| Pseudomonas aeruginosa | 1.2421 | 1.5–3 | 3–6 | 12–24 | 6–12 | >96 |
| Escherichia coli | 1.2389 | 3–6 | 3–6 | 12–24 | 6–12 | >96 |
| Acinetobacter baumannii | 1.6769 | 3–6 | 3–6 | 3–6 | 6–12 | 48–96 |
| Fungi | ||||||
| Crytococcus neoformans | 2.1563 | 6–12 | 6–12 | 24–48 | 3–6 | 6–12 |
| Fusarium graminearum | 3.4521 | 1.5–3 | 1.5–3 | 12–24 | 12–24 | 12–24 |
| Fusarium solani | 3.5840 | 1.5–3 | 3–6 | 12–24 | 12–24 | 24–48 |
| Aspergillus niger | 3.316 | 12–24 | 12–24 | 12–24 | 12–24 | 24–48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, M.; Chen, R.; Zhang, J.; Chen, F.; Wang, K.-J. A Novel Antimicrobial Peptide Spampcin56–86 from Scylla paramamosain Exerting Rapid Bactericidal and Anti-Biofilm Activity In Vitro and Anti-Infection In Vivo. Int. J. Mol. Sci. 2022, 23, 13316. https://doi.org/10.3390/ijms232113316
Jiang M, Chen R, Zhang J, Chen F, Wang K-J. A Novel Antimicrobial Peptide Spampcin56–86 from Scylla paramamosain Exerting Rapid Bactericidal and Anti-Biofilm Activity In Vitro and Anti-Infection In Vivo. International Journal of Molecular Sciences. 2022; 23(21):13316. https://doi.org/10.3390/ijms232113316
Chicago/Turabian StyleJiang, Manyu, Roushi Chen, Jingrong Zhang, Fangyi Chen, and Ke-Jian Wang. 2022. "A Novel Antimicrobial Peptide Spampcin56–86 from Scylla paramamosain Exerting Rapid Bactericidal and Anti-Biofilm Activity In Vitro and Anti-Infection In Vivo" International Journal of Molecular Sciences 23, no. 21: 13316. https://doi.org/10.3390/ijms232113316





