Revealing Differential Expression of Phytohormones in Sorghum in Response to Aphid Attack Using the Metabolomics Approach
Abstract
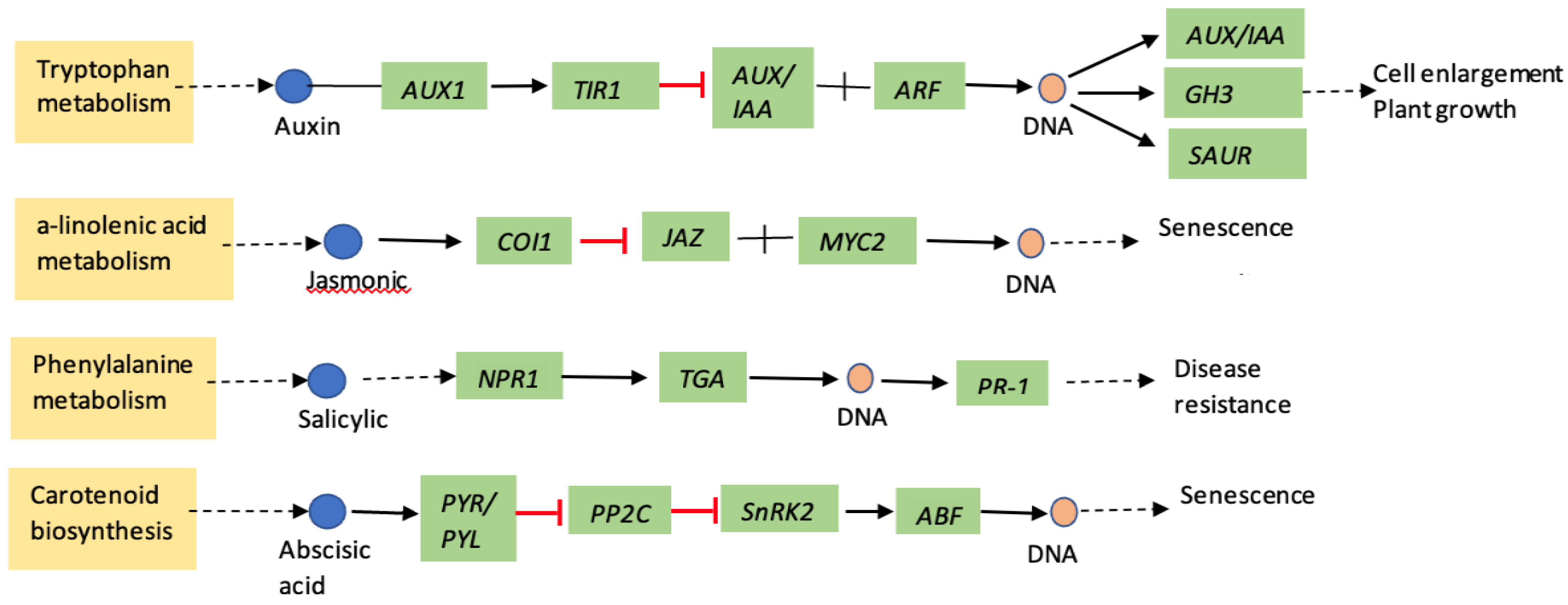
1. Introduction
2. Results
2.1. Differential Response to Aphid between Resistant and Susceptible Genotypes
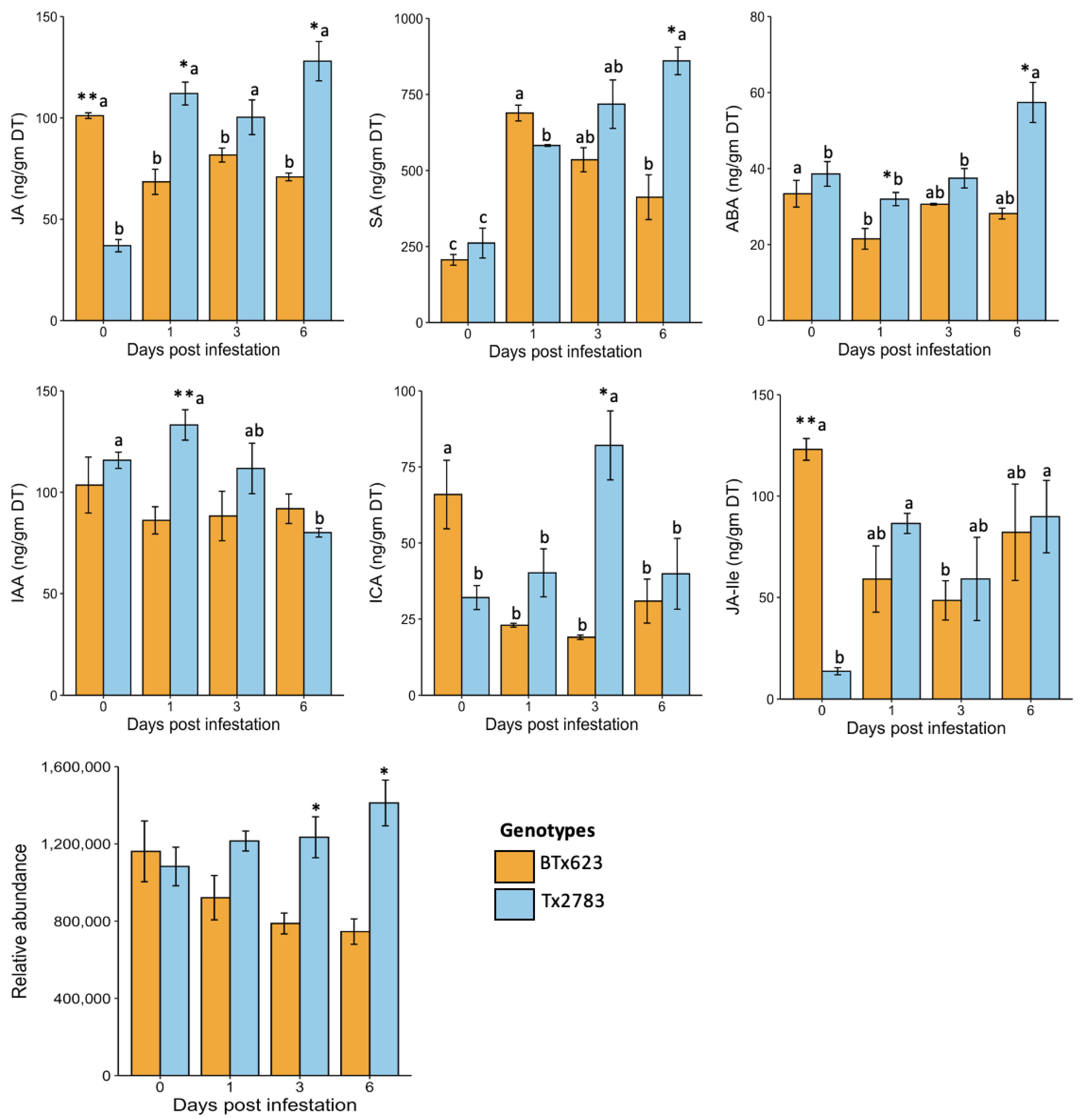
2.2. Phytohormone Profiles in Sorghum Seedlings during Plant-Aphid Interaction
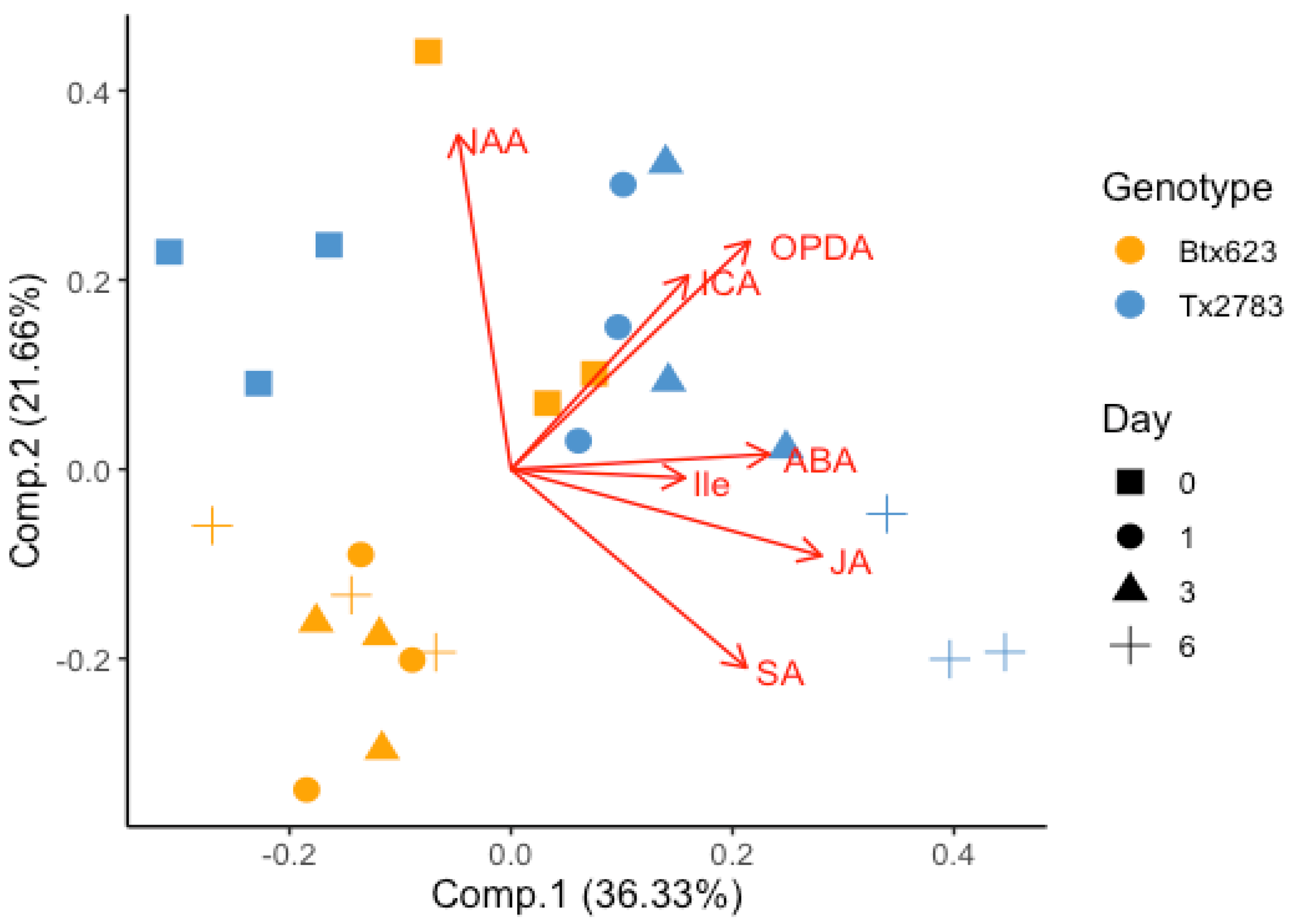
2.3. PCA Indicated the Correlation between Phytohormonal Level and Plant Resistance
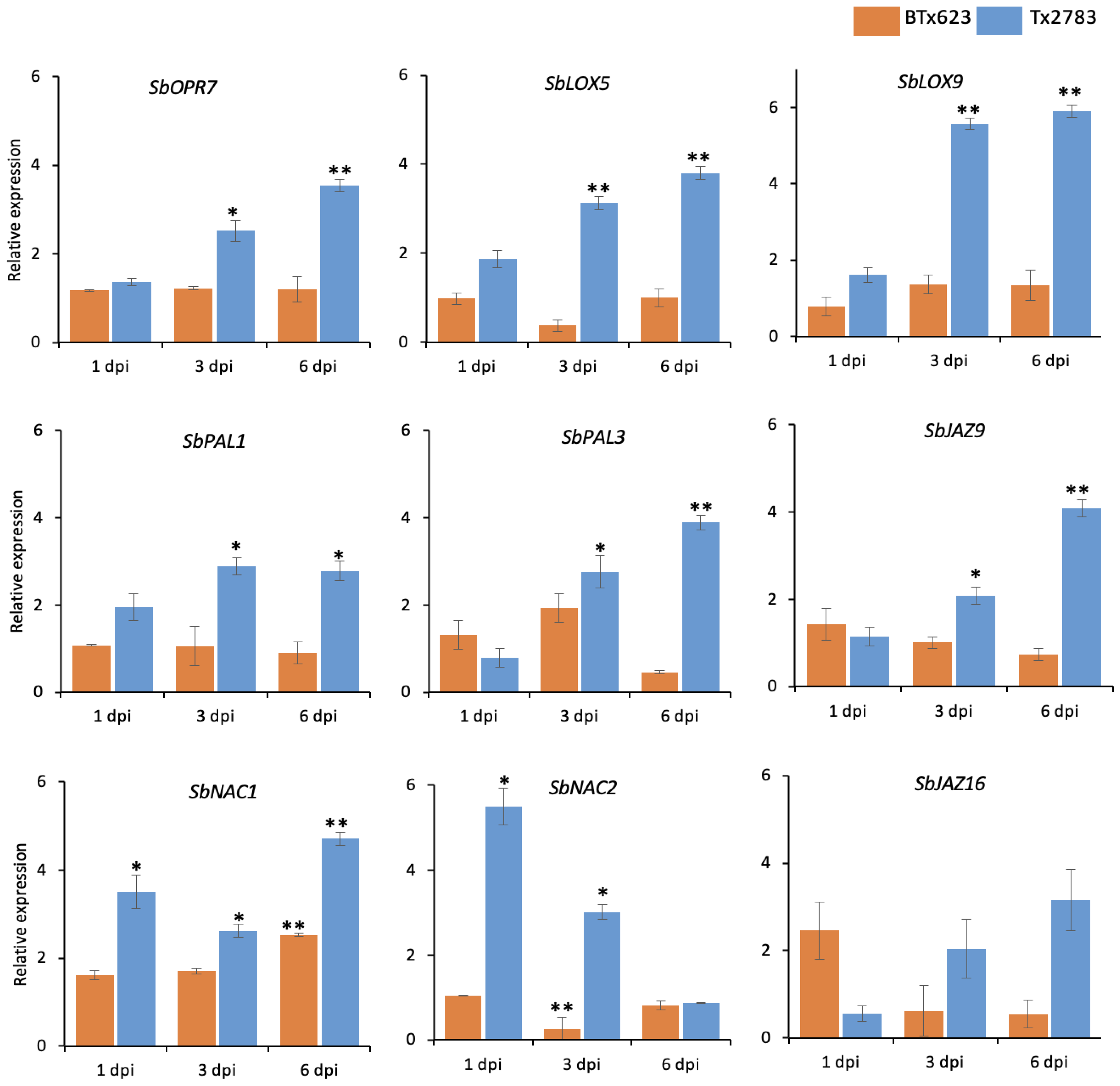
2.4. Differential Expression of Phytohormone Genes
2.5. Phytohormonal Treatment Induces Resistance in Susceptible Genotypes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Sugarcane Aphids
4.2. Sample Preparation, Phytohormone Extraction and Quantification
4.3. Quantification of Phytohormones by LC-MS/Phytohormone Quantification
4.4. RNA Extraction and Quantitative Real-Time PCR Analysis
4.5. Analysis of Phytohormonal Effect on Host Plant Resistance to SCA
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balakrishna, D.; Vinodh, R.; Madhu, P.; Avinash, S.; Rajappa, P.; Bhat, B.V. Tissue culture and genetic transformation in Sorghum bicolor. In Breeding Sorghum for Diverse End Uses; Elsevier: Amsterdam, The Netherlands, 2019; pp. 115–130. [Google Scholar]
- Teferra, T.F.; Awika, J.M. Sorghum as a healthy global food security crop: Opportunities and challenges. Cereal Foods World 2019, 64, 1–8. [Google Scholar]
- Chaturvedi, P.; Govindaraj, M.; Govindan, V.; Weckwerth, W. Sorghum and Pearl Millet as Climate Resilient Crops for Food and Nutrition Security. Front. Plant Sci. 2022, 13, 851970. [Google Scholar] [CrossRef] [PubMed]
- Bowling, R.D.; Brewer, M.J.; Kerns, D.L.; Gordy, J.; Seiter, N.; Elliott, N.E.; Buntin, G.D.; Way, M.; Royer, T.; Biles, S. Sugarcane aphid (Hemiptera: Aphididae): A new pest on sorghum in North America. J. Integr. Pest Manag. 2016, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, R.; Brewer, M.; Way, M.; Biles, S.; Sekula, D.; Bynum, E.; Swart, J.; Crumley, C.; Knutson, A.; Porter, P. Sugarcane Aphid: A New Pest of Sorghum. Texas A&M Agrilife Extension, Ento-035. 2014. Available online: http://denton.agrilife.org/files/2013/08/ENTO-035-The-Sugarcane-Aphid-2014.pdf (accessed on 15 September 2021).
- Armstrong, S.J.; Rooney, W.L.; Peterson, G.C.; Villenueva, R.T.; Brewer, M.J.; Sekula-Ortiz, D. Sugarcane aphid (Hemiptera: Aphididae): Host range and sorghum resistance including cross-resistance from greenbug sources. J. Econ. Entomol. 2015, 108, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y. Host plant defense against sugarcane aphid in sorghum and genetic mechanism of resistance to the new pest. In 2016 International Congress of Entomology; ESA: Paris, France, 2016. [Google Scholar]
- Pant, S.; Huang, Y. Elevated production of reactive oxygen species is related to host plant resistance to sugarcane aphid in sorghum. Plant Signal. Behav. 2021, 16, 1849523. [Google Scholar] [CrossRef]
- Park, S.-J.; Huang, Y.; Ayoubi, P. Identification of expression profiles of sorghum genes in response to greenbug phloem-feeding using cDNA subtraction and microarray analysis. Planta 2006, 223, 932–947. [Google Scholar] [CrossRef]
- Shrestha, K.; Pant, S.; Huang, Y. Genome-wide identification and classification of Lipoxygenase gene family and their roles in sorghum-aphid interaction. Plant Mol. Biol. 2021, 105, 527–541. [Google Scholar] [CrossRef]
- Shrestha, K.; Huang, Y. Genome-wide characterization of the sorghum JAZ gene family and their responses to phytohormone treatments and aphid infestation. Sci. Rep. 2022, 12, 3238. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Pramanik, K.; Bera, A. Response of biofertilizers and phytohormone on growth and yield of chickpea (Cicer arietinium L.). J. Crop Weed 2012, 8, 45–49. [Google Scholar]
- Cabello-Conejo, M.; Prieto-Fernández, Á.; Kidd, P. Exogenous treatments with phytohormones can improve growth and nickel yield of hyperaccumulating plants. Sci. Total Environ. 2014, 494, 1–8. [Google Scholar] [CrossRef]
- Wu, C.; Tang, S.; Li, G.; Wang, S.; Fahad, S.; Ding, Y. Roles of phytohormone changes in the grain yield of rice plants exposed to heat: A review. PeerJ 2019, 7, e7792. [Google Scholar] [CrossRef]
- Erb, M.; Meldau, S.; Howe, G.A. Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 2012, 17, 250–259. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Dicke, M. Plant interactions with microbes and insects: From molecular mechanisms to ecology. Trends Plant Sci. 2007, 12, 564–569. [Google Scholar] [CrossRef]
- Walling, L.L. The myriad plant responses to herbivores. J. Plant Growth Regul. 2000, 19, 195–216. [Google Scholar] [CrossRef]
- Mewis, I.; Tokuhisa, J.G.; Schultz, J.C.; Appel, H.M.; Ulrichs, C.; Gershenzon, J. Gene expression and glucosinolate accumulation in Arabidopsis thaliana in response to generalist and specialist herbivores of different feeding guilds and the role of defense signaling pathways. Phytochemistry 2006, 67, 2450–2462. [Google Scholar] [CrossRef]
- Broekgaarden, C.; Voorrips, R.E.; Dicke, M.; Vosman, B. Transcriptional responses of Brassica nigra to feeding by specialist insects of different feeding guilds. Insect Sci. 2011, 18, 259–272. [Google Scholar] [CrossRef]
- Kessler, A.; Baldwin, I.T. Plant responses to insect herbivory: The emerging molecular analysis. Annu. Rev. Plant Biol. 2002, 53, 299–328. [Google Scholar] [CrossRef]
- De Vos, M.; Van Oosten, V.R.; Van Poecke, R.M.; Van Pelt, J.A.; Pozo, M.J.; Mueller, M.J.; Buchala, A.J.; Métraux, J.-P.; Van Loon, L.C.; Dicke, M. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant-Microbe Interact. 2005, 18, 923–937. [Google Scholar] [CrossRef]
- Heidel-Fischer, H.; Musser, R.; Vogel, H. Annual Plant Reviews; John Wiley & Sons, Ltd.: West Sussex, UK, 2014. [Google Scholar]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Anderson, J.P.; Badruzsaufari, E.; Schenk, P.M.; Manners, J.M.; Desmond, O.J.; Ehlert, C.; Maclean, D.J.; Ebert, P.R.; Kazan, K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 2004, 16, 3460–3479. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed]
- Quandahor, P.; Gou, Y.; Lin, C.; Mujitaba Dawuda, M.; Coulter, J.A.; Liu, C. Phytohormone cross-talk synthesizes glycoalkaloids in potato (Solanum tuberosum L.) in response to aphid (Myzus persicae Sulzer) infestation under drought stress. Insects 2020, 11, 724. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekhar, K.; Shavit, R.; Distelfeld, A.; Christensen, S.; Tzin, V. Exploring the metabolic variation between domesticated and wild tetraploid wheat genotypes in response to corn leaf aphid infestation. Plant Signal. Behav. 2018, 13, e1486148. [Google Scholar] [CrossRef] [PubMed]
- Paudyal, S.; Armstrong, J.S.; Giles, K.L.; Payton, M.E.; Opit, G.P.; Limaje, A. Categories of resistance to sugarcane aphid (Hemiptera: Aphididae) among sorghum genotypes. J. Econ. Entomol. 2019, 112, 1932–1940. [Google Scholar] [CrossRef] [PubMed]
- Limaje, A.; Hayes, C.; Armstrong, J.S.; Hoback, W.; Zarrabi, A.; Paudyal, S.; Burke, J. Antibiosis and tolerance discovered in USDA-ARS sorghums resistant to the sugarcane aphid (Hemiptera: Aphididae). J. Entomol. Sci. 2018, 53, 230–241. [Google Scholar] [CrossRef]
- Gao, L.-L.; Anderson, J.P.; Klingler, J.P.; Nair, R.M.; Edwards, O.R.; Singh, K.B. Involvement of the octadecanoid pathway in bluegreen aphid resistance in Medicago truncatula. Mol. Plant-Microbe Interact. 2007, 20, 82–93. [Google Scholar] [CrossRef]
- Morkunas, I.; Mai, V.C.; Gabryś, B. Phytohormonal signaling in plant responses to aphid feeding. Acta Physiol. Plant. 2011, 33, 2057–2073. [Google Scholar] [CrossRef]
- Korth, K.L.; Thompson, G.A. Chemical signals in plants: Jasmonates and the role of insect-derived elicitors in responses to herbivores. In Multigenic and Induced Systemic Resistance in Plants; Springer: Berlin/Heidelberg, Germany, 2006; pp. 259–278. [Google Scholar]
- Chapman, K.M.; Marchi-Werle, L.; Hunt, T.E.; Heng-Moss, T.M.; Louis, J. Abscisic and jasmonic acids contribute to soybean tolerance to the soybean aphid (Aphis glycines Matsumura). Sci. Rep. 2018, 8, 15148. [Google Scholar] [CrossRef]
- Grover, S.; Agpawa, E.; Sarath, G.; Sattler, S.E.; Louis, J. Interplay of phytohormones facilitate sorghum tolerance to aphids. Plant Mol. Biol. 2020, 109, 639–650. [Google Scholar] [CrossRef]
- Upadhyay, R.K.; Mattoo, A.K. Genome-wide identification of tomato (Solanum lycopersicum L.) lipoxygenases coupled with expression profiles during plant development and in response to methyl-jasmonate and wounding. J. Plant Physiol. 2018, 231, 318–328. [Google Scholar] [CrossRef]
- Moran, P.J.; Thompson, G.A. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 2001, 125, 1074–1085. [Google Scholar] [CrossRef]
- Ogunola, O.F.; Hawkins, L.K.; Mylroie, E.; Kolomiets, M.V.; Borrego, E.; Tang, J.D.; Williams, W.P.; Warburton, M.L. Characterization of the maize lipoxygenase gene family in relation to aflatoxin accumulation resistance. PLoS ONE 2017, 12, e0181265. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, J.; Cheng, D.; Sun, J.; Liu, Y.; Tian, Z. Biochemical and molecular characterizations of Sitobion avenae-induced wheat defense responses. Crop Prot. 2009, 28, 435–442. [Google Scholar] [CrossRef]
- Kawazu, K.; Mochizuki, A.; Sato, Y.; Sugeno, W.; Murata, M.; Seo, S.; Mitsuhara, I. Different expression profiles of jasmonic acid and salicylic acid inducible genes in the tomato plant against herbivores with various feeding modes. Arthropod-Plant Interact. 2012, 6, 221–230. [Google Scholar] [CrossRef]
- Kim, D.S.; Hwang, B.K. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [CrossRef]
- Shine, M.; Yang, J.W.; El-Habbak, M.; Nagyabhyru, P.; Fu, D.Q.; Navarre, D.; Ghabrial, S.; Kachroo, P.; Kachroo, A. Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol. 2016, 212, 627–636. [Google Scholar] [CrossRef]
- Mohase, L.; van der Westhuizen, A.J. Salicylic acid is involved in resistance responses in the Russian wheat aphid-wheat interaction. J. Plant Physiol. 2002, 159, 585–590. [Google Scholar] [CrossRef]
- Chaman, M.E.; Copaja, S.V.; Argandoña, V.H. Relationships between salicylic acid content, phenylalanine ammonia-lyase (PAL) activity, and resistance of barley to aphid infestation. J. Agric. Food Chem. 2003, 51, 2227–2231. [Google Scholar] [CrossRef]
- Khoshfarman-Borji, H.; Yali, M.P.; Bozorg-Amirkalaee, M. Induction of resistance against Brevicoryne brassicae by Pseudomonas putida and salicylic acid in canola. Bull. Entomol. Res. 2020, 110, 597–610. [Google Scholar] [CrossRef]
- Flors, V.; Ton, J.; Mauch-Mani, B. Role of abscisic acid in disease resistance. In Signal Crosstalk Plant Stress Responses; John Wiley & Sons: Hoboken, NJ, USA, 2009; pp. 1–22. [Google Scholar]
- Zhu-Salzman, K.; Salzman, R.A.; Ahn, J.-E.; Koiwa, H. Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiol. 2004, 134, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Lackman, P.; González-Guzman, M.; Tilleman, S.; Carqueijeiro, I.; Perez, A.C.; Moses, T.; Seo, M.; Kanno, Y.; Häkkinen, S.T.; Van Montagu, M.C.; et al. Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc. Natl. Acad. Sci. USA 2011, 108, 5891–5896. [Google Scholar] [CrossRef] [PubMed]
- Koo, Y.M.; Heo, A.Y.; Choi, H.W. Salicylic acid as a safe plant protector and growth regulator. Plant Pathol. J. 2020, 36, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Pajerowska-Mukhtar, K.; Culler, A.H.; Dong, X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 2007, 17, 1784–1790. [Google Scholar] [CrossRef] [PubMed]
- Machado, R.A.; Robert, C.A.; Arce, C.C.; Ferrieri, A.P.; Xu, S.; Jimenez-Aleman, G.H.; Baldwin, I.T.; Erb, M. Auxin is rapidly induced by herbivore attack and regulates a subset of systemic, jasmonate-dependent defenses. Plant Physiol. 2016, 172, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Perez-Alonso, M.; Pollmann, S. How Auxin May Contribute to the Regulation of Plant Defense Responses against Herbivory. Austin J. Plant Biol 2018, 4, 02–05. [Google Scholar]
- Vos, I.A.; Moritz, L.; Pieterse, C.M.; Van Wees, S.C. Impact of hormonal crosstalk on plant resistance and fitness under multi-attacker conditions. Front. Plant Sci. 2015, 6, 639. [Google Scholar] [CrossRef]
- Guo-Zhang, K.; Zheng-Xun, W.; Gu-Chou, S. Participation of H2O2 in enhancement of cold chilling by salicylic acid in banana seedlings. J. Integr. Plant Biol. 2003, 45, 567. [Google Scholar]
- Sheflin, A.M.; Kirkwood, J.S.; Wolfe, L.M.; Jahn, C.E.; Broeckling, C.D.; Schachtman, D.P.; Prenni, J.E. High-throughput quantitative analysis of phytohormones in sorghum leaf and root tissue by ultra-performance liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 2019, 411, 4839–4848. [Google Scholar] [CrossRef]
- Scully, E.D.; Gries, T.; Sarath, G.; Palmer, N.A.; Baird, L.; Serapiglia, M.J.; Dien, B.S.; Boateng, A.A.; Ge, Z.; Funnell-Harris, D.L. Overexpression of SbMyb60 impacts phenylpropanoid biosynthesis and alters secondary cell wall composition in Sorghum bicolor. Plant J. 2016, 85, 378–395. [Google Scholar] [CrossRef]
- Le Thanh, T.; Thumanu, K.; Wongkaew, S.; Boonkerd, N.; Teaumroong, N.; Phansak, P.; Buensanteai, N. Salicylic acid-induced accumulation of biochemical components associated with resistance against Xanthomonas oryzae pv. oryzae in rice. J. Plant Interact. 2017, 12, 108–120. [Google Scholar] [CrossRef]
- Ma, F.; Yang, X.; Shi, Z.; Miao, X. Novel crosstalk between ethylene-and jasmonic acid-pathway responses to a piercing–sucking insect in rice. New Phytol. 2020, 225, 474–487. [Google Scholar] [CrossRef]
- Li, X.; Li, S.; Wang, J.; Lin, J. Exogenous abscisic acid alleviates harmful effect of salt and alkali stresses on wheat seedlings. Int. J. Environ. Res. Public Health 2020, 17, 3770. [Google Scholar] [CrossRef]


| dpi | Aphid Count | Damage Ratings | |||
|---|---|---|---|---|---|
| Btx623 | Tx2783 | p-Value | Btx623 | Tx2783 | |
| 1 | 24 ± 2.23 | 14 ± 1.11 | *** | 0 | 0 |
| 3 | 41 ± 4.23 | 26 ± 2.79 | ** | 1 | 0 |
| 6 | 192 ± 21.1 | 65 ± 4.66 | *** | 1 | 0 |
| 9 | 542 ± 41.6 | 153 ± 9.80 | *** | 2 | 1 |
| 12 | 913 ± 54.3 | 309± 21.8 | *** | 4 | 2 |
| 15 | - | - | - | 6 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Shrestha, K.; Huang, Y. Revealing Differential Expression of Phytohormones in Sorghum in Response to Aphid Attack Using the Metabolomics Approach. Int. J. Mol. Sci. 2022, 23, 13782. https://doi.org/10.3390/ijms232213782
Huang J, Shrestha K, Huang Y. Revealing Differential Expression of Phytohormones in Sorghum in Response to Aphid Attack Using the Metabolomics Approach. International Journal of Molecular Sciences. 2022; 23(22):13782. https://doi.org/10.3390/ijms232213782
Chicago/Turabian StyleHuang, Jian, Kumar Shrestha, and Yinghua Huang. 2022. "Revealing Differential Expression of Phytohormones in Sorghum in Response to Aphid Attack Using the Metabolomics Approach" International Journal of Molecular Sciences 23, no. 22: 13782. https://doi.org/10.3390/ijms232213782
APA StyleHuang, J., Shrestha, K., & Huang, Y. (2022). Revealing Differential Expression of Phytohormones in Sorghum in Response to Aphid Attack Using the Metabolomics Approach. International Journal of Molecular Sciences, 23(22), 13782. https://doi.org/10.3390/ijms232213782





