Microbiota Dysbiosis and Gut Barrier Dysfunction Associated with Non-Alcoholic Fatty Liver Disease Are Modulated by a Specific Metabolic Cofactors’ Combination
Abstract
1. Introduction
2. Results
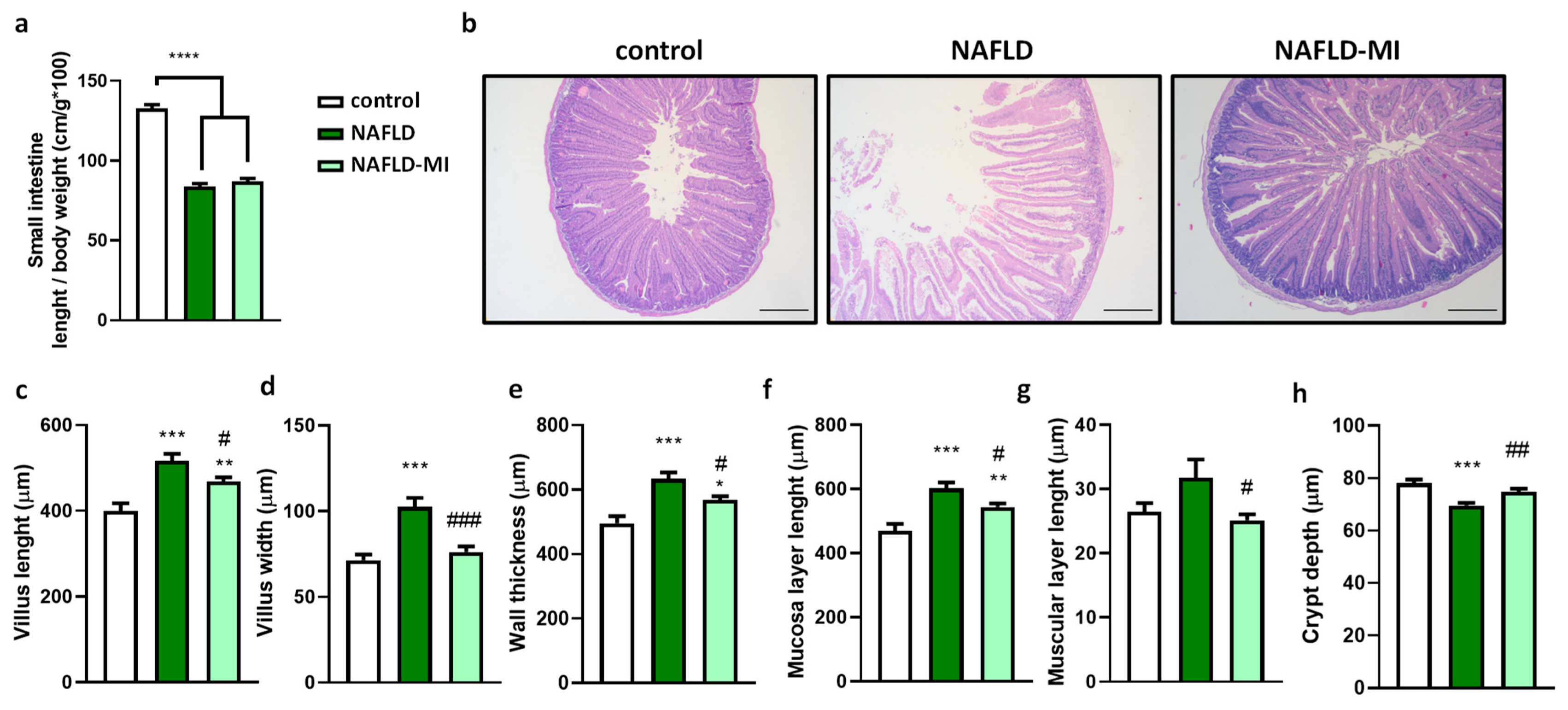
2.1. Multi-Ingredient Treatment Modified Epithelium Morphology in the Small Intestine in a NAFLD
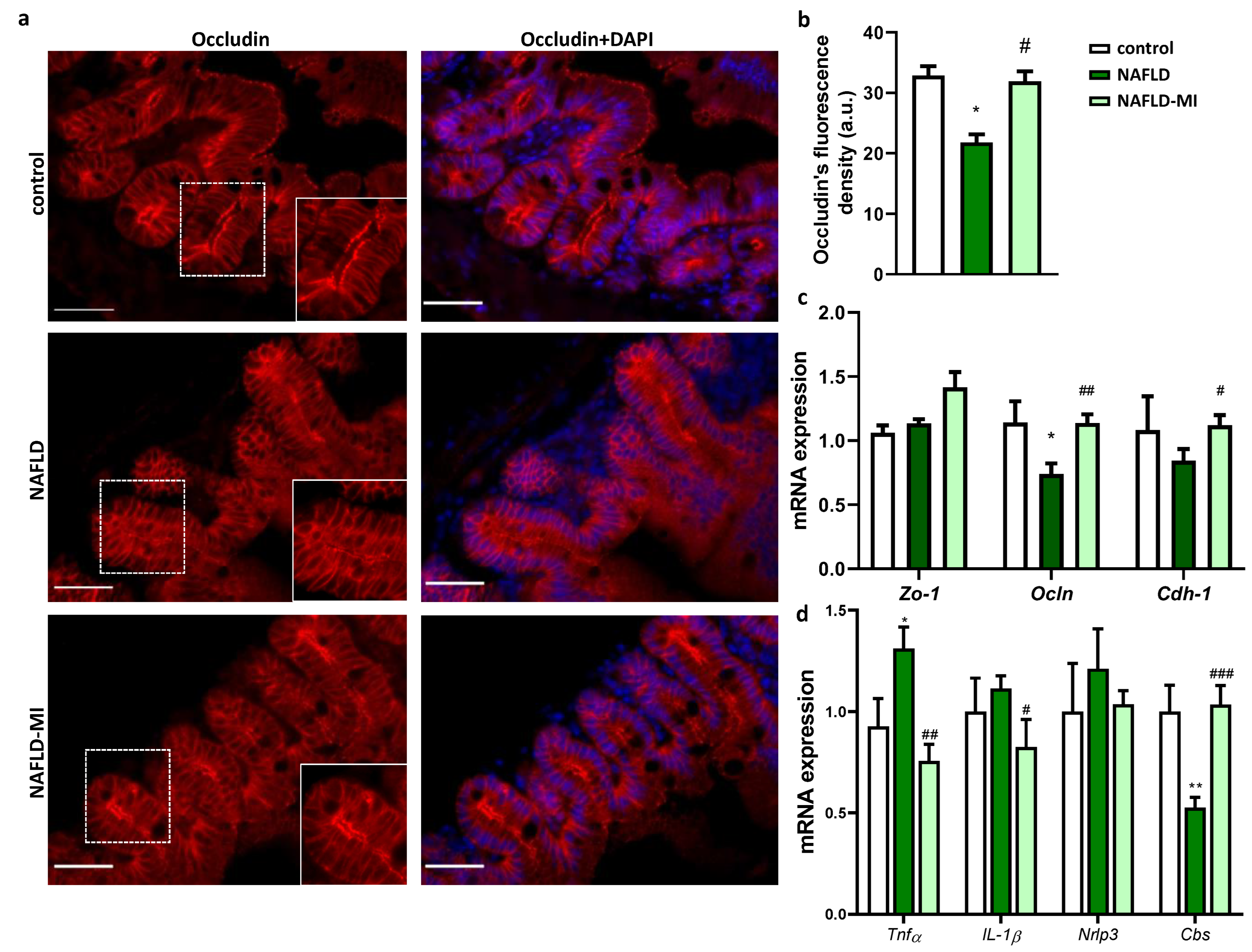
2.2. MI Supplementation Ameliorates Intestinal Permeability and Inflammation
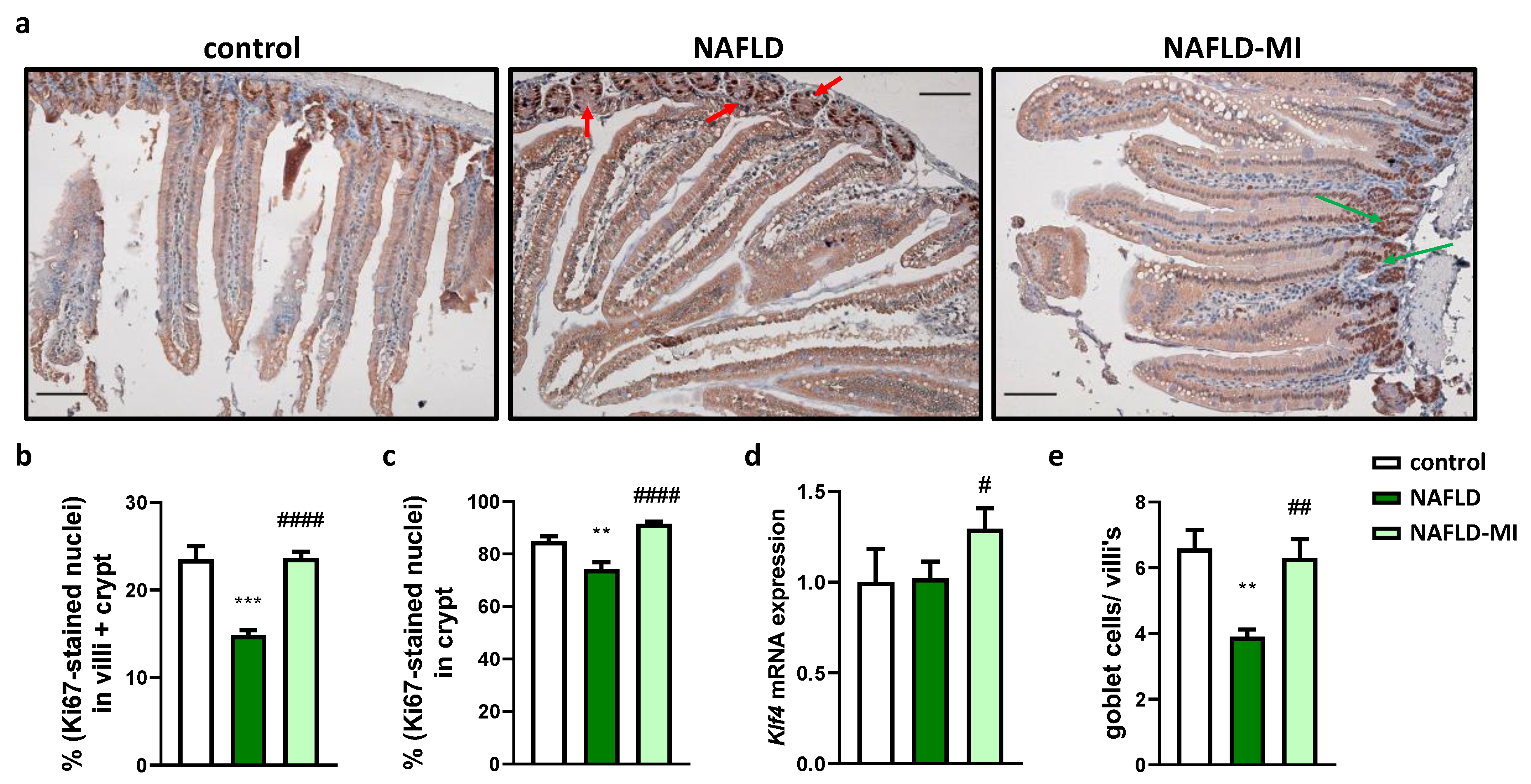
2.3. MI Supplementation Recovers Proliferative Cells Localization in the Small Intestine
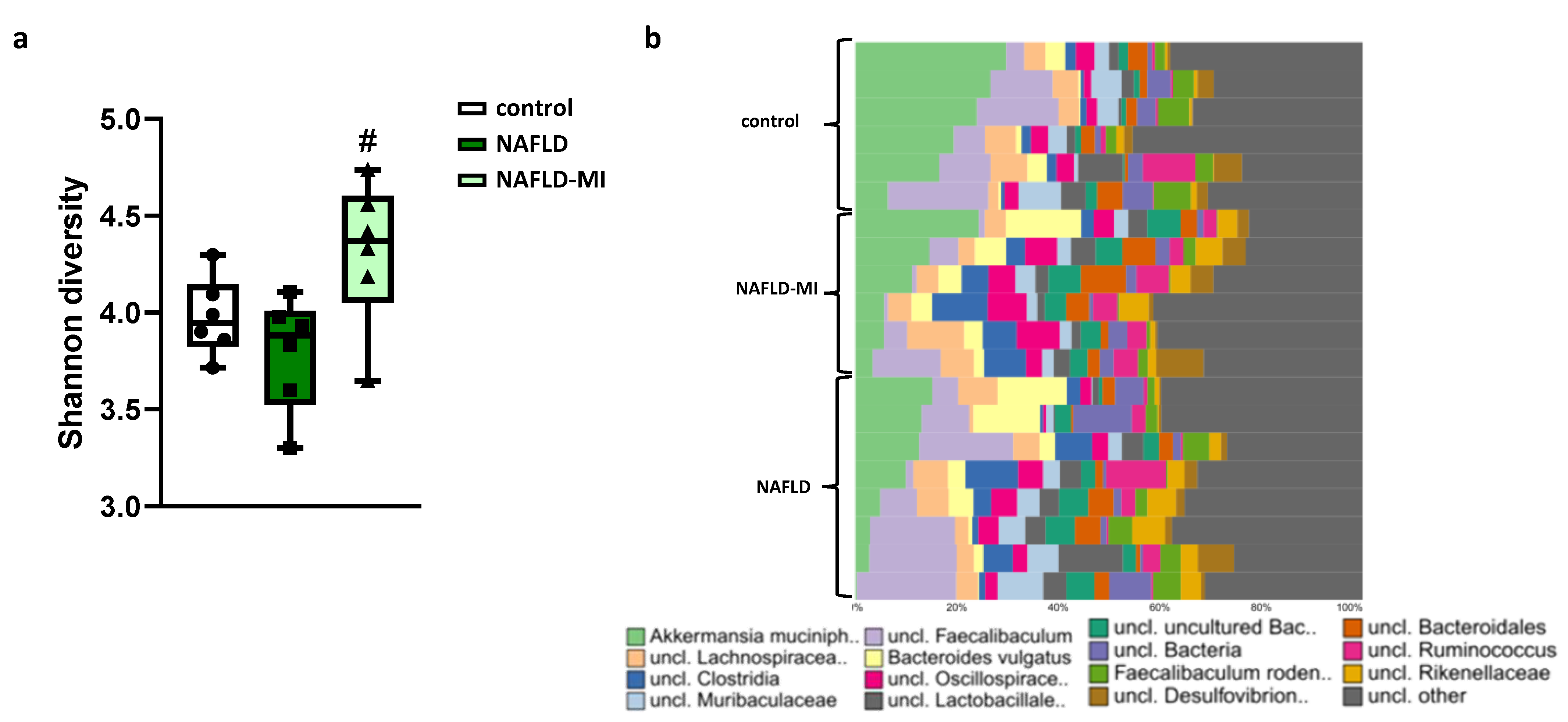
2.4. Microbiota Dysbiosis Present in NAFLD Is Attenuated after MI Supplementation
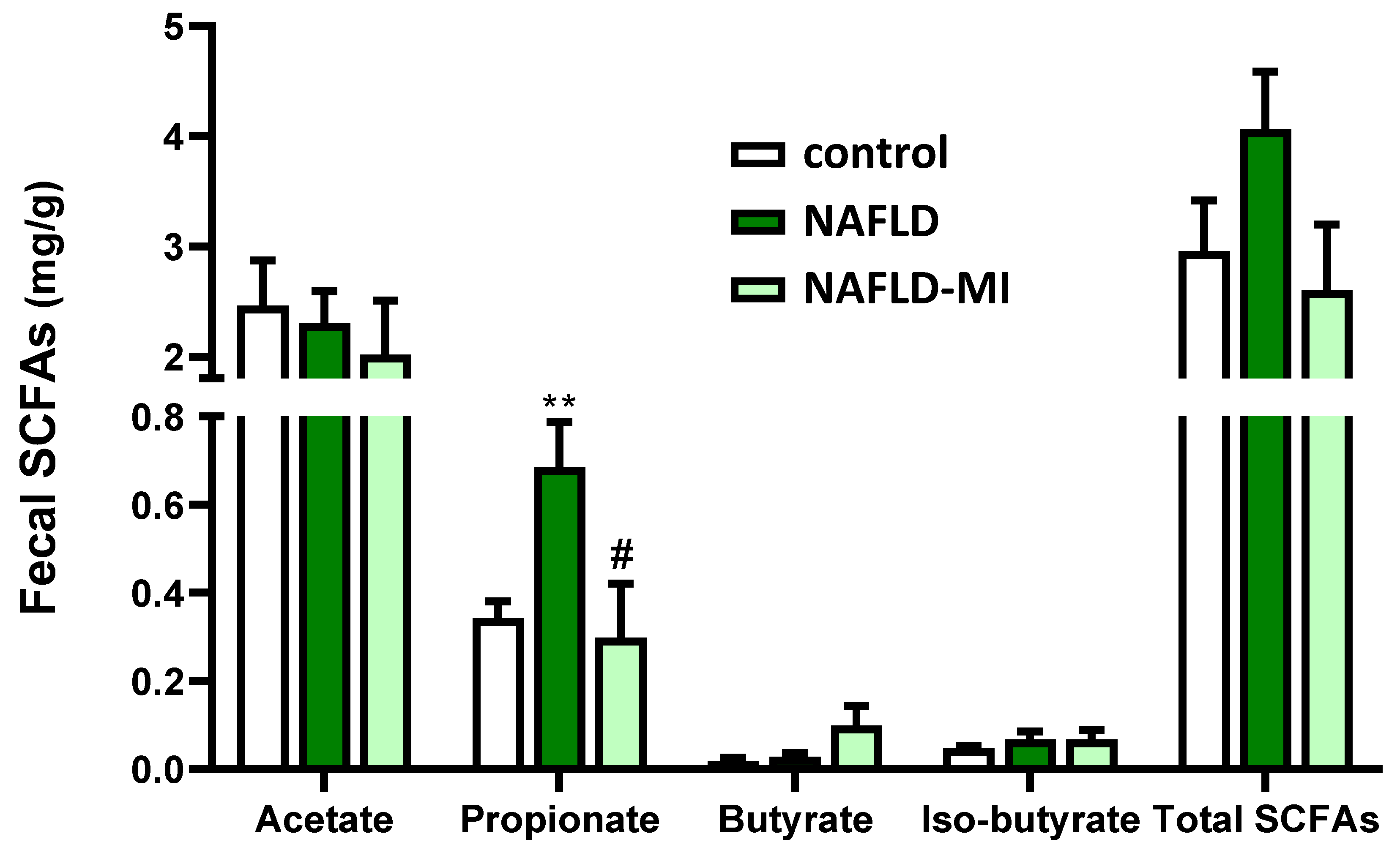
2.5. MI Supplementation Modulates the SCFAs Fecal Concentrations
2.6. Fecal SCFAs Levels Are Correlated with Changes in Specific Fecal Genera Bacteria
3. Discussion
4. Materials and Methods
4.1. MI Treatment Composition
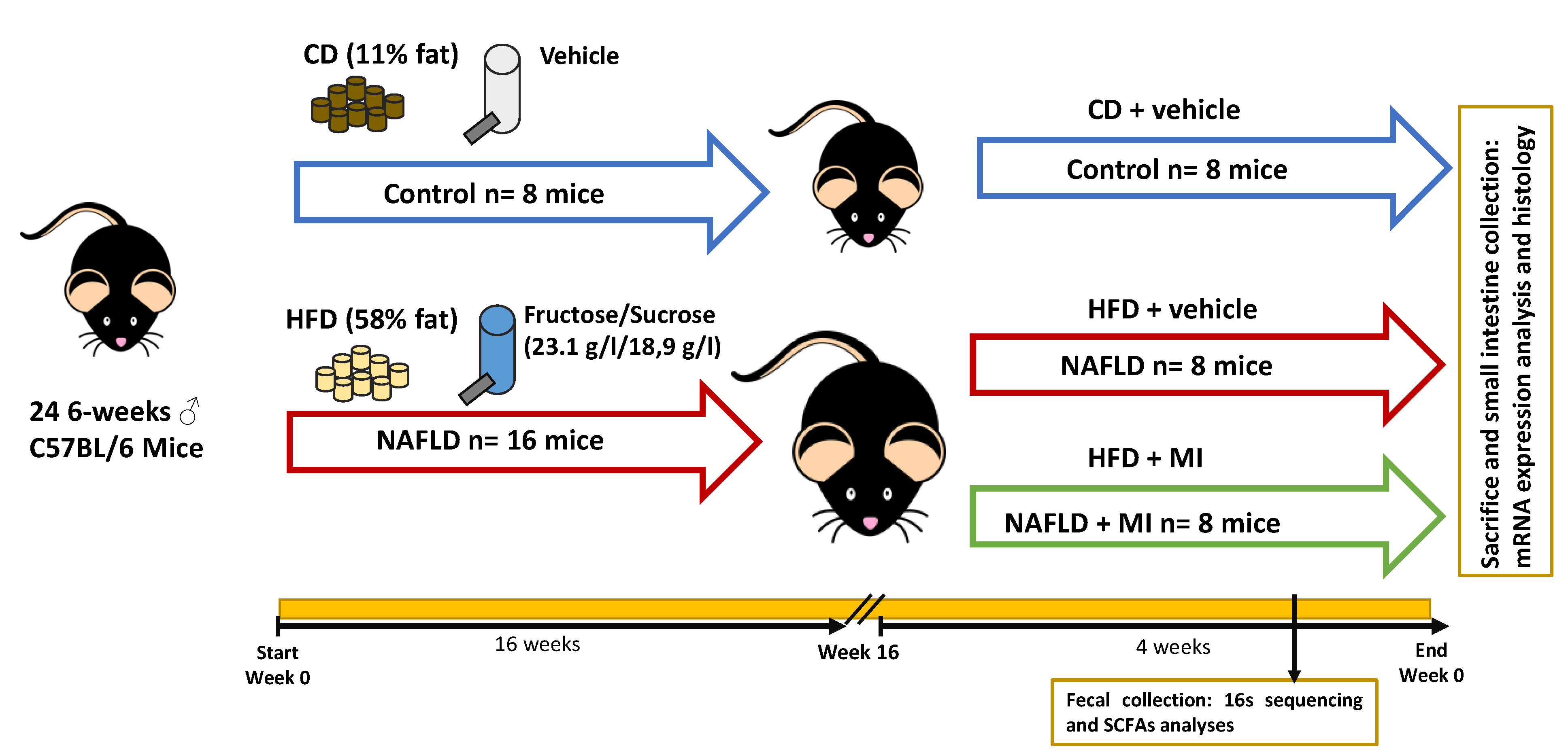
4.2. Animal Model and Experimental Design
4.3. Histological Staining Analysis of Intestinal Sections
4.4. Immunofluorescence Analysis of Intestinal Sections
4.5. Immunohistochemistry Analysis of Intestinal Sections
4.6. Quantification of Short Chain Fatty Acids in Fecal Samples
4.7. mRNA Extraction for Quantitative Polymerase Chain Reaction
4.8. Bacterial Genomic DNA Isolation and 16s rRNA Sequencing
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ji, Y.; Yin, Y.; Sun, L.; Zhang, W. The Molecular and Mechanistic Insights Based on Gut–Liver Axis: Nutritional Target for Non-Alcoholic Fatty Liver Disease (NAFLD) Improvement. Int. J. Mol. Sci. 2020, 21, 3066. [Google Scholar] [CrossRef] [PubMed]
- Yao, N.; Yang, Y.; Li, X.; Wang, Y.; Guo, R.; Wang, X.; Li, J.; Xie, Z.; Li, B.; Cui, W. Effects of Dietary Nutrients on Fatty Liver Disease Associated With Metabolic Dysfunction (MAFLD): Based on the Intestinal-Hepatic Axis. Front. Nutr. 2022, 9, 906511. [Google Scholar] [CrossRef] [PubMed]
- Quesada-Vázquez, S.; Aragonès, G.; del Bas, J.M.; Escoté, X. Diet, Gut Microbiota and Non-Alcoholic Fatty Liver Disease: Three Parts of the Same Axis. Cells 2020, 9, 176. [Google Scholar] [CrossRef]
- Isaacs-Ten, A.; Echeandia, M.; Moreno-Gonzalez, M.; Brion, A.; Goldson, A.; Philo, M.; Patterson, A.M.; Parker, A.; Galduroz, M.; Baker, D.; et al. Intestinal Microbiome-Macrophage Crosstalk Contributes to Cholestatic Liver Disease by Promoting Intestinal Permeability in Mice. Hepatology 2020, 72, 2090–2108. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Y.G. Intestinal Epithelial Plasticity and Regeneration via Cell Dedifferentiation. Cell Regen. 2020, 9, 14. [Google Scholar] [CrossRef]
- Portincasa, P.; Bonfrate, L.; Khalil, M.; de Angelis, M.; Calabrese, F.M.; D’amato, M.; Wang, D.Q.H.; di Ciaula, A. Intestinal Barrier and Permeability in Health, Obesity and NAFLD. Biomedicines 2022, 10, 83. [Google Scholar] [CrossRef]
- Festi, D.; Schiumerini, R.; Birtolo, C.; Marzi, L.; Montrone, L.; Scaioli, E.; di Biase, A.R.; Colecchia, A. Gut Microbiota and Its Pathophysiology in Disease Paradigms. Dig. Dis. 2011, 29, 518–524. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Reddy, D.N. Role of the Normal Gut Microbiota. World J. Gastroenterol. 2015, 21, 8836–8847. [Google Scholar] [CrossRef]
- Fianchi, F.; Liguori, A.; Gasbarrini, A.; Grieco, A.; Miele, L. Nonalcoholic Fatty Liver Disease (Nafld) as Model of Gut–Liver Axis Interaction: From Pathophysiology to Potential Target of Treatment for Personalized Therapy. Int. J. Mol. Sci. 2021, 22, 6485. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The Role of Short-Chain Fatty Acids in Health and Disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [CrossRef]
- Jiang, W.; Wu, N.; Wang, X.; Chi, Y.; Zhang, Y.; Qiu, X.; Hu, Y.; Li, J.; Liu, Y. Dysbiosis Gut Microbiota Associated with Inflammation and Impaired Mucosal Immune Function in Intestine of Humans with Non-Alcoholic Fatty Liver Disease. Sci. Rep. 2015, 5, 8096. [Google Scholar] [CrossRef] [PubMed]
- Lanthier, N.; Delzenne, N. Targeting the Gut Microbiome to Treat Metabolic Dysfunction-Associated Fatty Liver Disease: Ready for Prime Time? Cells 2022, 11, 2718. [Google Scholar] [CrossRef] [PubMed]
- Houghton, D.; Stewart, C.J.; Day, C.P.; Trenell, M. Gut Microbiota and Lifestyle Interventions in NAFLD. Int. J. Mol. Sci. 2016, 17, 447. [Google Scholar] [CrossRef] [PubMed]
- Bauer, K.C.; Littlejohn, P.T.; Ayala, V.; Creus-Cuadros, A.; Finlay, B.B. Nonalcoholic Fatty Liver Disease and the Gut-Liver Axis: Exploring an Undernutrition Perspective. Gastroenterology 2022, 162, 1858–1875.e2. [Google Scholar] [CrossRef]
- Quesada-Vázquez, S.; Colom-Pellicer, M.; Navarro-Masip, È.; Aragonès, G.; del Bas, J.M.; Caimari, A.; Escoté, X. Supplementation with a Specific Combination of Metabolic Cofactors Ameliorates Non-Alcoholic Fatty Liver Disease and, Hepatic Fibrosis, and Insulin Resistance in Mice. Nutrients 2021, 13, 3532. [Google Scholar] [CrossRef]
- Yang, H.; Mayneris-Perxachs, J.; Boqué, N.; del Bas, J.M.; Arola, L.; Yuan, M.; Türkez, H.; Uhlén, M.; Borén, J.; Zhang, C.; et al. Combined Metabolic Activators Decrease Liver Steatosis by Activating Mitochondrial Metabolism in Hamsters Fed with a High-Fat Diet. Biomedicines 2021, 9, 1440. [Google Scholar] [CrossRef]
- Mardinoglu, A.; Ural, D.; Zeybel, M.; Yuksel, H.H.; Uhlén, M.; Borén, J. The Potential Use of Metabolic Cofactors in Treatment of NAFLD. Nutrients 2019, 11, 1578. [Google Scholar] [CrossRef]
- Zhang, C.; Bjornson, E.; Arif, M.; Tebani, A.; Lovric, A.; Benfeitas, R.; Ozcan, M.; Juszczak, K.; Kim, W.; Kim, J.T.; et al. The Acute Effect of Metabolic Cofactor Supplementation: A Potential Therapeutic Strategy against Non-alcoholic Fatty Liver Disease. Mol. Syst. Biol. 2020, 16, e9495. [Google Scholar] [CrossRef]
- Mardinoglu, A.; Bjornson, E.; Zhang, C.; Klevstig, M.; Söderlund, S.; Ståhlman, M.; Adiels, M.; Hakkarainen, A.; Lundbom, N.; Kilicarslan, M.; et al. Personal Model-assisted Identification of NAD + and Glutathione Metabolism as Intervention Target in NAFLD. Mol. Syst. Biol. 2017, 13, 916. [Google Scholar] [CrossRef]
- Xia, Y.; Li, Q.; Zhong, W.; Dong, J.; Wang, Z.; Wang, C. L-Carnitine Ameliorated Fatty Liver in High-Calorie Diet/STZ-Induced Type 2 Diabetic Mice by Improving Mitochondrial Function. Diabetol. Metab. Syndr. 2011, 3, 31. [Google Scholar] [CrossRef]
- Dludla, P.V.; Nkambule, B.B.; Mazibuko-Mbeje, S.E.; Nyambuya, T.M.; Marcheggiani, F.; Cirilli, I.; Ziqubu, K.; Shabalala, S.C.; Johnson, R.; Louw, J.; et al. N-Acetyl Cysteine Targets Hepatic Lipid Accumulation to Curb Oxidative Stress and Inflammation in Nafld: A Comprehensive Analysis of the Literature. Antioxidants 2020, 9, 1283. [Google Scholar] [CrossRef] [PubMed]
- Khodayar, M.J.; Kalantari, H.; Khorsandi, L.; Rashno, M.; Zeidooni, L. Betaine Protects Mice against Acetaminophen Hepatotoxicity Possibly via Mitochondrial Complex II and Glutathione Availability. Biomed. Pharmacother. 2018, 103, 1436–1445. [Google Scholar] [CrossRef] [PubMed]
- Cantó, C.; Houtkooper, R.H.; Pirinen, E.; Youn, D.Y.; Oosterveer, M.H.; Cen, Y.; Fernandez-Marcos, P.J.; Yamamoto, H.; Andreux, P.A.; Cettour-Rose, P.; et al. The NAD+ Precursor Nicotinamide Riboside Enhances Oxidative Metabolism and Protects against High-Fat Diet-Induced Obesity. Cell Metab. 2012, 15, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, K.; Deng, Y.; Chen, R.; Liang, S.; Xie, H.; He, Y.; Chen, Y.; Yang, Q. Effects of Shenling Baizhu Powder Herbal Formula on Intestinal Microbiota in High-Fat Diet-Induced NAFLD Rats. Biomed. Pharmacother. 2018, 102, 1025–1036. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, L.; Zhao, Y.; Wang, C.; Duan, C.; Yang, G.; Niu, C.; Li, S. Lactobacillus Plantarum NA136 Ameliorates Nonalcoholic Fatty Liver Disease by Modulating Gut Microbiota, Improving Intestinal Barrier Integrity, and Attenuating Inflammation. Appl. Microbiol. Biotechnol. 2020, 104, 5273–5282. [Google Scholar] [CrossRef]
- Wu, S.; Hu, R.; Nakano, H.; Chen, K.; Liu, M.; He, X.; Zhang, H.; He, J.; Hou, D.X. Modulation of Gut Microbiota by Lonicera Caerulea l. Berry Polyphenols in a Mouse Model of Fatty Liver Induced by High Fat Diet. Molecules 2018, 23, 3213. [Google Scholar] [CrossRef]
- Jung, Y.; Kim, I.; Mannaa, M.; Kim, J.; Wang, S.; Park, I.; Kim, J.; Seo, Y.S. Effect of Kombucha on Gut-Microbiota in Mouse Having Non-Alcoholic Fatty Liver Disease. Food Sci. Biotechnol. 2019, 28, 261–267. [Google Scholar] [CrossRef]
- Suárez, M.; Boqué, N.; del Bas, J.M.; Mayneris-Perxachs, J.; Arola, L.; Caimari, A. Mediterranean Diet and Multi-Ingredient-Based Interventions for the Management of Non-Alcoholic Fatty Liver Disease. Nutrients 2017, 9, 1052. [Google Scholar] [CrossRef]
- Taylor, S.R.; Ramsamooj, S.; Liang, R.J.; Katti, A.; Pozovskiy, R.; Vasan, N.; Hwang, S.K.; Nahiyaan, N.; Francoeur, N.J.; Schatoff, E.M.; et al. Dietary Fructose Improves Intestinal Cell Survival and Nutrient Absorption. Nature 2021, 597, 263–267. [Google Scholar] [CrossRef]
- Soares, A.; Beraldi, E.J.; Ferreira, P.E.B.; Bazotte, R.B.; Buttow, N.C. Intestinal and Neuronal Myenteric Adaptations in the Small Intestine Induced by a High-Fat Diet in Mice. BMC Gastroenterol. 2015, 15, 3. [Google Scholar] [CrossRef]
- Igarashi, M.; Miura, M.; Williams, E.; Jaksch, F.; Kadowaki, T.; Yamauchi, T.; Guarente, L. NAD+ Supplementation Rejuvenates Aged Gut Adult Stem Cells. Aging Cell 2019, 18, e12935. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, S.; Fang, S.; Yang, X.; Feng, J. Betaine Improves Intestinal Functions by Enhancing Digestive Enzymes, Ameliorating Intestinal Morphology, and Enriching Intestinal Microbiota in High-Salt Stressed Rats. Nutrients 2018, 10, 907. [Google Scholar] [CrossRef] [PubMed]
- Lauz Medeiros, S.H.; de Oliveira Menezes, A.; Zogbi, L.; Frasson de Souza Montero, E. N-Acetylcysteine Use in Hepatic Ischemia/Reperfusion in Rats Minimizing Bowel Injury. Transplant. Proc. 2016, 48, 2371–2374. [Google Scholar] [CrossRef] [PubMed]
- Febbraio, M.A.; Reibe, S.; Shalapour, S.; Ooi, G.J.; Watt, M.J.; Karin, M. Preclinical Models for Studying NASH-Driven HCC: How Useful Are They? Cell Metab. 2019, 29, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Strowig, T.; Henao-Mejia, J.; Elinav, E.; Flavell, R. Inflammasomes in Health and Disease. Nature 2012, 481, 278–286. [Google Scholar] [CrossRef]
- Schuppan, D.; Afdhal, N.H. Liver Cirrhosis. Lancet 2008, 371, 838–851. [Google Scholar] [CrossRef]
- Guo, X.; Li, J.; Tang, R.; Zhang, G.; Zeng, H.; Wood, R.J.; Liu, Z. High Fat Diet Alters Gut Microbiota and the Expression of Paneth Cell-Antimicrobial Peptides Preceding Changes of Circulating Inflammatory Cytokines. Mediat. Inflamm. 2017, 2017, 9474896. [Google Scholar] [CrossRef]
- Zhao, W.; Xiao, M.; Yang, J.; Zhang, L.; Ba, Y.; Xu, R.; Liu, Z.; Zou, H.; Yu, P.; Wu, X.; et al. The Combination of Ilexhainanoside D and Ilexsaponin A1 Reduces Liver Inflammation and Improves Intestinal Barrier Function in Mice with High-Fat Diet-Induced Non-Alcoholic Fatty Liver Disease. Phytomedicine 2019, 63, 153039. [Google Scholar] [CrossRef]
- Moeinian, M.; Ghasemi-Niri, S.F.; Mozaffari, S.; Abdolghaffari, A.H.; Baeeri, M.; Navaea-Nigjeh, M.; Abdollahi, M. Beneficial Effect of Butyrate, Lactobacillus Casei and L-Carnitine Combination in Preference to Each in Experimental Colitis. World J. Gastroenterol. 2014, 20, 10876–10885. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, Y.; Jiao, F.; Shi, C.; Pei, M.; Wang, L.; Gong, Z. Betaine Inhibits Toll-like Receptor 4 Responses and Restores Intestinal Microbiota in Acute Liver Failure Mice. Sci. Rep. 2020, 10, 21850. [Google Scholar] [CrossRef]
- Lee, S.I.; Kang, K.S. N-Acetylcysteine Modulates Lipopolysaccharide-Induced Intestinal Dysfunction. Sci. Rep. 2019, 9, 1004. [Google Scholar] [CrossRef]
- Lee, S.H. Intestinal Permeability Regulation by Tight Junction: Implication on Inflammatory Bowel Diseases. Intest. Res. 2015, 13, 11. [Google Scholar] [CrossRef] [PubMed]
- Werge, M.P.; McCann, A.; Galsgaard, E.D.; Holst, D.; Bugge, A.; Albrechtsen, N.J.W.; Gluud, L.L. The Role of the Transsulfuration Pathway in Non-Alcoholic Fatty Liver Disease. J. Clin. Med. 2021, 10, 1081. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Xu, Z.; Huang, K. Effects of Dietary Selenium on Inflammation and Hydrogen Sulfide in the Gastrointestinal Tract in Chickens. Biol. Trace Elem. Res. 2016, 174, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Maclean, K.N.; Sikora, J.; Kožich, V.; Jiang, H.; Greiner, L.S.; Kraus, E.; Krijt, J.; Crnic, L.S.; Allen, R.H.; Stabler, S.P.; et al. Cystathionine Beta-Synthase Null Homocystinuric Mice Fail to Exhibit Altered Hemostasis or Lowering of Plasma Homocysteine in Response to Betaine Treatment. Mol. Genet. Metab. 2010, 101, 163–171. [Google Scholar] [CrossRef]
- Rohr, M.W.; Narasimhulu, C.A.; Rudeski-Rohr, T.A.; Parthasarathy, S. Negative Effects of a High-Fat Diet on Intestinal Permeability: A Review. Adv. Nutr. 2020, 11, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Gotoh, Y.; Jennings, M.H.; Ann Rhoads, C.; Yee, T.A. Lipid Hydroperoxide-Induced Apoptosis in Human Colonic CaCo-2 Cells Is Associated with an Early Loss of Cellular Redox Balance. FASEB J. 2000, 14, 1567–1576. [Google Scholar] [CrossRef] [PubMed]
- Bressenot, A.; Pooya, S.; Bossenmeyer-Pourie, C.; Gauchotte, G.; Germain, A.; Chevaux, J.B.; Coste, F.; Vignaud, J.M.; Guéant, J.L.; Peyrin-Biroulet, L. Methyl Donor Deficiency Affects Small-Intestinal Differentiation and Barrier Function in Rats. Br. J. Nutr. 2013, 109, 667–677. [Google Scholar] [CrossRef]
- Kruidenier, L.; Verspaget, H.W. Oxidative Stress as a Pathogenic Factor in Inflammatory Bowel Disease. Aliment. Pharmacol. Ther. 2002, 16, 1997–2015. [Google Scholar] [CrossRef]
- Gulhane, M.; Murray, L.; Lourie, R.; Tong, H.; Sheng, Y.H.; Wang, R.; Kang, A.; Schreiber, V.; Wong, K.Y.; Magor, G.; et al. High Fat Diets Induce Colonic Epithelial Cell Stress and Inflammation That Is Reversed by IL-22. Sci. Rep. 2016, 6, 28990. [Google Scholar] [CrossRef]
- Katano, T.; Bialkowska, A.B.; Yang, V.W. KLF4 Regulates Goblet Cell Differentiation in BMI1+ Reserve Intestinal Stem Cell Lineage during Homeostasis. Int. J. Stem Cells 2020, 13, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Ommati, M.M.; Farshad, O.; Mousavi, K.; Jamshidzadeh, A.; Azmoon, M.; Heidari, S.; Azarpira, N.; Niknahad, H.; Heidari, R. Betaine Supplementation Mitigates Intestinal Damage and Decreases Serum Bacterial Endotoxin in Cirrhotic Rats. PharmaNutrition 2020, 12, 100179. [Google Scholar] [CrossRef]
- le Roy, T.; Llopis, M.; Lepage, P.; Bruneau, A.; Rabot, S.; Bevilacqua, C.; Martin, P.; Philippe, C.; Walker, F.; Bado, A.; et al. Intestinal Microbiota Determines Development of Non-Alcoholic Fatty Liver Disease in Mice. Gut 2013, 62, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Thevaranjan, N.; Puchta, A.; Schulz, C.; Naidoo, A.; Szamosi, J.C.; Verschoor, C.P.; Loukov, D.; Schenck, L.P.; Jury, J.; Foley, K.P.; et al. Age-Associated Microbial Dysbiosis Promotes Intestinal Permeability, Systemic Inflammation, and Macrophage Dysfunction. Cell Host Microbe 2017, 21, 455–466.e4. [Google Scholar] [CrossRef] [PubMed]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between Body Mass Index and Firmicutes/Bacteroidetes Ratio in an Adult Ukrainian Population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Yuan, X.; Zhang, C.; Jia, P.; Jiao, S.; Zhao, X.; Yin, H.; Du, Y.; Liu, H. N-Acetylcysteine Alleviates Gut Dysbiosis and Glucose Metabolic Disorder in High-Fat Diet-Fed Mice. J. Diabetes 2019, 11, 32–45. [Google Scholar] [CrossRef]
- Zeybel, M.; Altay, O.; Arif, M.; Li, X.; Yang, H.; Fredolini, C.; Akyildiz, M.; Saglam, B.; Gonenli, M.G.; Ural, D.; et al. Combined Metabolic Activators Therapy Ameliorates Liver Fat in Nonalcoholic Fatty Liver Disease Patients. Mol. Syst. Biol. 2021, 17, e10459. [Google Scholar] [CrossRef]
- Raman, M.; Ahmed, I.; Gillevet, P.M.; Probert, C.S.; Ratcliffe, N.M.; Smith, S.; Greenwood, R.; Sikaroodi, M.; Lam, V.; Crotty, P.; et al. Fecal Microbiome and Volatile Organic Compound Metabolome in Obese Humans with Nonalcoholic Fatty Liver Disease. Clin. Gastroenterol. Hepatol. 2013, 11, 868–875.e3. [Google Scholar] [CrossRef]
- Raimondi, S.; Musmeci, E.; Candeliere, F.; Amaretti, A.; Rossi, M. Identification of Mucin Degraders of the Human Gut Microbiota. Sci. Rep. 2021, 11, 11094. [Google Scholar] [CrossRef]
- Cui, H.; Li, Y.; Wang, Y.; Jin, L.; Yang, L.; Wang, L.; Liao, J.; Wang, H.; Peng, Y.; Zhang, Z.; et al. Da-Chai-Hu Decoction Ameliorates High Fat Diet-Induced Nonalcoholic Fatty Liver Disease Through Remodeling the Gut Microbiota and Modulating the Serum Metabolism. Front Pharmacol. 2020, 11, 584090. [Google Scholar] [CrossRef]
- Zhang, X.; Coker, O.O.; Chu, E.S.H.; Fu, K.; Lau, H.C.H.; Wang, Y.X.; Chan, A.W.H.; Wei, H.; Yang, X.; Sung, J.J.Y.; et al. Dietary Cholesterol Drives Fatty Liver-Associated Liver Cancer by Modulating Gut Microbiota and Metabolites. Gut 2021, 70, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Kudo, C.; Kessoku, T.; Kamata, Y.; Hidaka, K.; Kurihashi, T.; Iwasaki, T.; Takashiba, S.; Kodama, T.; Tamura, T.; Nakajima, A.; et al. Relationship between Non-Alcoholic Fatty Liver Disease and Periodontal Disease: A Review and Study Protocol on the Effect of Periodontal Treatment on Non-Alcoholic Fatty Liver Disease. J. Transl. Sci. 2016, 2, 340–345. [Google Scholar] [CrossRef]
- Yin, X.; Peng, J.; Zhao, L.; Yu, Y.; Zhang, X.; Liu, P.; Feng, Q.; Hu, Y.; Pang, X. Structural Changes of Gut Microbiota in a Rat Non-Alcoholic Fatty Liver Disease Model Treated with a Chinese Herbal Formula. Syst. Appl. Microbiol. 2013, 36, 188–196. [Google Scholar] [CrossRef]
- John, U.V.; Carvalho, J. Enterococcus: Review of Its Physiology, Pathogenesis, Diseases and the Challenges It Poses for Clinical Microbiology. Front. Biol. (Beijing) 2011, 6, 357–366. [Google Scholar] [CrossRef]
- Fukui, H. Role of Gut Dysbiosis in Liver Diseases: What Have We Learned So Far? Diseases 2019, 7, 58. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.; Zhou, Q.; Yang, R.; Zeng, J.; Li, X.; Zhang, R.; Tang, W.; Li, H.; Wang, S.; Shen, T.; et al. Naringin Attenuates High Fat Diet Induced Non-Alcoholic Fatty Liver Disease and Gut Bacterial Dysbiosis in Mice. Front. Microbiol. 2020, 11, 585066. [Google Scholar] [CrossRef]
- Rau, M.; Rehman, A.; Dittrich, M.; Groen, A.K.; Hermanns, H.M.; Seyfried, F.; Beyersdorf, N.; Dandekar, T.; Rosenstiel, P.; Geier, A. Fecal SCFAs and SCFA-Producing Bacteria in Gut Microbiome of Human NAFLD as a Putative Link to Systemic T-Cell Activation and Advanced Disease. United Eur. Gastroenterol. J. 2018, 6, 1496–1507. [Google Scholar] [CrossRef]
- Schwiertz, A.; Taras, D.; Schäfer, K.; Beijer, S.; Bos, N.A.; Donus, C.; Hardt, P.D. Microbiota and SCFA in Lean and Overweight Healthy Subjects. Obesity 2010, 18, 190–195. [Google Scholar] [CrossRef]
- Upadhyaya, B.; McCormack, L.; Fardin-Kia, A.R.; Juenemann, R.; Nichenametla, S.; Clapper, J.; Specker, B.; Dey, M. Impact of Dietary Resistant Starch Type 4 on Human Gut Microbiota and Immunometabolic Functions. Sci. Rep. 2016, 6, 28797. [Google Scholar] [CrossRef]
- Feng, Y.; Huang, Y.; Wang, Y.; Wang, P.; Wang, F. Severe Burn Injury Alters Intestinal Microbiota Composition and Impairs Intestinal Barrier in Mice. Burns Trauma 2019, 7, s41038-019-0156-1. [Google Scholar] [CrossRef]
- Killingsworth, J.; Sawmiller, D.; Shytle, R.D. Propionate and Alzheimer’s Disease. Front. Aging Neurosci. 2021, 12, 580001. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; You, H.J.; Bajaj, J.S.; Joo, S.K.; Yu, J.; Park, S.; Kang, H.; Park, J.H.; Kim, J.H.; Lee, D.H.; et al. Distinct Signatures of Gut Microbiome and Metabolites Associated with Significant Fibrosis in Non-Obese NAFLD. Nat. Commun. 2020, 11, 4982. [Google Scholar] [CrossRef] [PubMed]
- Van Loo, P.L.P.; Kuin, N.; Sommer, R.; Avsaroglu, H.; Pham, T.; Baumans, V. Impact of “living Apart Together” on Postoperative Recovery of Mice Compared with Social and Individual Housing. Lab. Anim. 2007, 41, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Q.; Wang, H.; Zhao, X.; Li, N.; Zhang, H.; Chen, G.; Liu, Z. Fish Oil Alleviates Circadian Bile Composition Dysregulation in Male Mice with NAFLD. J. Nutr. Biochem. 2019, 69, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Salic, K.; Gart, E.; Seidel, F.; Verschuren, L.; Caspers, M.; van Duyvenvoorde, W.; Wong, K.E.; Keijer, J.; Bobeldijk-Pastorova, I.; Wielinga, P.Y.; et al. Combined Treatment with L-Carnitine and Nicotinamide Riboside Improves Hepatic Metabolism and Attenuates Obesity and Liver Steatosis. Int. J. Mol. Sci. 2019, 20, 4359. [Google Scholar] [CrossRef]
- Martínez-Fernández, L.; González-Muniesa, P.; Sáinz, N.; Laiglesia, L.M.; Escoté, X.; Martínez, J.A.; Moreno-Aliaga, M.J. Maresin 1 Regulates Hepatic FGF21 in Diet-Induced Obese Mice and in Cultured Hepatocytes. Mol. Nutr. Food Res. 2019, 63, e1900358. [Google Scholar] [CrossRef]
- Cardiff, R.D.; Miller, C.H.; Munn, R.J. Manual Hematoxylin and Eosin Staining of Mouse Tissue Sections. Cold Spring Harb. Protoc. 2014, 2014, 655–658. [Google Scholar] [CrossRef]
- Mir, H.; Meena, A.S.; Chaudhry, K.K.; Shukla, P.K.; Gangwar, R.; Manda, B.; Padala, M.K.; Shen, L.; Turner, J.R.; Dietrich, P.; et al. Occludin Deficiency Promotes Ethanol-Induced Disruption of Colonic Epithelial Junctions, Gut Barrier Dysfunction and Liver Damage in Mice. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 765–774. [Google Scholar] [CrossRef]
- García-Villalba, R.; Giménez-Bastida, J.A.; García-Conesa, M.T.; Tomás-Barberán, F.A.; Carlos Espín, J.; Larrosa, M. Alternative Method for Gas Chromatography-Mass Spectrometry Analysis of Short-Chain Fatty Acids in Faecal Samples. J. Sep. Sci. 2012, 35, 1906–1913. [Google Scholar] [CrossRef]
- Zhao, G.; Nyman, M.; Jönsson, J.Å. Rapid Determination of Short-Chain Fatty Acids in Colonic Contents and Faeces of Humans and Rats by Acidified Water-Extraction and Direct-Injection Gas Chromatography. Biomed. Chromatogr. 2006, 20, 674–682. [Google Scholar] [CrossRef]
- Saha, S.; Day-Walsh, P.; Shehata, E.; Kroon, P.A. Development and Validation of a Lc-Ms/Ms Technique for the Analysis of Short Chain Fatty Acids in Tissues and Biological Fluids without Derivatisation Using Isotope Labelled Internal Standards. Molecules 2021, 26, 6444. [Google Scholar] [CrossRef] [PubMed]
- Dunoyer-Geindre, S.; Kruithof, E.K.O. Epigenetic Control of Tissue-Type Plasminogen Activator Synthesis in Human Endothelial Cells. Cardiovasc. Res. 2011, 90, 457–463. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shynlova, O.; Dorogin, A.; Li, Y.; Lye, S. Inhibition of Infection-Mediated Preterm Birth by Administration of Broad Spectrum Chemokine Inhibitor in Mice. J. Cell Mol. Med. 2014, 18, 1816–1829. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, M.; Neyrinck, A.M.; Olivares, M.; Rodriguez, J.; de Rocca Serra, A.; Roumain, M.; Bindels, L.B.; Cani, P.D.; Evenepoel, P.; Muccioli, G.G.; et al. The Gut Microbiota Metabolite Indole Alleviates Liver Inflammation in Mice. FASEB J. 2018, 32, 6681–6693. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhao, C.; Dong, Y.; Zhang, M.; Wang, Y.; Li, F.; Li, X.; McClain, C.; Yang, S.; Feng, W. Inhibition of MiR122a by Lactobacillus Rhamnosus GG Culture Supernatant Increases Intestinal Occludin Expression and Protects Mice from Alcoholic Liver Disease. Toxicol. Lett. 2015, 234, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Moscovitz, J.E.; Kong, B.; Buckley, K.; Buckley, B.; Guo, G.L.; Aleksunes, L.M. Restoration of Enterohepatic Bile Acid Pathways in Pregnant Mice Following Short Term Activation of Fxr by GW4064. Toxicol. Appl. Pharmacol. 2016, 310, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, R.; Liu, Y.; Zhang, X.; Zhang, M.; Zeng, Z.; Wu, L.; Gao, X.; Lan, T.; Wang, Y. Polydatin Protects against Carbon Tetrachloride-Induced Liver Fibrosis in Mice. Arch. Biochem. Biophys. 2017, 629, 1–7. [Google Scholar] [CrossRef]
- Özkurt, E.; Fritscher, J.; Soranzo, N.; Ng, D.Y.K.; Davey, R.P.; Bahram, M.; Hildebrand, F. LotuS2: An Ultrafast and Highly Accurate Tool for Amplicon Sequencing Analysis. bioRxiv 2021, 12, 474111. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Edgar, R.C. UNOISE2: Improved Error-Correction for Illumina 16S and ITS Amplicon Sequencing. bioRxiv 2016, 081257. [Google Scholar] [CrossRef]
- Bedarf, J.R.; Beraza, N.; Khazneh, H.; Özkurt, E.; Baker, D.; Borger, V.; Wüllner, U.; Hildebrand, F. Much Ado about Nothing? Off-Target Amplification Can Lead to False-Positive Bacterial Brain Microbiome Detection in Healthy and Parkinson’s Disease Individuals. Microbiome 2021, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Saary, P.; Forslund, K.; Bork, P.; Hildebrand, F. RTK: Efficient Rarefaction Analysis of Large Datasets. Bioinformatics 2017, 33, 2594–2595. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, F.; Moitinho-Silva, L.; Blasche, S.; Jahn, M.T.; Gossmann, T.I.; Huerta-Cepas, J.; Hercog, R.; Luetge, M.; Bahram, M.; Pryszlak, A.; et al. Antibiotics-Induced Monodominance of a Novel Gut Bacterial Order. Gut 2019, 68, 1781–1790. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y. Controlling the False Discovery Rate-A Practical And Powerful Approach To Multiple Testing. J. R. Stat. Soc. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]

| Testing against Control, NAFLD, and MI; Requested Test(s): Wilcox; Used Matrix Type: Normed; Used Test Statistic Is: Kruskal-Wallis Test. Only Genera Are Listed When the p-Value < 0.05.Genus | Phylum | Direction | p-Value | q-Value |
|---|---|---|---|---|
| Anaerotruncus | Firmicutes | NAFLD > MI >> control | 0.0008 | 0.0291 |
| Butyricicoccus | Firmicutes | MI > control = NAFLD | 0.0036 | 0.0515 |
| Christensenella | Firmicutes | NAFLD > MI = control | 0.0350 | 0.0920 |
| Eggerthellaceae | Actinobacteria | NAFLD > MI = control | 0.0139 | 0.0679 |
| Enterococcus | Firmicutes | NAFLD > control = MI | 0.0065 | 0.0557 |
| Eubacterium nodatum | Firmicutes | NAFLD > MI > control | 0.0011 | 0.0317 |
| Escherichia/Shigella | Proteobacteria | NAFLD > MI > control | 0.0015 | 0.0374 |
| Faecalibaculum | Firmicutes | NAFLD > MI = control | 0.0270 | 0.0920 |
| Faecalibacterium | Firmicutes | NAFLD > MI = control | 0.0167 | 0.0753 |
| Lachnoclostridium | Firmicutes | NAFLD > MI > control | 0.0040 | 0.05 |
| Lachnospiraceae UCG-001 | Firmicutes | NAFLD > MI > control | 0.0065 | 0.05 |
| Peptococcus | Firmicutes | MI > control = NAFLD | 0.0349 | 0.0920 |
| Ruminiclostridium | Firmicutes | MI > control = NAFLD | 0.021 | 0.088 |
| Eubacterium | Bifidobacterium | Eggerthellaceae;? | Parvibacter | Bacteroides | Odoribacter | Alloprevotella | Enterococcus | Lactobacillus | Lachnoclostridium | Lachnospiraceae UCG-001 | Butyricicoccus | Anaerotruncus | Caproiciproducens | Faecalibacterium | Ruminococcaceae;UBA1819 | Peptococcus | Romboutsia | Terrisporobacter | Parasutterella | Citrobacter | Enterobacter | Faecalibaculum | Ruminiclostridium | Christensenella | Escherichia-Shigella | Akkermansia | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Propionate | All animals | ** | ||||||||||||||||||||||||||
| control | * | * | * | * | * | |||||||||||||||||||||||
| NAFLD | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | |||||||||||||||||
| NAFLD-MI | *** | |||||||||||||||||||||||||||
| Acetate | All animals | * | * | ** | ||||||||||||||||||||||||
| control | * | * | ||||||||||||||||||||||||||
| NAFLD | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | |||||||||||||||||
| NAFLD-MI | ||||||||||||||||||||||||||||
| Butyrate | All animals | |||||||||||||||||||||||||||
| control | * | ** | ** | |||||||||||||||||||||||||
| NAFLD | *** | *** | *** | *** | *** | *** | ||||||||||||||||||||||
| NAFLD-MI | ||||||||||||||||||||||||||||
| Iso-butyrate | All animals | |||||||||||||||||||||||||||
| control | * | |||||||||||||||||||||||||||
| NAFLD | ||||||||||||||||||||||||||||
| NAFLD-MI | ||||||||||||||||||||||||||||
| Positive correlation | Negative correlation | |||||||||||||||||||||||||||
| Primers | Forward | Reverse | Reference |
|---|---|---|---|
| Cdh-1 | 5′-CATCCCAGAACCTCGAAACA-3′ | 5′-TGGGTTAGCTCAGCAGTAA-3′ | This study |
| Cbs | 5′-GCAGCGCTGTGTGGTCATC-3′ | 5′-CATCCATTTGTCACTCAGGAACTT-3′ | [15] |
| Il-1β | 5′-GGACCCCAAAAGATGAAGGGCTGC-3′ | 5′-GCTCTTGTTGATGTGCTGCTGCG-3′ | [83] |
| Klf4 | 5′-AGCCACCCACACTTGTGACTATG-3′ | 5′-CAGTGGTAAGGTTTCTCGCCTGTG-3′ | [50] |
| Nlrp3 | 5′-GCCCAAGGAGGAAGAAGAAG-3′ | 5′-AGAAGAGACCACGGCAGAAG-3′ | [84] |
| Ocln | 5′-ACCCGAAGAAAGATGGATCG-3′ | 5′-CATAGTCAGATGGGGGTGGA-3′ | [85] |
| Ostα | 5′-CACTGGCTCAGTTGCCATTT-3′ | 5′-GCATACGGCATAAAACGAGGT-3′ | [86] |
| Tnfα | 5′-AGGGTCTGGGCCATAGAACT-3′ | 5′-CCACCACGCTCTTCTGTCTAC-3′ | [87] |
| Zo-1 | 5′-TGGGAACAGCACACAGTGAC-3′ | 5′-GCTGGCCCTCCTTTTAACAC-3′ | [85] |
| 36b4 | 5′-AGTCCCTGCCCTTTGTACACA-3′ | 5′-CGATCCGAGGGCCTCACTA-3′ | [82] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quesada-Vázquez, S.; Bone, C.; Saha, S.; Triguero, I.; Colom-Pellicer, M.; Aragonès, G.; Hildebrand, F.; del Bas, J.M.; Caimari, A.; Beraza, N.; et al. Microbiota Dysbiosis and Gut Barrier Dysfunction Associated with Non-Alcoholic Fatty Liver Disease Are Modulated by a Specific Metabolic Cofactors’ Combination. Int. J. Mol. Sci. 2022, 23, 13675. https://doi.org/10.3390/ijms232213675
Quesada-Vázquez S, Bone C, Saha S, Triguero I, Colom-Pellicer M, Aragonès G, Hildebrand F, del Bas JM, Caimari A, Beraza N, et al. Microbiota Dysbiosis and Gut Barrier Dysfunction Associated with Non-Alcoholic Fatty Liver Disease Are Modulated by a Specific Metabolic Cofactors’ Combination. International Journal of Molecular Sciences. 2022; 23(22):13675. https://doi.org/10.3390/ijms232213675
Chicago/Turabian StyleQuesada-Vázquez, Sergio, Caitlin Bone, Shikha Saha, Iris Triguero, Marina Colom-Pellicer, Gerard Aragonès, Falk Hildebrand, Josep M. del Bas, Antoni Caimari, Naiara Beraza, and et al. 2022. "Microbiota Dysbiosis and Gut Barrier Dysfunction Associated with Non-Alcoholic Fatty Liver Disease Are Modulated by a Specific Metabolic Cofactors’ Combination" International Journal of Molecular Sciences 23, no. 22: 13675. https://doi.org/10.3390/ijms232213675
APA StyleQuesada-Vázquez, S., Bone, C., Saha, S., Triguero, I., Colom-Pellicer, M., Aragonès, G., Hildebrand, F., del Bas, J. M., Caimari, A., Beraza, N., & Escoté, X. (2022). Microbiota Dysbiosis and Gut Barrier Dysfunction Associated with Non-Alcoholic Fatty Liver Disease Are Modulated by a Specific Metabolic Cofactors’ Combination. International Journal of Molecular Sciences, 23(22), 13675. https://doi.org/10.3390/ijms232213675








