Whole Genome Analyses Accurately Identify Neisseria spp. and Limit Taxonomic Ambiguity
Abstract
:1. Introduction
2. Results
2.1. Species Identification
2.2. Genome Sequencing and Genome Properties of the Lebanese Isolates
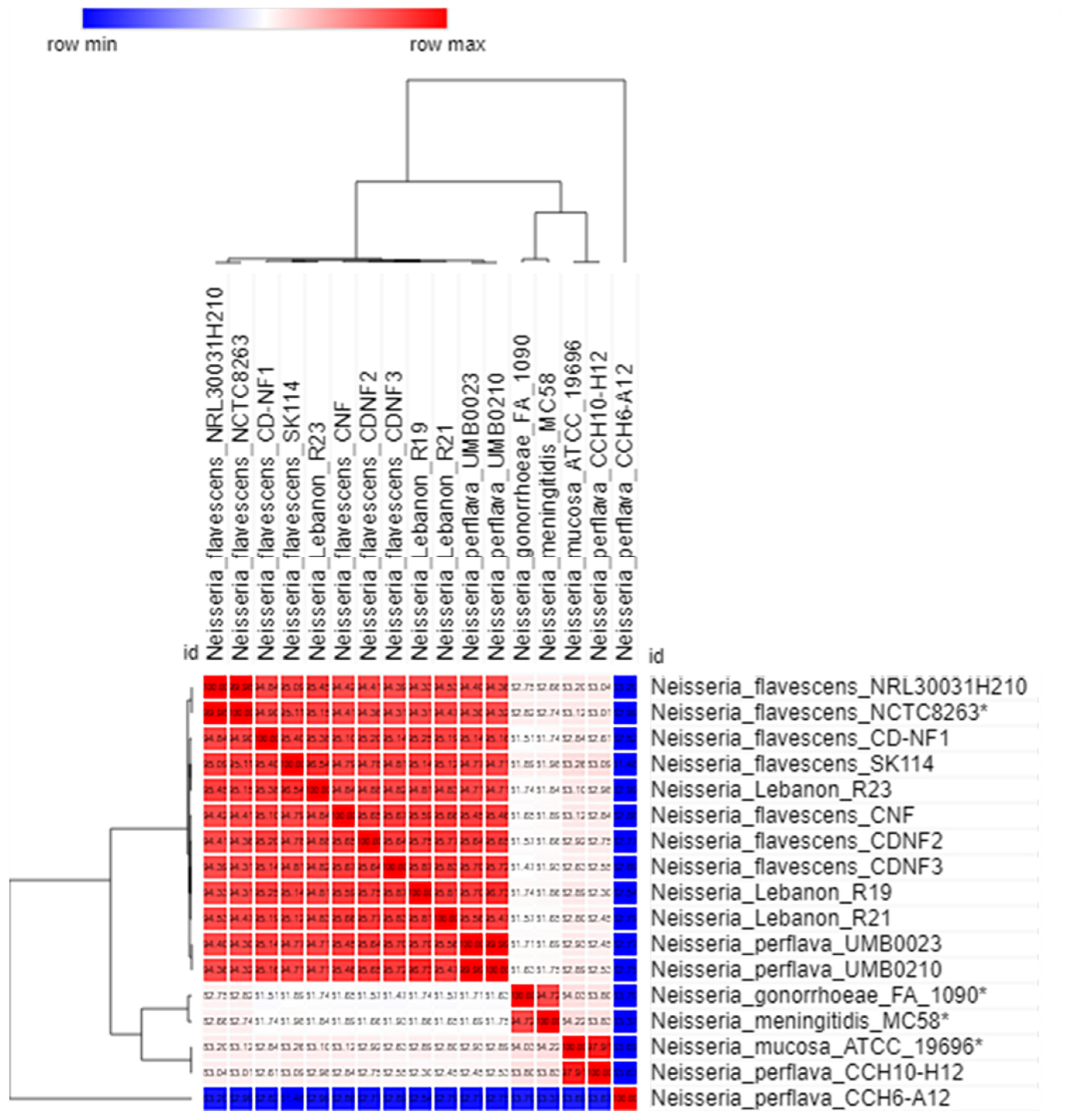
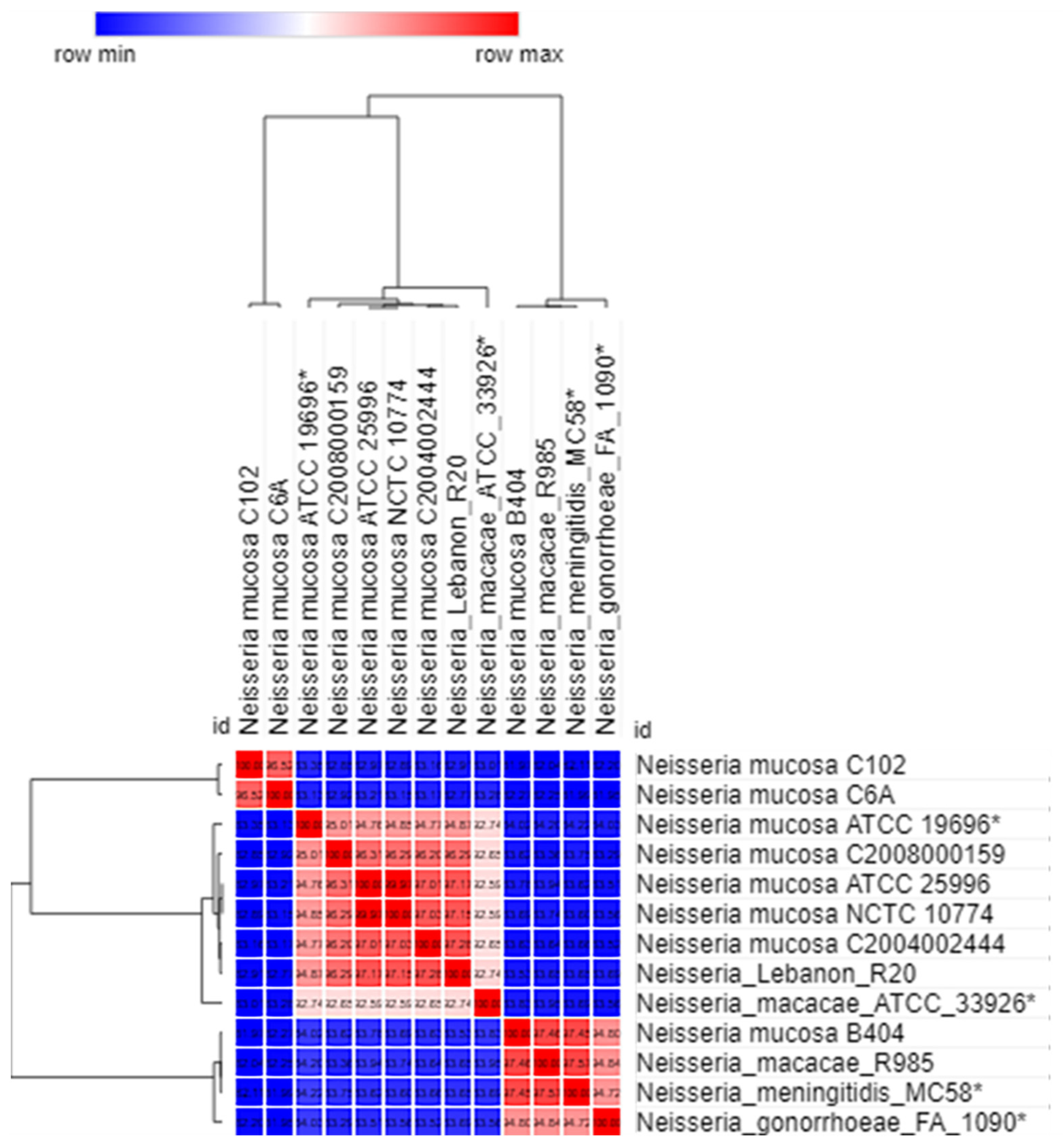
2.3. Genome Comparison between the Lebanese Isolates and Other Neisseria Strains from the NCBI GenBank Database
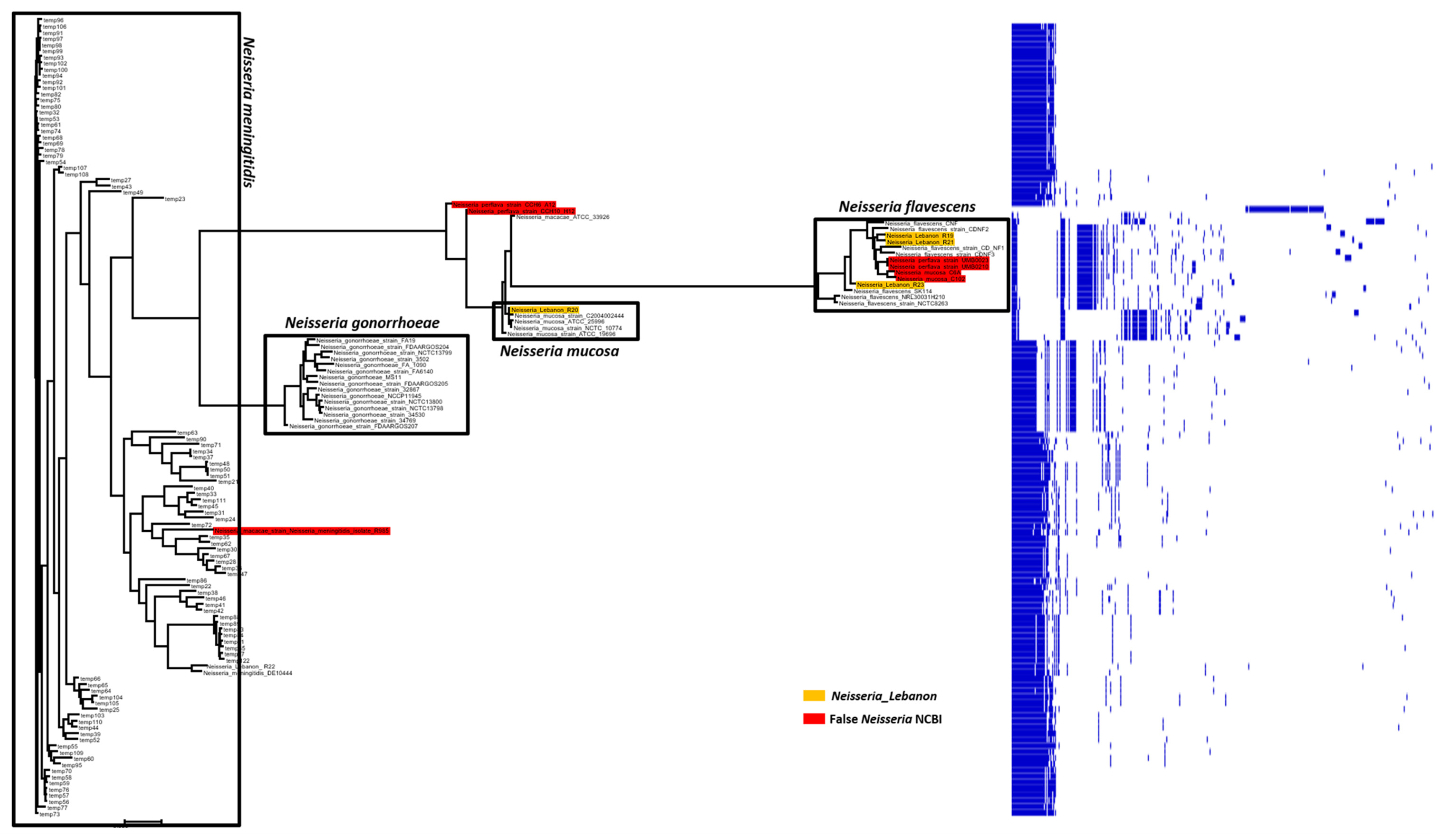
2.4. Pangenome and Phylogenetic Analysis of the Lebanese Isolates with Other Neisseria Strains Available in NCBI GenBank Database
3. Discussion
4. Materials and Methods
4.1. Isolation of Strains
4.2. Genomic DNA Preparation and Genome Sequencing
4.3. Genome Annotation and Genome Comparisons
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdallah, R.A.; Beye, M.; Diop, A.; Bakour, S.; Raoult, D.; Fournier, P.-E. The impact of culturomics on taxonomy in clinical microbiology. Antonie Van Leeuwenhoek 2017, 110, 1327–1337. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [Green Version]
- Janda, J.M.; Abbott, S.L. 16S rRNA Gene Sequencing for Bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi-Tamisier, M.; Benamar, S.; Raoult, D.; Fournier, P.E. Cautionary tale of using 16S rRNA gene sequence similarity values in identification of human-associated bacterial species. Int. J. Syst. Evol. Microbiol. 2015, 65 Pt 6, 1929–1934. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, D.; Mishra, A.K.; Lagier, J.C.; Padhmanabhan, R.; Rossi, M.; Sentausa, E.; Raoult, D.; Fournier, P.E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 2, 384–391. [Google Scholar] [CrossRef]
- Gupta, R.S. Impact of genomics on the understanding of microbial evolution and classification: The importance of Darwin’s views on classification. FEMS Microbiol. Rev. 2016, 40, 520–553. [Google Scholar] [CrossRef] [PubMed]
- Raven, K.E.; Girgis, S.T.; Akram, A.; Blane, B.; Leek, D.; Brown, N.; Peacock, S.J. A common protocol for the simultaneous processing of multiple clinically relevant bacterial species for whole genome sequencing. Sci. Rep. 2021, 11, 193. [Google Scholar] [CrossRef]
- Uelze, L.; Grützke, J.; Borowiak, M.; Hammerl, J.A.; Juraschek, K.; Deneke, C.; Tausch, S.H.; Malorny, B. Typing methods based on whole genome sequencing data. One Health Outlook 2020, 2, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quainoo, S.; Coolen, J.P.M.; van Hijum, S.; Huynen, M.A.; Melchers, W.J.G.; van Schaik, W.; Wertheim, H.F.L. Whole-genome sequencing of bacterial pathogens: The future of nosocomial outbreak analysis. Clin. Microbiol. Rev. 2017, 30, 1015–1063. [Google Scholar] [CrossRef] [Green Version]
- Bennett, J.S.; Jolley, K.A.; Earle, S.G.; Corton, C.; Bentley, S.D.; Parkhill, J.; Maiden, M.C.J. A genomic approach to bacterial taxonomy: An examination and proposed reclassification of species within the genus Neisseria. Microbiology 2012, 158 Pt 6, 1570–1580. [Google Scholar] [CrossRef]
- Harrison, O.B.; Schoen, C.; Retchless, A.C.; Wang, X.; Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Neisseria genomics: Current status and future perspectives. Pathog. Dis. 2017, 75, ftx060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoder, M.; Osman, M.; Diene, S.M.; Okdah, L.; Lalaoui, R.; Al Achkar, M.; Mallat, H.; Hamze, M.; Rolain, J.M. Evaluation of different testing tools for the identification of non-gonococcal Neisseria spp. isolated from Lebanese male semen: A strong and significant association with infertility. J. Med. Microbiol. 2019, 68, 1012–1020. [Google Scholar] [CrossRef] [PubMed]
- Khoder, M.; Rafei, R.; Osman, M.; Kassem, I.I.; Shahin, A.; Hamze, M.; Rolain, J.-M. Emergence of a Neisseria flavescens clinical strain with a high level of third-generation cephalosporins resistance in Lebanon. Diagn. Microbiol. Infect. Dis. 2022, 103, 115660. [Google Scholar] [CrossRef] [PubMed]
- Humbert, M.V.; Christodoulides, M. Atypical, yet not infrequent, infections with Neisseria species. Pathogens 2019, 9, 10. [Google Scholar] [CrossRef] [Green Version]
- Weyand, N.J. Neisseria models of infection and persistence in the upper respiratory tract. Pathog. Dis. 2017, 75, ftx031. [Google Scholar] [CrossRef] [PubMed]
- Morel, F.; Jacquier, H.; Desroches, M.; Fihman, V.; Kumanski, S.; Cambau, E.; Decousser, J.-W.; Berçot, B. Use of Andromas and Bruker MALDI-TOF MS in the identification of Neisseria. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2273–2277. [Google Scholar] [CrossRef]
- Ilina, E.N.; Borovskaya, A.D.; Malakhova, M.M.; Vereshchagin, V.A.; Kubanova, A.A.; Kruglov, A.N.; Svistunova, T.S.; Gazarian, A.O.; Maier, T.; Kostrzewa, M.; et al. Direct bacterial profiling by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry for identification of pathogenic Neisseria. J. Mol. Diagn. 2009, 11, 75–86. [Google Scholar] [CrossRef] [Green Version]
- Kawahara-Matsumizu, M.; Yamagishi, Y.; Mikamo, H. Misidentification of Neisseria cinerea as Neisseria meningitidis by Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS). Jpn. J. Infect. Dis. 2018, 71, 85–87. [Google Scholar] [CrossRef] [Green Version]
- Laumen, J.G.E.; Van Dijck, C.; Abdellati, S.; De Baetselier, I.; Serrano, G.; Manoharan-Basil, S.S.; Bottieau, E.; Martiny, D.; Kenyon, C. Antimicrobial susceptibility of commensal Neisseria in a general population and men who have sex with men in Belgium. Sci. Rep. 2022, 12, 9. [Google Scholar] [CrossRef]
- Khachatryan, L.; de Leeuw, R.H.; Kraakman, M.E.M.; Pappas, N.; te Raa, M.; Mei, H.; de Knijff, P.; Laros, J.F.J. Taxonomic classification and abundance estimation using 16S and WGS—A comparison using controlled reference samples. Forensic Sci. Int. Genet. 2020, 46, 102257. [Google Scholar] [CrossRef]
- Shaskolskiy, B.; Kravtsov, D.; Kandinov, I.; Gorshkova, S.; Kubanov, A.; Solomka, V.; Deryabin, D.; Dementieva, E.; Gryadunov, D. Comparative whole-genome analysis of Neisseria gonorrhoeae isolates revealed changes in the gonococcal genetic island and specific genes as a link to antimicrobial resistance. Front. Cell. Infect. Microbiol. 2022, 12, 831336. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Busó, L.; Yeats, C.A.; Taylor, B.; Goater, R.J.; Underwood, A.; Abudahab, K.; Argimón, S.; Ma, K.C.; Mortimer, T.D.; Golparian, D.; et al. A community-driven resource for genomic epidemiology and antimicrobial resistance prediction of Neisseria gonorrhoeae at Pathogenwatch. Genome Med. 2021, 13, 61. [Google Scholar] [CrossRef] [PubMed]
- Honskus, M.; Okonji, Z.; Musilek, M.; Krizova, P. Whole genome sequencing of Neisseria meningitidis Y isolates collected in the Czech Republic in 1993–2018. PLoS ONE 2022, 17, e0265066. [Google Scholar] [CrossRef] [PubMed]
- Whaley, M.J.; Joseph, S.J.; Retchless, A.C.; Kretz, C.B.; Blain, A.; Hu, F.; Chang, H.Y.; Mbaeyi, S.A.; MacNeil, J.R.; Read, T.D.; et al. Whole genome sequencing for investigations of meningococcal outbreaks in the United States: A retrospective analysis. Sci. Rep. 2018, 8, 15803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Busó, L.; Cole, M.J.; Spiteri, G.; Day, M.; Jacobsson, S.; Golparian, D.; Sajedi, N.; Yeats, C.A.; Abudahab, K.; Underwood, A.; et al. Europe-wide expansion and eradication of multidrug-resistant Neisseria gonorrhoeae lineages: A genomic surveillance study. Lancet Microbe 2022, 3, e452–e463. [Google Scholar] [CrossRef]
- Peng, J.-P.; Yin, Y.-P.; Chen, S.-C.; Yang, J.; Dai, X.-Q.; Zheng, H.-P.; Gu, W.-M.; Zhu, B.-Y.; Yong, G.; Zhong, N.; et al. A Whole-genome sequencing analysis of Neisseria gonorrhoeae isolates in China: An observational study. eClinicalMedicine 2019, 7, 47–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Block, T.; Laumen, J.G.E.; Van Dijck, C.; Abdellati, S.; De Baetselier, I.; Manoharan-Basil, S.S.; Van den Bossche, D.; Kenyon, C. WGS of commensal Neisseria reveals acquisition of a new ribosomal protection protein (MsrD) as a possible explanation for high level azithromycin resistance in Belgium. Pathogens 2021, 10, 384. [Google Scholar] [CrossRef] [PubMed]
- Hanage, W.P.; Fraser, C.; Spratt, B.G. Fuzzy species among recombinogenic bacteria. BMC Biol. 2005, 3, 6. [Google Scholar] [CrossRef] [Green Version]
- Bennett, J.S.; Jolley, K.A.; Maiden, M.C.J. Genome sequence analyses show that Neisseria oralis is the same species as ‘Neisseria mucosa var. heidelbergensis’. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 10, 3920–3926. [Google Scholar] [CrossRef]
- Bennett, J.S.; Watkins, E.R.; Jolley, K.A.; Harrison, O.B.; Maiden, M.C. Identifying Neisseria species by use of the 50S ribosomal protein L6 (rplF) gene. J. Clin. Microbiol. 2014, 52, 1375–1381. [Google Scholar] [CrossRef]
- Caputo, A.; Merhej, V.; Georgiades, K.; Fournier, P.E.; Croce, O.; Robert, C.; Raoult, D. Pan-genomic analysis to redefine species and subspecies based on quantum discontinuous variation: The Klebsiella paradigm. Biol. Direct. 2015, 10, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walcher, M.; Skvoretz, R.; Montgomery-Fullerton, M.; Jonas, V.; Brentano, S. Description of an unusual Neisseria meningitidis isolate containing and expressing Neisseria gonorrhoeae-Specific 16S rRNA gene sequences. J. Clin. Microbiol. 2013, 51, 3199–3206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2014, 31, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
| Genome | Isolate | Species | Isolation Date | Genes (N) | CDS (N) | RNA (N) |
|---|---|---|---|---|---|---|
| R19 | CMUL013 | N. flavescens | 2014 | 2160 | 2091 | tmRNA: 2 rRNA: 2 tRNA: 54 misc_RNA: 11 |
| R20 | CMUL032 | N. mucosa | 2015 | 2358 | 2288 | tmRNA: 1 tRNA: 54 rRNA: 2 misc_RNA: 13 |
| R21 | CMUL057 | N. flavescens | 2016 | 2207 | 2121 | tmRNA: 1 tRNA: 55 rRNA: 2 misc_RNA: 28 |
| R23 | CMUL078 | N. flavescens | 2017 | 2206 | 2100 | tmRNA: 1 tRNA: 52 rRNA: 3 misc_RNA: 50 |
| Genome | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Neisseria_flavescens_NRL30031H210 | 100 | ||||||||||||||
| 2 | Neisseria_flavescens_SK114 | 60.6 | 100 | |||||||||||||
| 3 | Neisseria_flavescens_CD-NF1 | 59.4 | 62.9 | 100 | ||||||||||||
| 4 | Neisseria_flavssescens_CDNF2 | 57.4 | 59.6 | 61.4 | 100 | |||||||||||
| 5 | Neisseria_flavescens_CDNF3 | 57.2 | 59.8 | 60.8 | 64.9 | 100 | ||||||||||
| 6 | Neisseria_ flavescens_CNF | 57 | 59.3 | 60.9 | 63.7 | 63.3 | 100 | |||||||||
| 7 | Neisseria_flavescens_NCTC8263 * | 93.1 | 60.6 | 59.4 | 57.4 | 57.1 | 57.1 | 100 | ||||||||
| 8 | Neisseria_gonorrhoeae_FA_1090 * | 31.9 | 30.3 | 29.7 | 29.5 | 29.4 | 29.8 | 32 | 100 | |||||||
| 9 | Neisseria_meningitidis_MC58 * | 33.6 | 31.7 | 31 | 30.5 | 31.4 | 31.5 | 33.8 | 57.6 | 100 | ||||||
| 10 | Neisseria_mucosa_ATCC_19696 * | 30.2 | 31.5 | 29.1 | 29.9 | 29.2 | 29.6 | 30.6 | 33.5 | 35 | 100 | |||||
| 11 | Neisseria_perflava_CCH6-A12 | 14.5 | 14.7 | 14.8 | 14.7 | 0 | 14.5 | 14.5 | 15.6 | 0 | 13.6 | 100 | ||||
| 12 | Neisseria_perflava_CCH10-H12 | 29.8 | 30.7 | 29.5 | 28.9 | 29.1 | 29.2 | 30.2 | 33.3 | 34.8 | 80.7 | 14.9 | 100 | |||
| 13 | Neisseria_perflava_UMB0023 | 56.4 | 58.8 | 60.6 | 63.6 | 64.2 | 62.2 | 56.5 | 30 | 30.6 | 29.3 | 14.5 | 29.1 | 100 | ||
| 14 | Neisseria_perflava_UMB0210 | 56.5 | 58.9 | 60.6 | 63.6 | 64.2 | 62.2 | 56.5 | 30 | 30.6 | 29.3 | 14.5 | 29 | 99.2 | 100 | |
| 15 | Neisseria_Lebanon_R19 | 56.6 | 59.1 | 61.6 | 64.9 | 65.7 | 63.6 | 56.6 | 29.7 | 30.9 | 28.9 | 16.4 | 29.3 | 64.2 | 64.2 | 100 |
| 15 | Neisseria_Lebanon_R21 | 57.1 | 59.1 | 60.5 | 64.4 | 65.1 | 63.3 | 57.1 | 29.9 | 31.2 | 29.3 | 0 | 29.4 | 62.6 | 62.6 | 100 |
| 15 | Neisseria_Lebanon_R23 | 60.8 | 69.9 | 62.6 | 59.8 | 59.5 | 58.7 | 60.8 | 30.1 | 31.6 | 31.2 | 0 | 30.7 | 58.2 | 58.3 | 100 |
| Genome | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Neisseria_mucosa_C102 | 100 | |||||||||||||
| 2 | Neisseria_mucosa_ATCC_19696 * | 29.5 | 100 | ||||||||||||
| 3 | Neisseria_mucosa_ATCC_25996 | 29 | 58.4 | 100 | |||||||||||
| 4 | Neisseria_mucosa_C6A | 69.4 | 30.2 | 29.4 | 100 | ||||||||||
| 5 | Neisseria_mucosa_C2004002444 | 29 | 58.7 | 75.3 | 29.3 | 100 | |||||||||
| 6 | Neisseria_mucosa_C2008000159 | 44.4 | 59.5 | 67.8 | 29.2 | 67.6 | 100 | ||||||||
| 7 | Neisseria_mucosa_B404 | 28.9 | 34.6 | 34 | 30.7 | 33.9 | 33.7 | 100 | |||||||
| 8 | Neisseria_mucosa_NCTC_10774 | 29.2 | 58.4 | 89.4 | 29.6 | 74.4 | 67.5 | 34.2 | 100 | ||||||
| 9 | Neisseria_mucosa_CCH7-A10 | 28.5 | 63.4 | 0 | 27.1 | 100 | 100 | 0 | 59.7 | 100 | |||||
| 10 | Neisseria_macacae_ATCC_33926 * | 29.4 | 63.5 | 53.7 | 29.7 | 54 | 54.5 | 34.3 | 53.9 | 52.6 | 100 | ||||
| 11 | Neisseria_macacae_R985 | 30.5 | 34.6 | 33.9 | 30.6 | 34.1 | 33.4 | 76.6 | 34 | 0 | 33.9 | 100 | |||
| 12 | Neisseria_meningitidis_MC58 * | 30.9 | 35 | 34.3 | 31.1 | 34.6 | 34.1 | 76.3 | 34.5 | 35.8 | 34.2 | 78 | 100 | ||
| 13 | Neisseria_gonorrhoeae_FA_1090 * | 30 | 33.5 | 32.9 | 30 | 32.9 | 33.2 | 58.2 | 33.1 | 33.9 | 33.4 | 58.4 | 57.6 | 100 | |
| 14 | Neisseria_Lebanon_R20 | 28.8 | 58.9 | 74.3 | 29.2 | 76.4 | 68 | 33.6 | 74.3 | 60.5 | 54.2 | 33.6 | 34.2 | 32.8 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khoder, M.; Osman, M.; Kassem, I.I.; Rafei, R.; Shahin, A.; Fournier, P.E.; Rolain, J.-M.; Hamze, M. Whole Genome Analyses Accurately Identify Neisseria spp. and Limit Taxonomic Ambiguity. Int. J. Mol. Sci. 2022, 23, 13456. https://doi.org/10.3390/ijms232113456
Khoder M, Osman M, Kassem II, Rafei R, Shahin A, Fournier PE, Rolain J-M, Hamze M. Whole Genome Analyses Accurately Identify Neisseria spp. and Limit Taxonomic Ambiguity. International Journal of Molecular Sciences. 2022; 23(21):13456. https://doi.org/10.3390/ijms232113456
Chicago/Turabian StyleKhoder, May, Marwan Osman, Issmat I. Kassem, Rayane Rafei, Ahmad Shahin, Pierre Edouard Fournier, Jean-Marc Rolain, and Monzer Hamze. 2022. "Whole Genome Analyses Accurately Identify Neisseria spp. and Limit Taxonomic Ambiguity" International Journal of Molecular Sciences 23, no. 21: 13456. https://doi.org/10.3390/ijms232113456
APA StyleKhoder, M., Osman, M., Kassem, I. I., Rafei, R., Shahin, A., Fournier, P. E., Rolain, J.-M., & Hamze, M. (2022). Whole Genome Analyses Accurately Identify Neisseria spp. and Limit Taxonomic Ambiguity. International Journal of Molecular Sciences, 23(21), 13456. https://doi.org/10.3390/ijms232113456








