mRNA Localization to the Endoplasmic Reticulum in Plant Endosperm Cells
Abstract
:1. Introduction
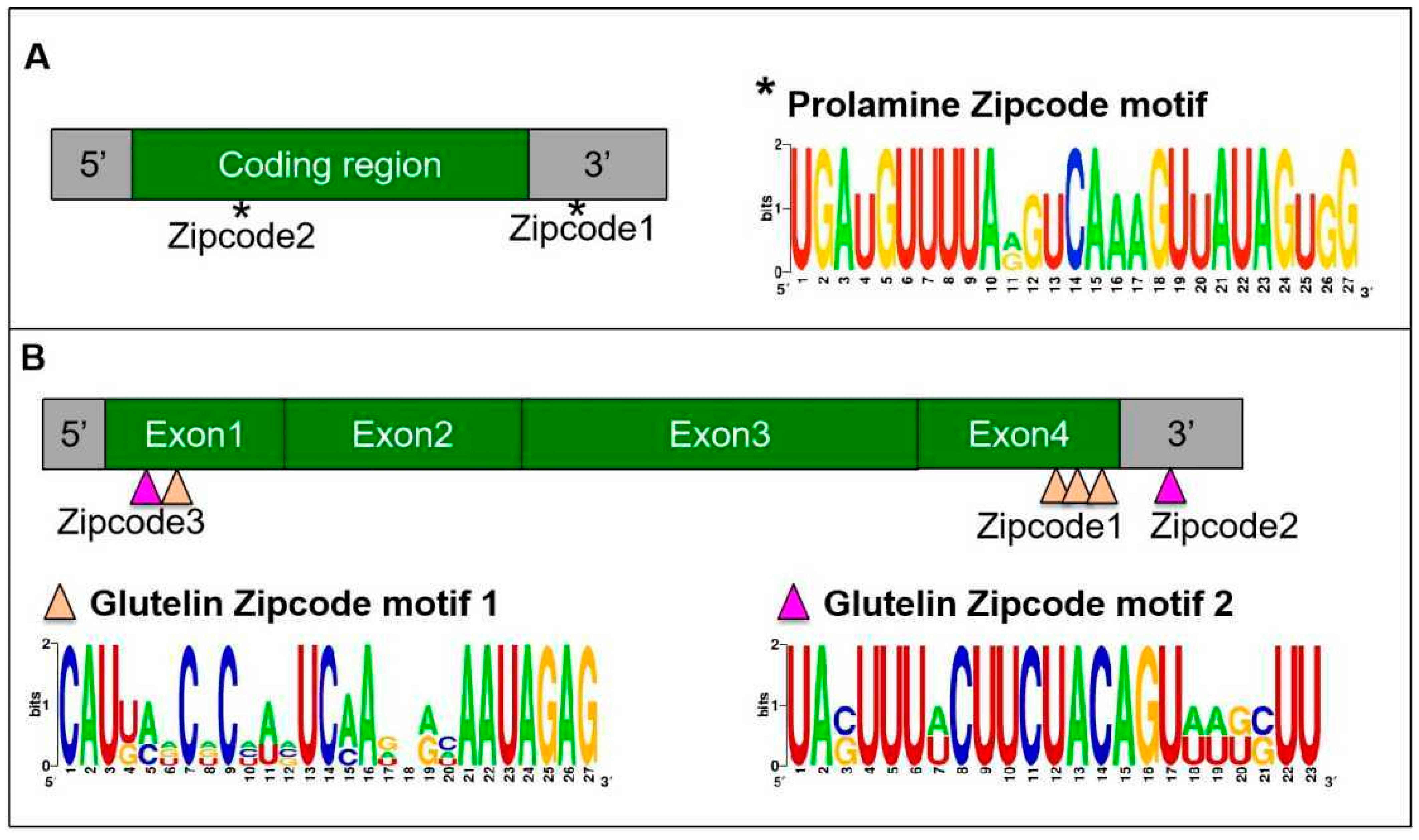
2. mRNA Targeting to the ER Subdomains Is Driven by Specific RNA Zipcodes
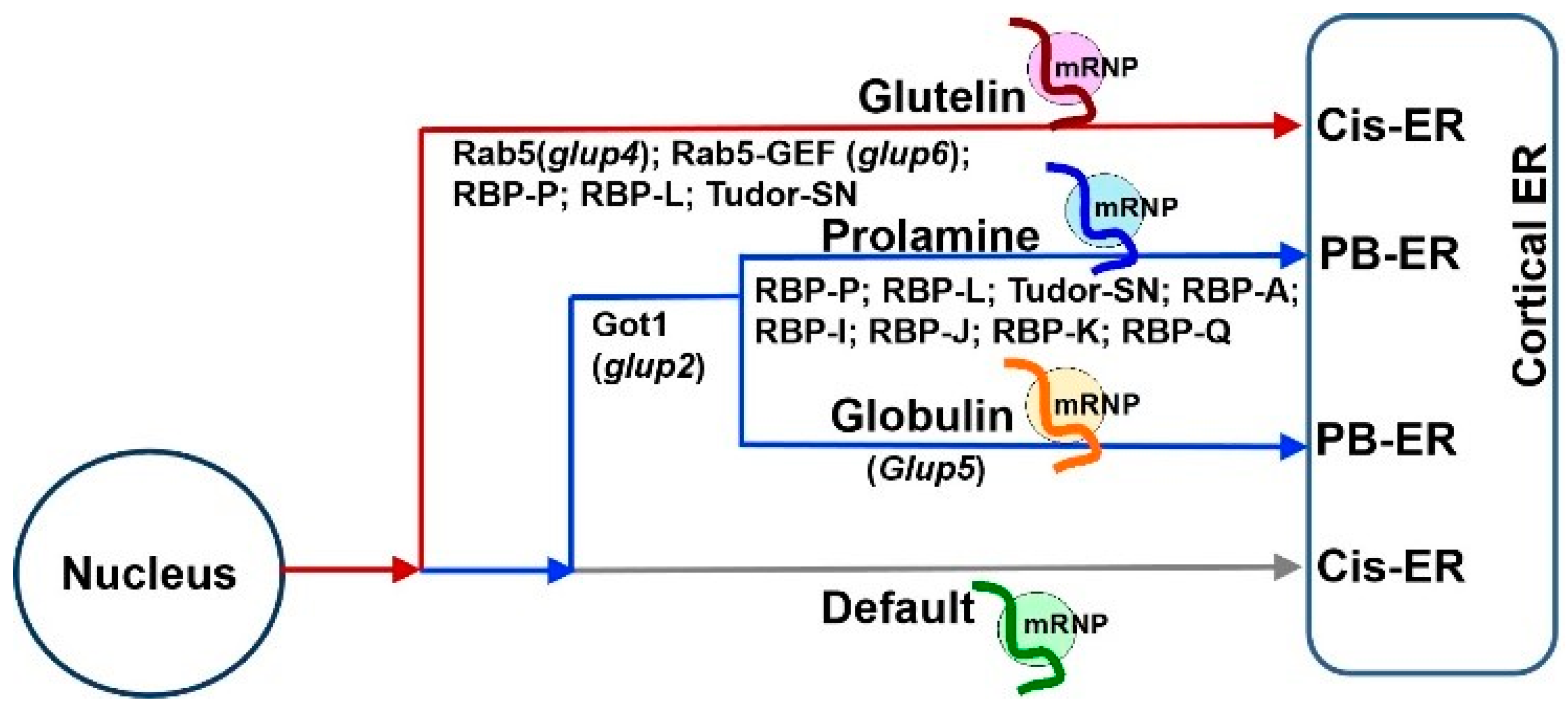
3. mRNA Targeting to the ER Subdomains Requires a Set of Trans-Acting RBPs
4. A Possible Role for Myosin Motor Protein Driving mRNA Transport to the ER Subdomains on Actin Filaments
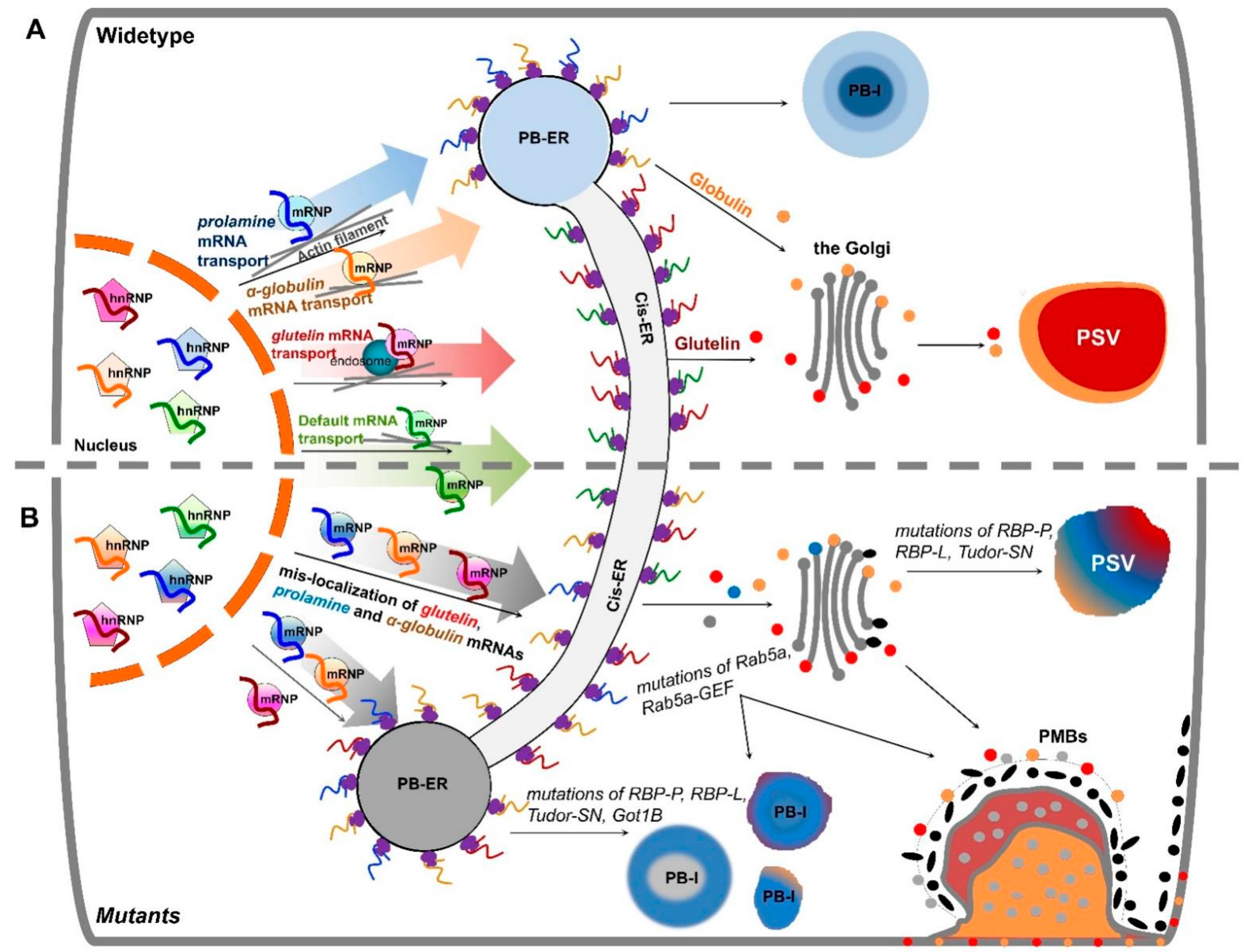
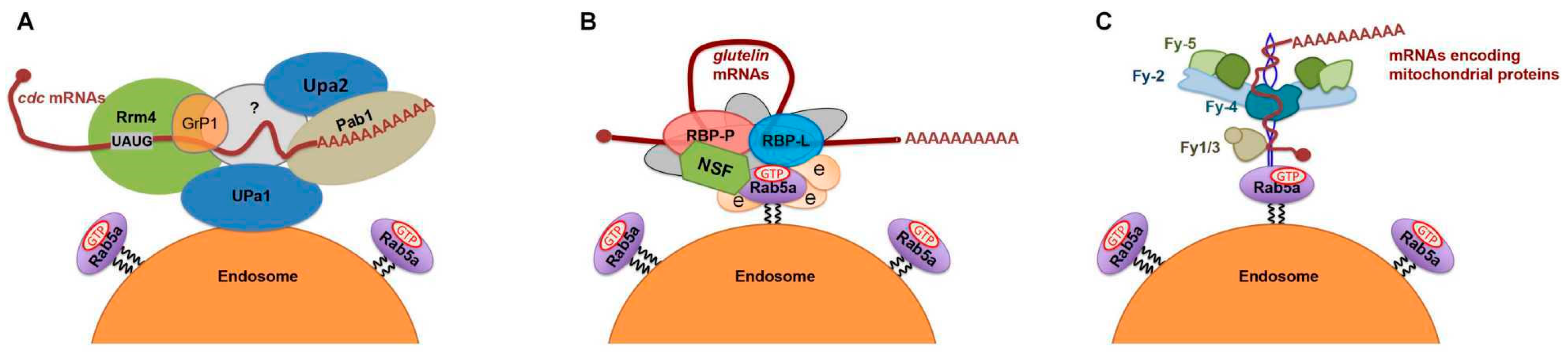
5. mRNA Transport to the ER Subdomains Meets Membrane Trafficking
6. mRNA Localization Plays a Determinant Role in Storage Organelle Biogenesis in Cereal Grains
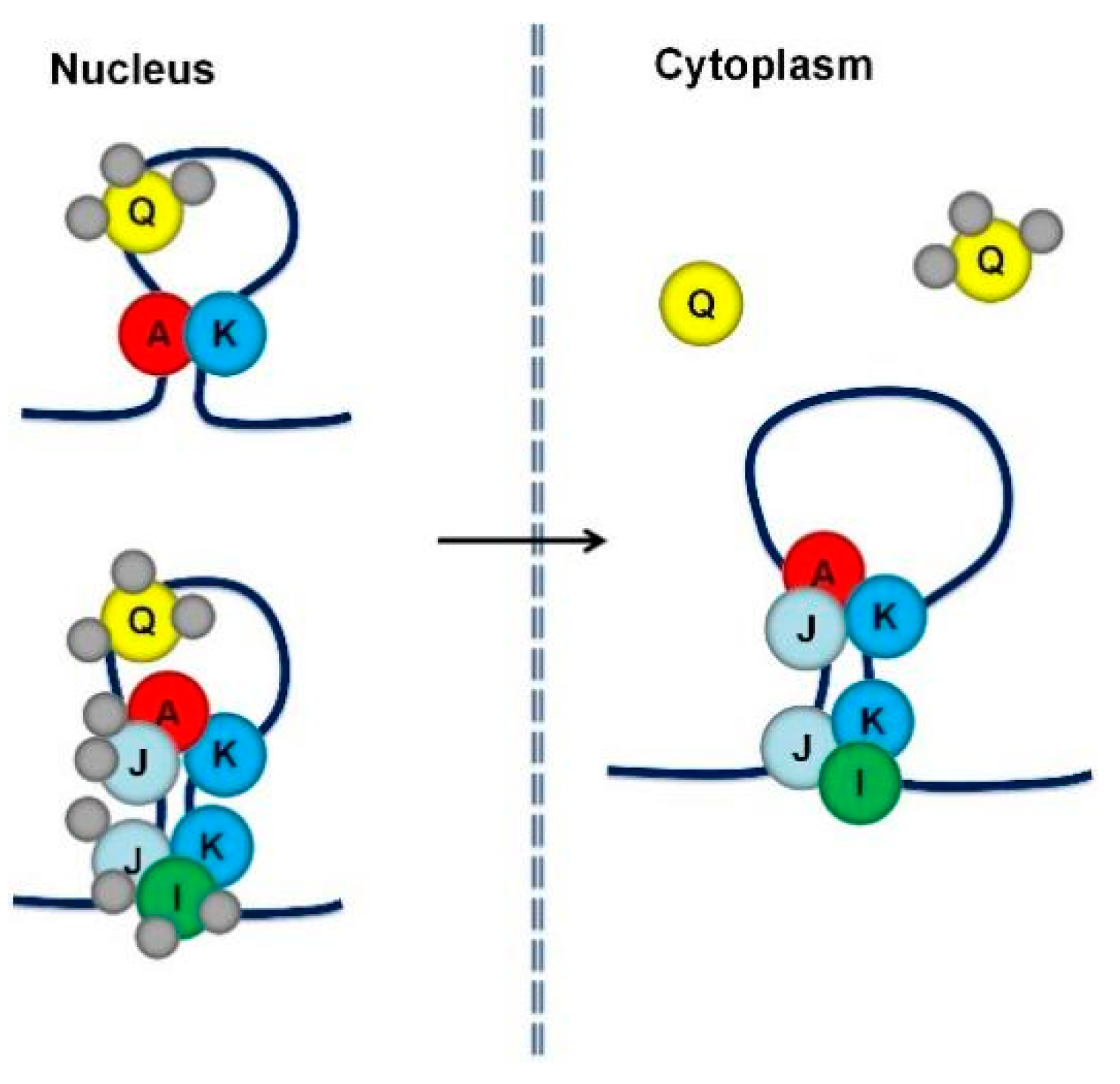
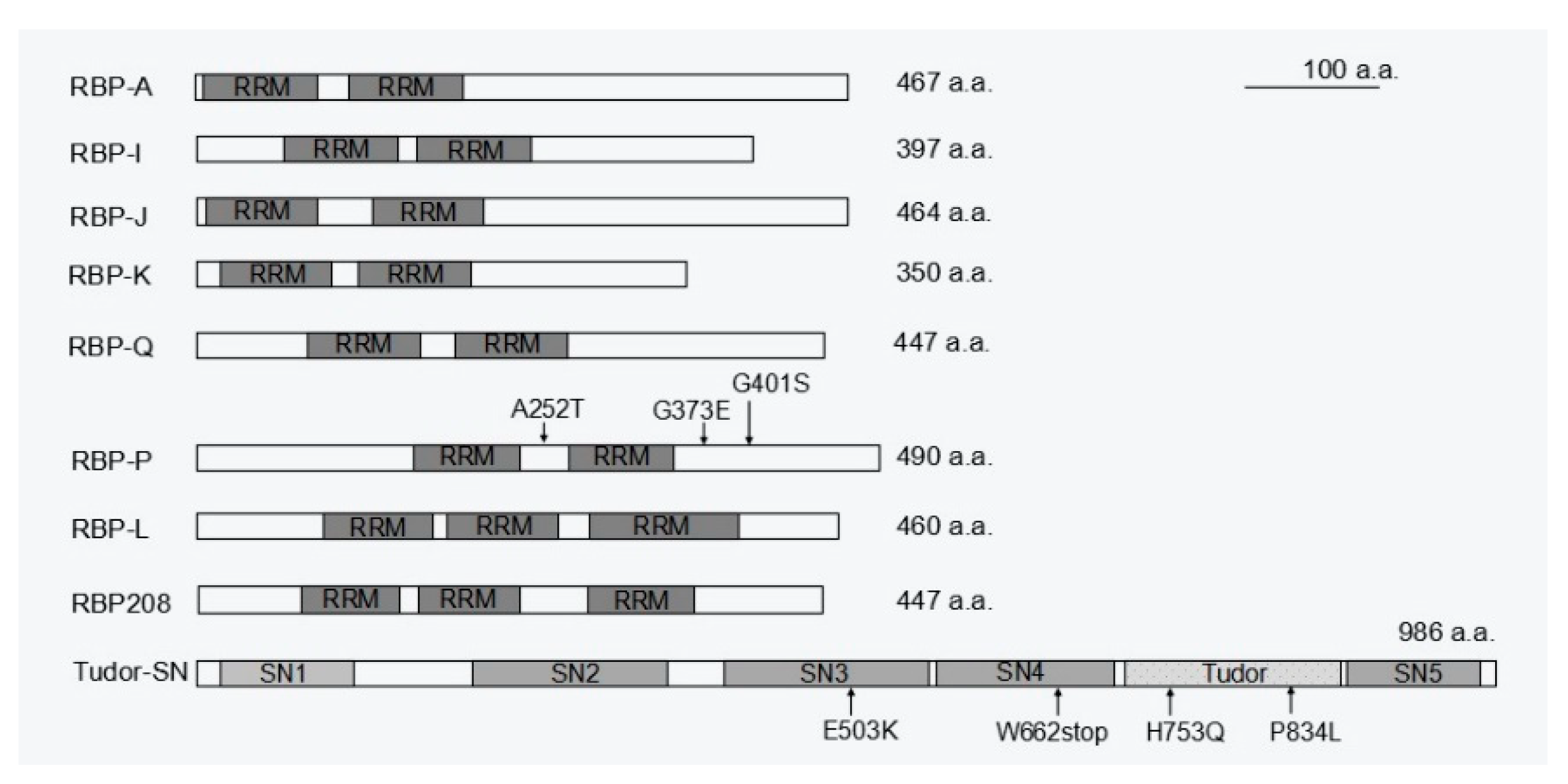
7. Accessory RBPs Is Required for mRNA Localization as well as Other Functions
8. Transport of mRNAs Is Highly Selective and as Large Regulons
9. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jeffery, W.R.; Tomlinson, C.R.; Brodeur, R.D. Localization of actin messenger RNA during early ascidian development. Dev. Biol. 1983, 99, 408–417. [Google Scholar] [CrossRef]
- Jeffery, W.R. Requirement of cell division for muscle actin expression in the primary muscle cell lineage of ascidian embryos. Development 1989, 105, 75–84. [Google Scholar] [CrossRef]
- Berleth, T.; Burri, M.; Thoma, G.; Bopp, D.; Richstein, S.; Frigerio, G.; Noll, M.; Nusslein-Volhard, C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988, 7, 1749–1756. [Google Scholar] [CrossRef]
- Frigerio, G.; Burri, M.; Bopp, D.; Baumgartner, S.; Noll, M. Structure of the segmentation gene paired and the Drosophila PRD gene set as part of a gene network. Cell 1986, 47, 735–746. [Google Scholar] [CrossRef]
- Rebagliati, M.R.; Weeks, D.L.; Harvey, R.P.; Melton, D.A. Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell 1985, 42, 769–777. [Google Scholar] [CrossRef]
- Kandler-Singer, I.; Kalthoff, K. RNase sensitivity of an anterior morphogenetic determinant in an insect egg (Smittia sp., Chironomidae, Diptera). Proc. Natl. Acad. Sci. USA 1976, 73, 3739–3743. [Google Scholar] [CrossRef] [Green Version]
- Davidson, E.H. Note on the control of gene expression during development. J. Biol. 1971, 32, 123–130. [Google Scholar] [CrossRef]
- Lawrence, J.B.; Singer, R.H. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 1986, 45, 407–415. [Google Scholar] [CrossRef]
- Garner, C.C.; Tucker, R.P.; Matus, A. Selective localization of messenger RNA for cytoskeletal protein MAP2 in dendrites. Nature 1988, 336, 674–677. [Google Scholar] [CrossRef]
- Li, X.; Franceschi, V.R.; Okita, T.W. Segregation of storage protein mRNAs on the rough endoplasmic reticulum membranes of rice endosperm cells. Cell 1993, 72, 869–879. [Google Scholar] [CrossRef]
- Trapp, B.D.; Nishiyama, A.; Cheng, D.; Macklin, W. Differentiation and death of premyelinating oligodendrocytes in developing rodent brain. J. Cell Biol. 1997, 137, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Buskila, A.A.; Kannaiah, S.; Amster-Choder, O. RNA localization in bacteria. RNA Biol. 2014, 11, 1051–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, L.; Okita, T.W. mRNA-based protein targeting to the endoplasmic reticulum and chloroplasts in plant cells. Curr. Opin. Plant. Biol. 2014, 22, 77–85. [Google Scholar] [CrossRef]
- Margeot, A.; Garcia, M.; Wang, W.; Tetaud, E.; di Rago, J.P.; Jacq, C. Why are many mRNAs translated to the vicinity of mitochondria: A role in protein complex assembly? Gene 2005, 354, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Michaud, M.; Ubrig, E.; Filleur, S.; Erhardt, M.; Ephritikhine, G.; Marechal-Drouard, L.; Duchene, A.M. Differential targeting of VDAC3 mRNA isoforms influences mitochondria morphology. Proc. Natl. Acad. Sci. USA 2014, 111, 8991–8996. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, A.U.; Fisher, P.R. Import of nuclear-encoded mitochondrial proteins: A cotranslational perspective. Int. Rev. Cell Mol. Biol. 2009, 273, 49–68. [Google Scholar]
- Vincent, T.; Vingadassalon, A.; Ubrig, E.; Azeredo, K.; Srour, O.; Cognat, V.; Graindorge, S.; Salinas, T.; Marechal-Drouard, L.; Duchene, A.M. A genome-scale analysis of mRNAs targeting to plant mitochondria: Upstream AUGs in 5′ untranslated regions reduce mitochondrial association. Plant J. 2017, 92, 1132–1142. [Google Scholar] [CrossRef] [Green Version]
- Moriceau, L.; Jomat, L.; Bressanelli, S.; Alcaide-Loridan, C.; Jupin, I. Identification and Molecular Characterization of the Chloroplast Targeting Domain of Turnip yellow mosaic virus Replication Proteins. Front. Plant Sci. 2017, 8, 2138. [Google Scholar] [CrossRef]
- Gomez, G.; Pallas, V. Noncoding RNA mediated traffic of foreign mRNA into chloroplasts reveals a novel signaling mechanism in plants. PLoS ONE 2010, 5, e12269. [Google Scholar] [CrossRef]
- Budziszewska, M.; Obrepalska-Steplowska, A. The Role of the Chloroplast in the Replication of Positive-Sense Single-Stranded Plant RNA Viruses. Front. Plant Sci. 2018, 9, 1776. [Google Scholar] [CrossRef]
- Walter, P.; Blobel, G. Translocation of proteins across the endoplasmic reticulum. Oncodev. Biol. Med. 1982, 4, 137–146. [Google Scholar] [CrossRef]
- Meyer, D.I.; Krause, E.; Dobberstein, B. Secretory protein translocation across membranes-the role of the ‘docking protein’. Nature 1982, 297, 647–650. [Google Scholar] [CrossRef] [Green Version]
- Gilmore, R.; Kellaris, K.V. Translocation of proteins across and integration of membrane proteins into the rough endoplasmic reticulum. Ann. N. Y. Acad. Sci. 1992, 674, 27–37. [Google Scholar] [CrossRef]
- Yamagata, H.; Tanaka, K. The Site of Synthesis and Accumulation of Rice Storage Proteins. Plant Cell Physiol. 1986, 27, 135–145. [Google Scholar]
- Choi, S.B.; Wang, C.; Muench, D.G.; Ozawa, K.; Franceschi, V.R.; Wu, Y.; Okita, T.W. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 2000, 407, 765–767. [Google Scholar] [CrossRef]
- Hamada, S.; Ishiyama, K.; Sakulsingharoj, C.; Choi, S.B.; Wu, Y.; Wang, C.; Singh, S.; Kawai, N.; Messing, J.; Okita, T.W. Dual regulated RNA transport pathways to the cortical region in developing rice endosperm. Plant Cell 2003, 15, 2265–2272. [Google Scholar] [CrossRef] [Green Version]
- Haag, C.; Steuten, B.; Feldbrugge, M. Membrane-Coupled mRNA Trafficking in Fungi. Annu. Rev. Microbiol. 2015, 69, 265–281. [Google Scholar] [CrossRef]
- Niessing, D.; Jansen, R.P.; Pohlmann, T.; Feldbrugge, M. mRNA transport in fungal top models. Wiley Interdiscip. Rev. RNA 2018, 9, e1453. [Google Scholar] [CrossRef]
- Schmid, M.; Jaedicke, A.; Du, T.G.; Jansen, R.P. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr. Biol. 2006, 16, 1538–1543. [Google Scholar] [CrossRef] [Green Version]
- Jansen, R.P. mRNA localization: Message on the move. Nat. Rev. Mol. Cell Biol. 2001, 2, 247–256. [Google Scholar] [CrossRef]
- Washida, H.; Kaneko, S.; Crofts, N.; Sugino, A.; Wang, C.; Okita, T.W. Identification of cis-localization elements that target glutelin RNAs to a specific subdomain of the cortical endoplasmic reticulum in rice endosperm cells. Plant Cell Physiol. 2009, 50, 1710–1714. [Google Scholar] [CrossRef] [Green Version]
- Washida, H.; Sugino, A.; Doroshenk, K.A.; Satoh-Cruz, M.; Nagamine, A.; Katsube-Tanaka, T.; Ogawa, M.; Kumamaru, T.; Satoh, H.; Okita, T.W. RNA targeting to a specific ER sub-domain is required for efficient transport and packaging of alpha-globulins to the protein storage vacuole in developing rice endosperm. Plant J. 2012, 70, 471–479. [Google Scholar] [CrossRef]
- Washida, H.; Sugino, A.; Kaneko, S.; Crofts, N.; Sakulsingharoj, C.; Kim, D.; Choi, S.B.; Hamada, S.; Ogawa, M.; Wang, C.; et al. Identification of cis-localization elements of the maize 10-kDa delta-zein and their use in targeting RNAs to specific cortical endoplasmic reticulum subdomains. Plant J. 2009, 60, 146–155. [Google Scholar] [CrossRef]
- Tian, L.; Chou, H.L.; Zhang, L.; Okita, T.W. Targeted Endoplasmic Reticulum Localization of Storage Protein mRNAs Requires the RNA-Binding Protein RBP-L. Plant Physiol. 2019, 179, 1111–1131. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Chou, H.L.; Fukuda, M.; Kumamaru, T.; Okita, T.W. mRNA Localization in Plant Cells. Plant Physiol. 2020, 182, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Crofts, A.J.; Crofts, N.; Whitelegge, J.P.; Okita, T.W. Isolation and identification of cytoskeleton-associated prolamine mRNA binding proteins from developing rice seeds. Planta 2010, 231, 1261–1276. [Google Scholar] [CrossRef]
- Yang, Y.; Crofts, A.J.; Crofts, N.; Okita, T.W. Multiple RNA binding protein complexes interact with the rice prolamine RNA cis-localization zipcode sequences. Plant Physiol. 2014, 164, 1271–1282. [Google Scholar] [CrossRef] [Green Version]
- Doroshenk, K.A.; Tian, L.; Crofts, A.J.; Kumamaru, T.; Okita, T.W. Characterization of RNA binding protein RBP-P reveals a possible role in rice glutelin gene expression and RNA localization. Plant Mol. Biol. 2014, 85, 381–394. [Google Scholar] [CrossRef]
- Tian, L.; Chou, H.L.; Zhang, L.; Hwang, S.K.; Starkenburg, S.R.; Doroshenk, K.A.; Kumamaru, T.; Okita, T.W. RNA-Binding Protein RBP-P Is Required for Glutelin and Prolamine mRNA Localization in Rice Endosperm Cells. Plant Cell 2018, 30, 2529–2552. [Google Scholar] [CrossRef] [Green Version]
- Trcek, T.; Lehmann, R. Germ granules in Drosophila. Traffic 2019, 20, 650–660. [Google Scholar] [CrossRef] [Green Version]
- Lecuyer, E.; Yoshida, H.; Parthasarathy, N.; Alm, C.; Babak, T.; Cerovina, T.; Hughes, T.R.; Tomancak, P.; Krause, H.M. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 2007, 131, 174–187. [Google Scholar] [CrossRef] [Green Version]
- Forrest, K.M.; Gavis, E.R. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol. 2003, 13, 1159–1168. [Google Scholar] [CrossRef]
- Fei, J.; Sharma, C.M. RNA Localization in Bacteria. Microbiol. Spectr. 2018, 6, RWR-0024-2018. [Google Scholar] [CrossRef]
- Edelmann, F.T.; Schlundt, A.; Heym, R.G.; Jenner, A.; Niedner-Boblenz, A.; Syed, M.I.; Paillart, J.C.; Stehle, R.; Janowski, R.; Sattler, M.; et al. Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA-transport complex. Nat. Struct. Mol. Biol. 2017, 24, 152–161. [Google Scholar] [CrossRef]
- Bohl, F.; Kruse, C.; Frank, A.; Ferring, D.; Jansen, R.P. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000, 19, 5514–5524. [Google Scholar] [CrossRef] [Green Version]
- Gonsalvez, G.B.; Little, J.L.; Long, R.M. ASH1 mRNA anchoring requires reorganization of the Myo4p-She3p-She2p transport complex. J. Biol. Chem. 2004, 279, 46286–46294. [Google Scholar] [CrossRef] [Green Version]
- Takizawa, P.A.; Vale, R.D. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. USA 2000, 97, 5273–5278. [Google Scholar] [CrossRef] [Green Version]
- Takizawa, P.A.; DeRisi, J.L.; Wilhelm, J.E.; Vale, R.D. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 2000, 290, 341–344. [Google Scholar] [CrossRef] [Green Version]
- Shepard, K.A.; Gerber, A.P.; Jambhekar, A.; Takizawa, P.A.; Brown, P.O.; Herschlag, D.; DeRisi, J.L.; Vale, R.D. Widespread cytoplasmic mRNA transport in yeast: Identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. USA 2003, 100, 11429–11434. [Google Scholar] [CrossRef] [Green Version]
- Latham, V.M.; Yu, E.H.; Tullio, A.N.; Adelstein, R.S.; Singer, R.H. A Rho-dependent signaling pathway operating through myosin localizes beta-actin mRNA in fibroblasts. Curr. Biol. 2001, 11, 1010–1016. [Google Scholar] [CrossRef] [Green Version]
- Denes, L.T.; Kelley, C.P.; Wang, E.T. Microtubule-based transport is essential to distribute RNA and nascent protein in skeletal muscle. Nat. Commun. 2021, 12, 6079. [Google Scholar] [CrossRef]
- Pichon, X.; Moissoglu, K.; Coleno, E.; Wang, T.; Imbert, A.; Robert, M.C.; Peter, M.; Chouaib, R.; Walter, T.; Mueller, F.; et al. The kinesin KIF1C transports APC-dependent mRNAs to cell protrusions. RNA 2021, 27, 1528–1544. [Google Scholar] [CrossRef]
- Katz, Z.B.; Wells, A.L.; Park, H.Y.; Wu, B.; Shenoy, S.M.; Singer, R.H. beta-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev. 2012, 26, 1885–1890. [Google Scholar] [CrossRef] [Green Version]
- Oleynikov, Y.; Singer, R.H. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr. Biol. 2003, 13, 199–207. [Google Scholar] [CrossRef] [Green Version]
- Song, T.; Zheng, Y.; Wang, Y.; Katz, Z.; Liu, X.; Chen, S.; Singer, R.H.; Gu, W. Specific interaction of KIF11 with ZBP1 regulates the transport of beta-actin mRNA and cell motility. J. Cell Sci. 2015, 128, 1001–1010. [Google Scholar]
- Hamada, S.; Ishiyama, K.; Choi, S.B.; Wang, C.; Singh, S.; Kawai, N.; Franceschi, V.R.; Okita, T.W. The transport of prolamine RNAs to prolamine protein bodies in living rice endosperm cells. Plant Cell 2003, 15, 2253–2264. [Google Scholar] [CrossRef] [Green Version]
- Okita, T.W.; Choi, S.B. mRNA localization in plants: Targeting to the cell’s cortical region and beyond. Curr. Opin. Plant Biol. 2002, 5, 553–559. [Google Scholar] [CrossRef]
- Fukuda, M.; Kawagoe, Y.; Murakami, T.; Washida, H.; Sugino, A.; Nagamine, A.; Okita, T.W.; Ogawa, M.; Kumamaru, T. The Dual Roles of the Golgi Transport 1 (GOT1B): RNA Localization to the Cortical Endoplasmic Reticulum and the Export of Proglutelin and alpha-Globulin from the Cortical ER to the Golgi. Plant Cell Physiol. 2016, 57, 2380–2391. [Google Scholar] [CrossRef] [Green Version]
- Lorente-Rodriguez, A.; Heidtman, M.; Barlowe, C. Multicopy suppressor analysis of thermosensitive YIP1 alleles implicates GOT1 in transport from the ER. J. Cell Sci. 2009, 122 Pt 10, 1540–1550. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, F.; Ren, Y.; Wang, Y.; Liu, X.; Long, W.; Wang, D.; Zhu, J.; Zhu, X.; Jing, R.; et al. GOLGI TRANSPORT 1B Regulates Protein Export from the Endoplasmic Reticulum in Rice Endosperm Cells. Plant Cell 2016, 28, 2850–2865. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, M.; Satoh-Cruz, M.; Wen, L.; Crofts, A.J.; Sugino, A.; Washida, H.; Okita, T.W.; Ogawa, M.; Kawagoe, Y.; Maeshima, M.; et al. The small GTPase Rab5a is essential for intracellular transport of proglutelin from the Golgi apparatus to the protein storage vacuole and endosomal membrane organization in developing rice endosperm. Plant Physiol. 2011, 157, 632–644. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Chou, H.L.; Crofts, A.J.; Zhang, L.; Tian, L.; Washida, H.; Fukuda, M.; Kumamaru, T.; Oviedo, O.J.; Starkenburg, S.R.; et al. Selective sets of mRNAs localize to extracellular paramural bodies in a rice glup6 mutant. J. Exp. Bot. 2018, 69, 5045–5058. [Google Scholar] [CrossRef] [Green Version]
- Pohlmann, T.; Baumann, S.; Haag, C.; Albrecht, M.; Feldbrugge, M. A FYVE zinc finger domain protein specifically links mRNA transport to endosome trafficking. elife 2015, 4, e06041. [Google Scholar] [CrossRef] [PubMed]
- Muntjes, K.; Devan, S.K.; Reichert, A.S.; Feldbrugge, M. Linking transport and translation of mRNAs with endosomes and mitochondria. EMBO Rep. 2021, 22, e52445. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Doroshenk, K.A.; Zhang, L.; Fukuda, M.; Washida, H.; Kumamaru, T.; Okita, T. Zipcode RNA-Binding Proteins and Membrane Trafficking Proteins Cooperate to Transport Glutelin mRNAs in Rice Endosperm. Plant Cell 2020, 32, 2566–2581. [Google Scholar] [CrossRef]
- Quentin, D.; Schuhmacher, J.S.; Klink, B.U.; Lauer, J.; Shaikh, T.R.; Huis, P.J.; Welp, L.M.; Urlaub, H.; Zerial, M.; Raunser, S. Structure of the human FERRY Rab5 effector complex. bioRxiv 2021. [Google Scholar] [CrossRef]
- Schuhmacher, J.S.; Dieck, S.T.; Christoforidis, S.; Landerer, C.; Hersemann, L.; Seifert, S.; Giner, A.; Toth-Petroczy, A.; Kalaidzidis, Y.; Schumann, E.M.; et al. The novel Rab5 effector FERRY links early endosomes with the translation machinery. bioRxiv 2021. [Google Scholar] [CrossRef]
- Guo, Y.; Yue, Q.; Gao, J.; Wang, Z.; Chen, Y.R.; Blissard, G.W.; Liu, T.X.; Li, Z. Roles of Cellular NSF Protein in Entry and Nuclear Egress of Budded Virions of Autographa californica Multiple Nucleopolyhedrovirus. J. Virol. 2017, 91, e01111-17. [Google Scholar] [CrossRef] [Green Version]
- Mastick, C.C.; Falick, A.L. Association of N-ethylmaleimide sensitive fusion (NSF) protein and soluble NSF attachment proteins-alpha and -gamma with glucose transporter-4-containing vesicles in primary rat adipocytes. Endocrinology 1997, 138, 2391–2397. [Google Scholar] [CrossRef]
- Zhao, C.; Matveeva, E.A.; Ren, Q.; Whiteheart, S.W. Dissecting the N-ethylmaleimide-sensitive factor: Required elements of the N and D1 domains. J. Biol. Chem. 2010, 285, 761–772. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Slevin, J.T.; Whiteheart, S.W. Cellular functions of NSF: Not just SNAPs and SNAREs. FEBS Lett. 2007, 581, 2140–2149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantegazza, A. Preprint Highlight: The novel Rab5 effector FERRY links early endosomes with the translation machinery. Mol. Biol. Cell 2022, 33, mbcP22041001. [Google Scholar] [CrossRef] [PubMed]
- Kloc, M.; Zearfoss, N.R.; Etkin, L.D. Mechanisms of subcellular mRNA localization. Cell 2002, 108, 533–544. [Google Scholar] [CrossRef]
- Hara-Nishimura, I.; Matsushima, R.; Shimada, T.; Nishimura, M. Diversity and formation of endoplasmic reticulum-derived compartments in plants. Are these compartments specific to plant cells? Plant Physiol. 2004, 136, 3435–3439. [Google Scholar] [CrossRef] [Green Version]
- Nagamine, A.; Matsusaka, H.; Ushijima, T.; Kawagoe, Y.; Ogawa, M.; Okita, T.W.; Kumamaru, T. A role for the cysteine-rich 10 kDa prolamin in protein body I formation in rice. Plant Cell Physiol. 2011, 52, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Chou, H.L.; Tian, L.; Fukuda, M.; Kumamaru, T.; Okita, T.W. The Role of RNA-Binding Protein OsTudor-SN in Post-Transcriptional Regulation of Seed Storage Proteins and Endosperm Development. Plant Cell Physiol. 2019, 60, 2193–2205. [Google Scholar] [CrossRef]
- Washida, H.; Sugino, A.; Messing, J.; Esen, A.; Okita, T.W. Asymmetric localization of seed storage protein RNAs to distinct subdomains of the endoplasmic reticulum in developing maize endosperm cells. Plant Cell Physiol. 2004, 45, 1830–1837. [Google Scholar] [CrossRef] [Green Version]
- Sami-Subbu, R.; Choi, S.B.; Wu, Y.; Wang, C.; Okita, T.W. Identification of a cytoskeleton-associated 120 kDa RNA-binding protein in developing rice seeds. Plant Mol. Biol. 2001, 46, 79–88. [Google Scholar] [CrossRef]
- Sami-Subbu, R.; Muench, D.G.; Okita, T.W. A cytoskeleton-associated RNA-binding protein binds to the untranslated regions of prolamine mRNA and to poly(A). Plant Sci. 2000, 152, 115–122. [Google Scholar] [CrossRef]
- Ponting, C.P. P100, a transcriptional coactivator, is a human homologue of staphylococcal nuclease. Protein Sci. 1997, 6, 459–463. [Google Scholar] [CrossRef] [Green Version]
- Chou, H.L.; Tian, L.; Kumamaru, T.; Hamada, S.; Okita, T.W. Multifunctional RNA Binding Protein OsTudor-SN in Storage Protein mRNA Transport and Localization. Plant Physiol. 2017, 175, 1608–1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keene, J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007, 8, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Samaj, J.; Salaj, J.; Obert, B.; Baluska, F.; Menzel, D.; Volkmann, D. Calreticulin mRNA and protein are localized to protein bodies in storage maize callus cells. Plant Cell Rep. 2008, 27, 231–239. [Google Scholar] [CrossRef]
- Urbinati, C.R.; Long, R.M. Techniques for following the movement of single RNAs in living cells. Wiley Interdiscip. Rev. RNA 2011, 2, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Chao, J.A.; Singer, R.H. Fluorescence fluctuation spectroscopy enables quantitative imaging of single mRNAs in living cells. Biophys. J. 2012, 102, 2936–2944. [Google Scholar] [CrossRef] [Green Version]
- Luo, K.R.; Huang, N.C.; Yu, T.S. Selective Targeting of Mobile mRNAs to Plasmodesmata for Cell-to-Cell Movement. Plant Physiol. 2018, 177, 604–614. [Google Scholar] [CrossRef] [Green Version]
- Bann, D.V.; Parent, L.J. Application of live-cell RNA imaging techniques to the study of retroviral RNA trafficking. Viruses 2012, 4, 963–979. [Google Scholar] [CrossRef] [Green Version]
- Dean, K.M.; Palmer, A.E. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat. Chem Biol. 2014, 10, 512–523. [Google Scholar] [CrossRef]
- Paige, J.S.; Wu, K.Y.; Jaffrey, S.R. RNA mimics of green fluorescent protein. Science 2011, 333, 642–646. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Si, Q.; Yang, K.; Zhang, W.; Okita, T.W.; Tian, L. mRNA Localization to the Endoplasmic Reticulum in Plant Endosperm Cells. Int. J. Mol. Sci. 2022, 23, 13511. https://doi.org/10.3390/ijms232113511
Zhang L, Si Q, Yang K, Zhang W, Okita TW, Tian L. mRNA Localization to the Endoplasmic Reticulum in Plant Endosperm Cells. International Journal of Molecular Sciences. 2022; 23(21):13511. https://doi.org/10.3390/ijms232113511
Chicago/Turabian StyleZhang, Laining, Qidong Si, Kejie Yang, Wenwei Zhang, Thomas W. Okita, and Li Tian. 2022. "mRNA Localization to the Endoplasmic Reticulum in Plant Endosperm Cells" International Journal of Molecular Sciences 23, no. 21: 13511. https://doi.org/10.3390/ijms232113511




