Abstract
Potato microtuber (MT) development through in vitro techniques are ideal propagules for producing high quality potato plants. MT formation is influenced by several factors, i.e., photoperiod, sucrose, hormones, and osmotic stress. We have previously developed a protocol of MT induction in medium with sucrose (8% w/v), gelrite (6g/L), and 2iP as cytokinin under darkness. To understand the molecular mechanisms involved, we performed a transcriptome-wide analysis. Here we show that 1715 up- and 1624 down-regulated genes were involved in this biological process. Through the protein–protein interaction (PPI) network analyses performed in the STRING database (v11.5), we found 299 genes tightly associated in 14 clusters. Two major clusters of up-regulated proteins fundamental for life growth and development were found: 29 ribosomal proteins (RPs) interacting with 6 PEBP family members and 117 cell cycle (CC) proteins. The PPI network of up-regulated transcription factors (TFs) revealed that at least six TFs–MYB43, TSF, bZIP27, bZIP43, HAT4 and WOX9–may be involved during MTs development. The PPI network of down-regulated genes revealed a cluster of 83 proteins involved in light and photosynthesis, 110 in response to hormone, 74 in hormone mediate signaling pathway and 22 related to aging.
1. Introduction
Potato (Solanum tuberosum L.) is the most important staple food worldwide just after maize, rice, and wheat with an approximate production of around 360 million tons in 2020 on a surface area of 16.5 million hectares [1]. Potato is a geophyte plant with a dual reproduction system, forming vegetative storage organs called tubers and a sexual propagation that is tightly regulated to ensure fitness in adverse climatic conditions [2].
The potato tubers are swollen underground storage organs differentiated from the subapical meristem of the stolon (modified stem) mediated by radial divisions, expanding to further differentiate into starch accumulating parenchyma. Tubers function in vegetative propagation (eyes; axillary meristems) and as storage enabling survival during winter months and/or prolonged periods of abiotic stress (perennialism) and are a means to bypass sexual reproduction.
The molecular mechanisms of the potato tuberization process have been well documented and depend on light (day length and light quality), temperature, and nutrient supply [3,4,5].
Plant biotechnology techniques have been applied to produce potato tubers in vitro called microtubers (MTs). The MTs process occurs when axillary buds are cultured in medium containing high content of sucrose, hormones, and different light quality/darkness. MTs have advantages in terms of storage, transport, and cultivation, due to their reduced size and weight, requiring less space compared to seedlings, with a higher multiplication rate producing seed potato faster and cheaper than other methods [6,7,8,9].
We developed a protocol for MTs induction in potato S. tuberosum var Alpha culturing stolon explants in MS medium supplemented with 8% SUC, high content of gelrite 6.0 g/L, and cytokinin (CK) 2iP 10 mg/L under darkness. The rationale of this protocol was that two-component and cytokinin (CK) signaling interacts with homeobox TFs; ribosomal proteins (RPs), cell cycle (CC), carbon metabolism, auxin responsive factors, and stem cell maintenance genes were involved in this process [10].
Several transcriptomic analyses during potato tuberization have revealed conserved molecular machinery during this process: PEBP members (florigen and tuberigen genes), photoperiod, transcriptional regulation, protein synthesis, cell cycle regulation, metabolic pathways, secondary metabolism, starch and sucrose metabolism, hormone signaling, among others [11,12,13,14,15,16].
Moreover, a few papers have published reliable gene ID in order to compare results. Sharma et al. [15] published the targets of StBEL5 in tuber development by a transcriptome analysis. A set of more than 2000 up-regulated genes can be read from the Supplementary Data Table S2. A PPI network in STRING data base v11.5 with the highest confidence (0.900) revealed that the up-regulated genes are involved in fundamental processes for life: RPs, CC, spliceosome, ribonucleoprotein complex, and basal TFs interacting with several clusters of genes involved in light, stress and metabolic pathways.
Light modulates the molecular mechanisms during potato tuberization (photomorphogenesis); little attention has been given to the molecular events in the dark. To elucidate the molecular mechanisms of MTs development under these conditions, we performed a transcriptomic-wide analysis.
Differential expressed genes (DEG) obtained from the analysis were screened in a bioinformatic tool STRING database v11.5 (https://string-db.org/cgi/input?sessionId=bbmDlM5pze5B&input_page_active_form=multiple_identifiers) with the highest confidence (0.860).
The PPI network of up-regulated TFs revealed that at least six TFs–MYB43, TSF, bZIP27, bZIP43, HAT4, and WOX9–may be involved during MTs development. These TFs were involved in vascularization, proliferation process, cell fate commitment, epigenetic regulation, meristem organization, cell cycle, and ribosomes.
The PPI network of down-regulated TFs demonstrated several TCP TFs families known as repressor of shoot morphogenesis interacting with MYB TFs involved in flower development.
The PPI network of up-regulated genes revealed two clusters–one made of RPs, that play an important role in the control of cell growth, division, and development [17].
PEBP family members, StSP6A, StSP3D, StSP5G, CEN1/BFT, ATC, and FD TF interact with RPL11. The second cluster belongs to CC, whose function is to correctly duplicate the DNA present in the genome and to segregate the resulting copies correctly into the two daughter cells [18]. The CC cluster interacts tightly with several metabolic pathways: one and carbon metabolism, CK response, citrate cycle, fatty acid metabolism, thylakoid, sulfur metabolism, disulfide isomerase, oxidative stress, and amino sugar and nucleotide sugar metabolism. Down-regulated genes were associated with photosynthesis, light harvesting complex, response to light, hormone signal transduction, response to hormone, flowering, and plant organ development.
In this manuscript we have made a deep analysis by text mining, using the STRING data base from which we proposed a new model of MTs development under darkness conditions.
2. Results
2.1. MTs Transcriptomic-Wide Analysis
In order to identify the genes involved in the development of MTs under dark conditions, a transcriptomic-wide analysis was performed. We sequenced cDNA libraries constructed from two treatments: MR8-G6-2iP inducing MTs and MR1-G3-2iP, and non-inducing MTs using the Illumina HiSeq 2000 platform. This produced a total of 397,834,274 reads, from all six cDNA libraries from MR8-G6-2iP and MR1-G3-2iP. A summary of mapping statistics obtained for each sample is given in Table 1.

Table 1.
Mapping statistics for quality filtered reads generated for lines with MTs and non-MTs induction.
Overall, 4839 DEG were found in MR8G62iP (MTs inducing treatment) versus control line (non-MTs); 2896 were up-regulated and 1843 down-regulated.
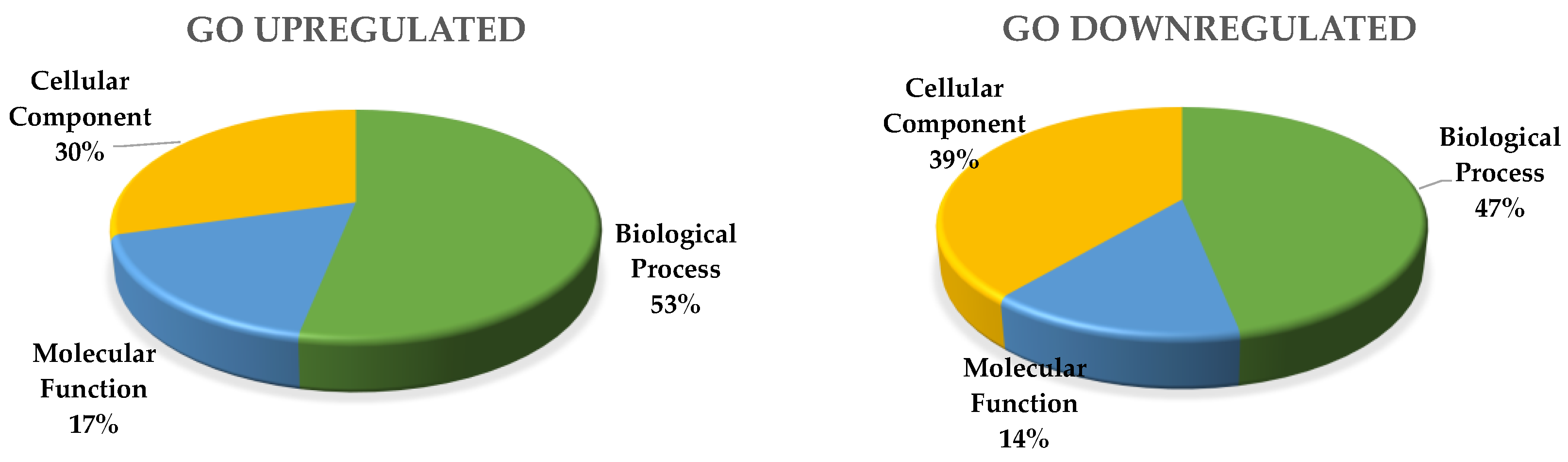
Gene Ontology (GO) enrichment analysis of up-regulated genes identified 2713 genes with annotated GO terms.
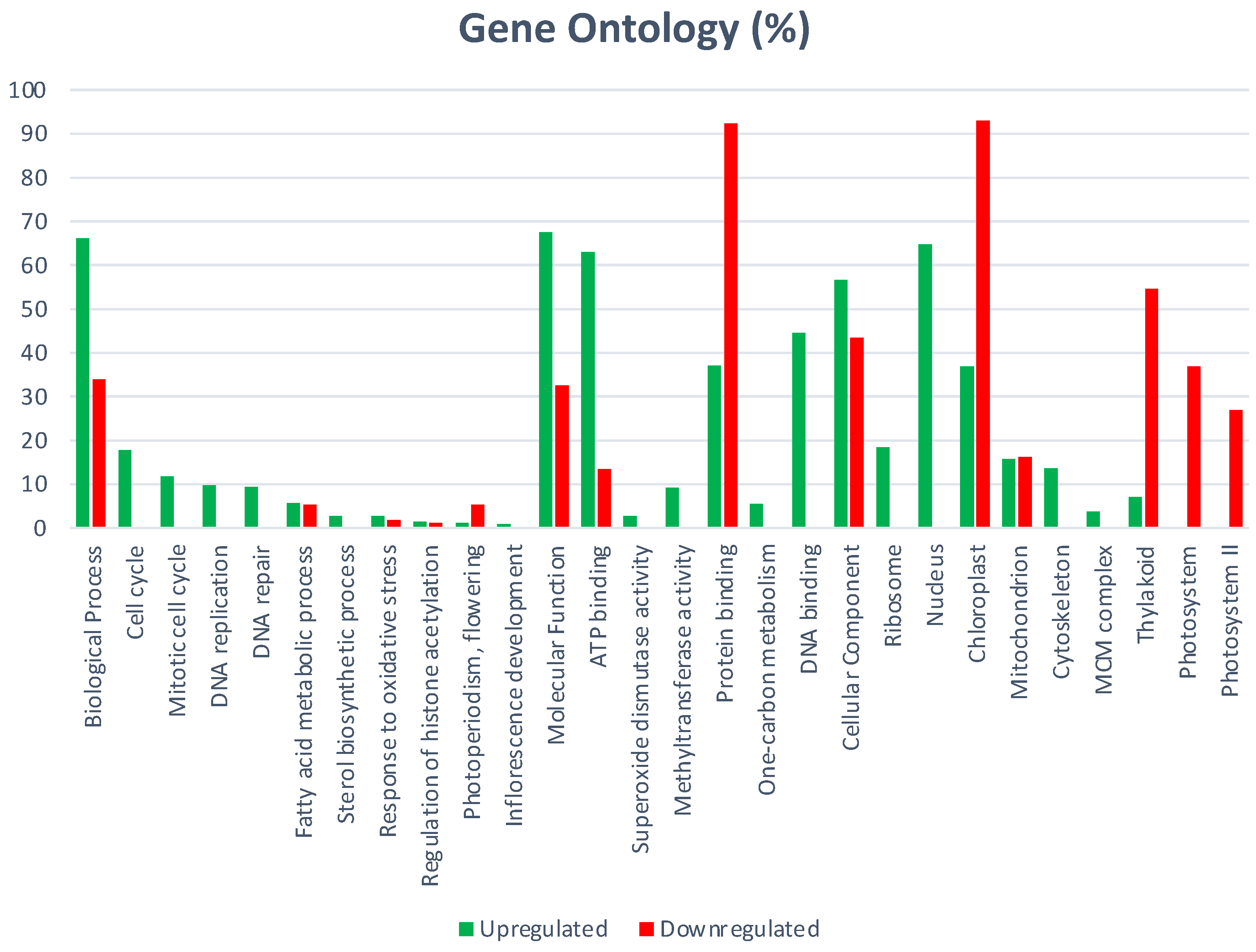
GO biological process includes 440 significantly enriched GO terms. Both up- and down-regulated biological process were related to protein phosphorylation, regulation of transcription, transmembrane transport, carbohydrate metabolic process, and proteolysis.
Specific up-regulated biological processes include: microtubule-based movement, translation, fatty acid biosynthetic process, lipid transport, DNA replication, gycolitic process, positive regulation of transcription, cell wall modification, DNA repair, defense response, carboxylic acid metabolic process, protein dephosphorylation, gluthatione metabolic process, and the amino acid metabolic process. Specific down-regulated biological processes include: photosynthesis, light harvesting, cell wall biogenesis, cellular glucan, and the xyloglucan metabolic process (Figure 1).

Figure 1.
Biological process regulated during MTs development of potato induced in dark.
GO cellular components include 82 significantly enriched GO-terms; 64 were up-regulated and those specific of MTs were: ribosome, nucleosome, microtubule, extracellular region, and cytoplasm. Specific down-regulated cellular components include: cell wall, extrinsic component of membrane, photosystem I and II (Figure 2).
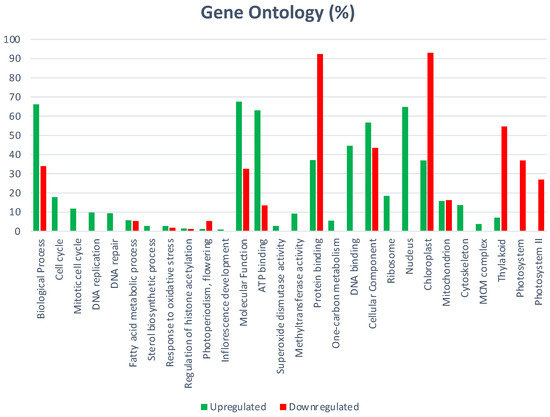
Figure 2.
Cellular components regulated during MTs development of potato induced in the dark.
GO molecular function includes 132 significantly enriched GO-terms; the seven main functional subgroups are: GO:0003824 catalytic activity (963), GO:0005488 binding (998), GO:00116491 oxidoreductase activity (243), GO:0043167 ion binding (604), GO:0016787 hydrolase activity, GO:0098772 molecular function regulator, and GO:0008092 cytoskeletal protein binding (67).
2.2. TFs
Overall, 112 up-regulated and 145 down-regulated TFs were found in the transcriptomic-wide analysis of MTs induced under dark conditions.
2.2.1. Up-Regulated TFs
Up-regulated TFs are related to C2H2-type zinc finger (20), F-box (15), bHLH (13), AP2 ERF (13), MYB (10), GATA (6), ZF-HD protein (3), Basic leucine zipper (5), bZiP (5), B3 DNA binding d2ggomain (5), NF-Y (4), WRKY (3), TCP (2).
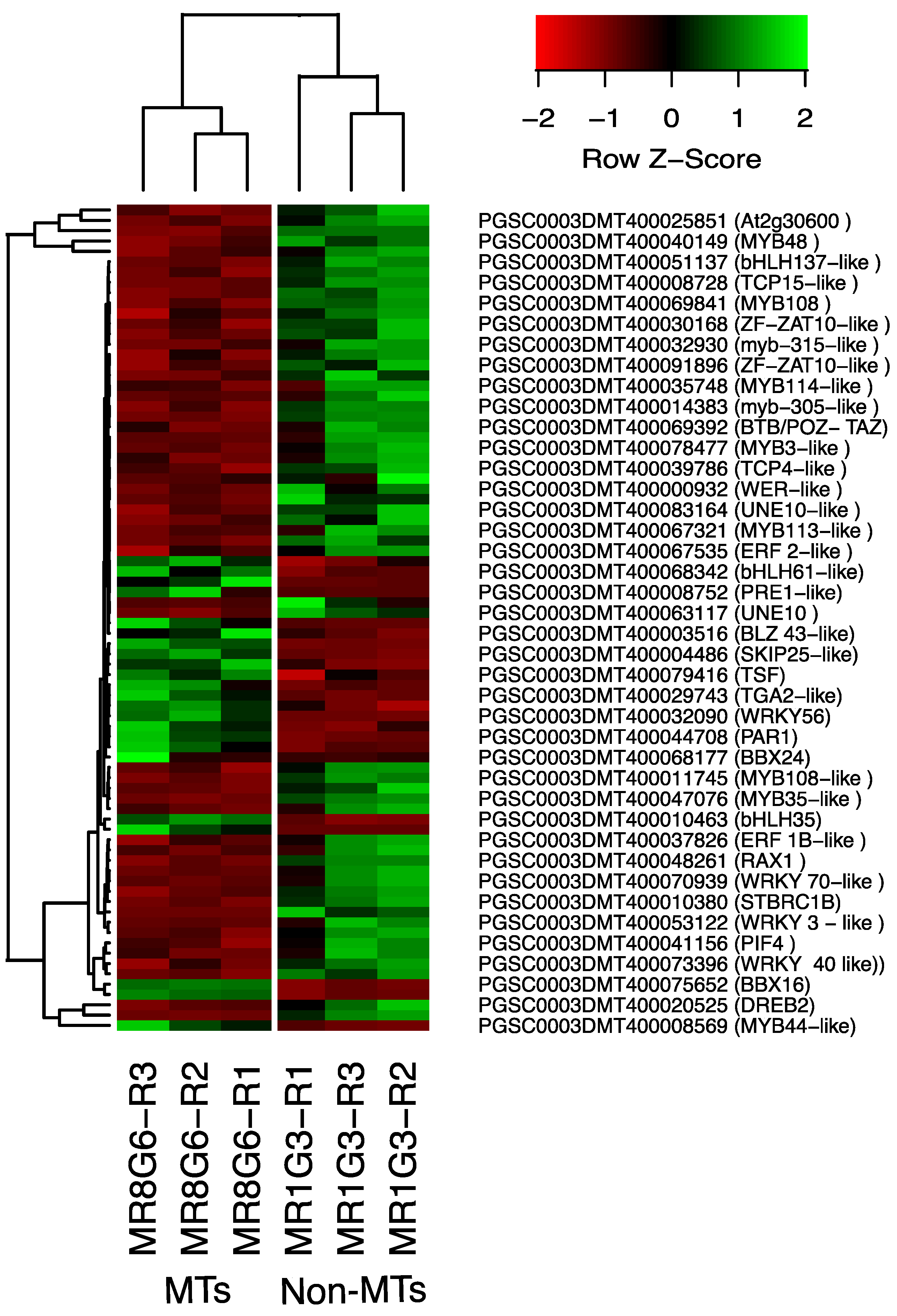
The PPI network of up-regulated TFs made on the STRING data base v11.5 with medium confidence revealed that 23 out of 112 TFs interact (Figure 3), while 14 interact with at least 1 TF. The list of up-regulated TFs interacting in the PPI network and levels of gene regulation are shown in the heat-map (Figure 4).
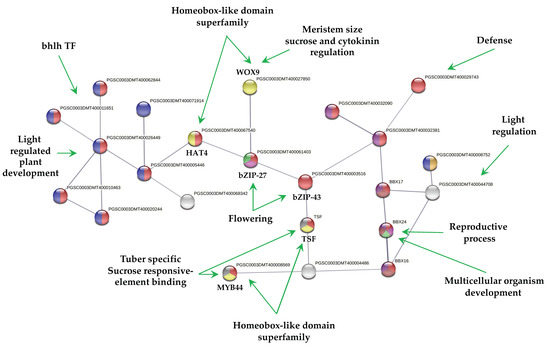
Figure 3.
PPI network of up-regulated TFs derived using STRING database from transcriptomic-wide analysis of potato S. tuberosum with medium confidence (0.400). The figure represents a full network; the edges indicate both functional and physical protein associations.
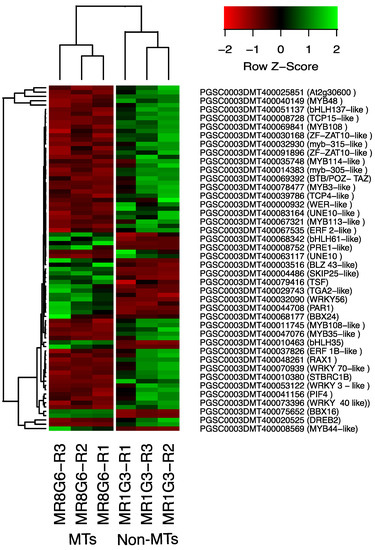
Figure 4.
Hierarchical clustering analyses (HCA) and heat map of up- and down-regulated TFs during MTs development under darks conditions, levels of up-regulation are present in Log2.
2.2.2. Down-Regulated TFs
Overall, 145 TFs were found to be down-regulated in MTs development. These TFs are related to bHLH (38), Homeobox (30), AP2/ERF (21), SANT/Myb domain (19), Myb domain (19), Zinc finger C2H2 superfamily (18), F-box (14), B-box-type zinc finger (11), WRKY (9), CCT domain (8), B3 DNA binding domain (7), Transcription factor TCP (6), BTB/POZ domain (6), and SKP1/BTB/POZ (6).
The PPI network of down-regulated TFs revealed that 58 out of 112 TFs interact (Figure 5), while 9 interact with at least 1 TF.

Figure 5.
The PPI network of down-regulated TFs derived using the STRING database from the transcriptomic-wide analysis of potato S. tuberosum with medium confidence (0.400). The figure represents a full network, the edges indicate both functional and physical protein associations.
The list of down-regulated TFs interacting in the network and levels of gene regulation are shown in the heat-map (Figure 4).
2.2.3. PPI Network of Up- and Down-Regulated TFs
The PPI network of up- and down-regulated TFs revealed both sets of TFs found interact in separated clusters–one forming MTs and the another corresponding to down-regulation TFs involved in shoot-stolon differentiation, gametophytes, plant organ development, and hormone signaling (Supplementary Figure S1).
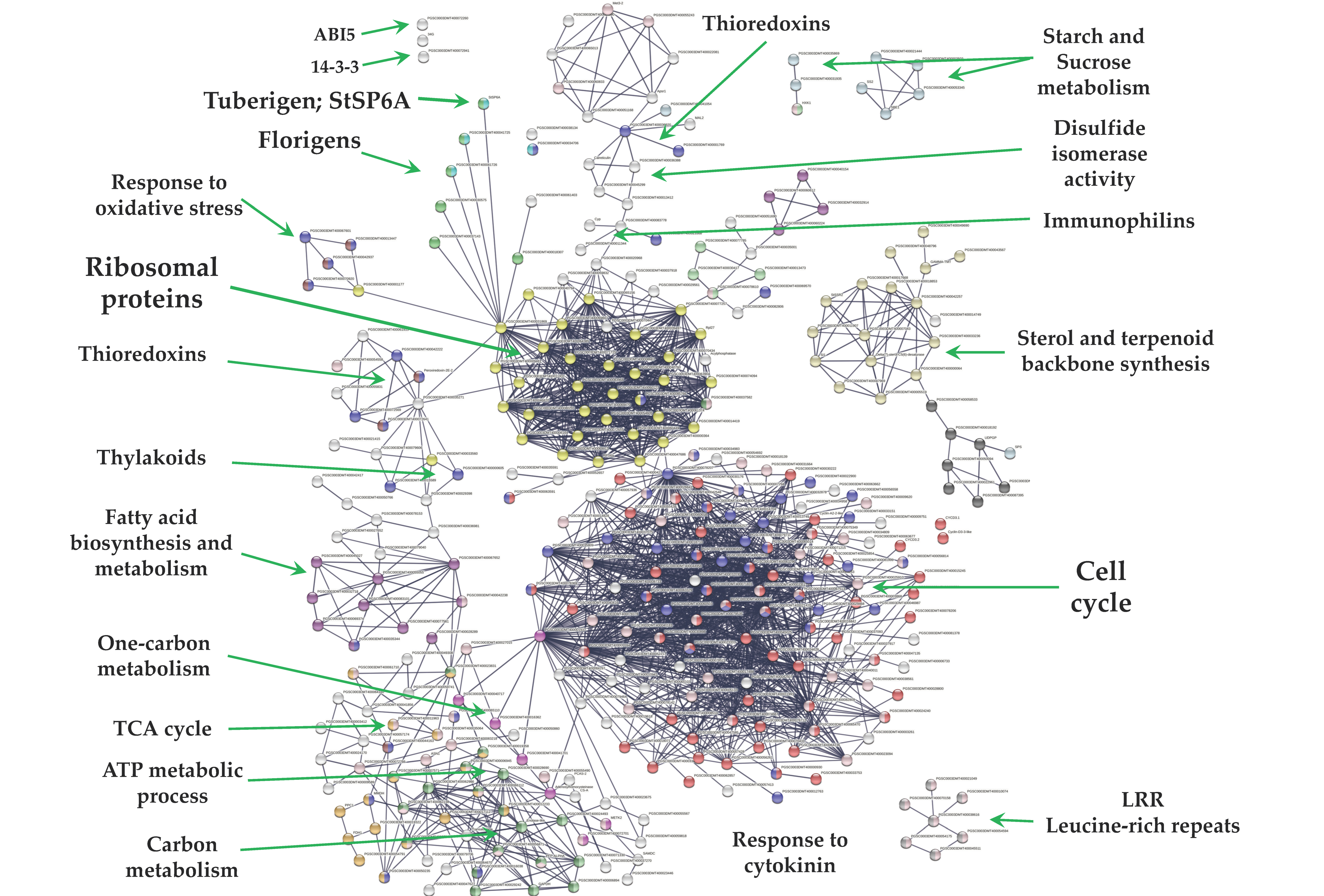
2.3. Up-Regulated Genes
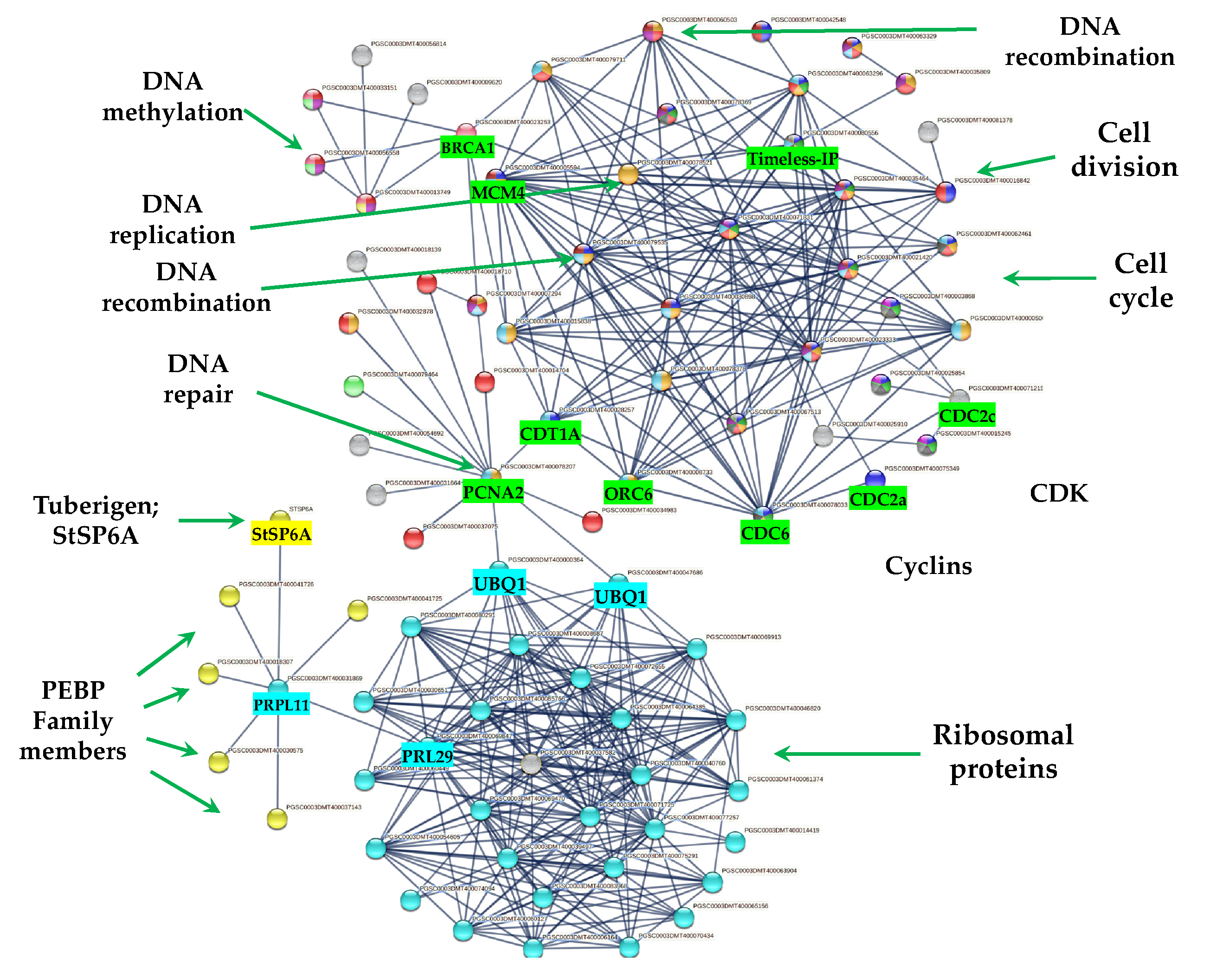
From 1699 up-regulated genes, 299 were tightly associated in two main clusters (Figure 6). Two fundamental processes for life were detected: RPs (29) and CC (117), interacting directly with proteins that sense the environment: osmotic stress (21), oxidative stress (9), CK response (23), one carbon metabolism (6), carbon metabolism (38), TCA cycle (16), acyl carrier proteins (6), fatty acid metabolism (14), thylakoid (13), redoxins (9), sulfur metabolism (8), disulfide isomerase activity (5), and immunophilins (12). Three clusters comprising 37 genes do not interact: sterol and terpenoid biosynthesis (16) and amino sugar and nucleotide sugar metabolism (7) (Figure 6).
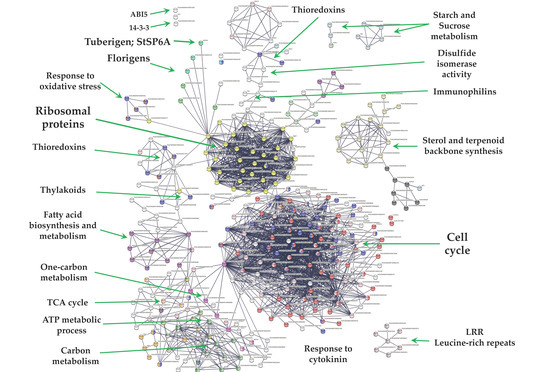
Figure 6.
PPI network of up-regulated genes derived from the STRING database of potato S. tuberosum from the transcriptomic-wide analysis with high confidence (0.860). Clusters are highlighted with the name of the function. The figure represents a full network, the edges indicate both functional and physical protein associations.
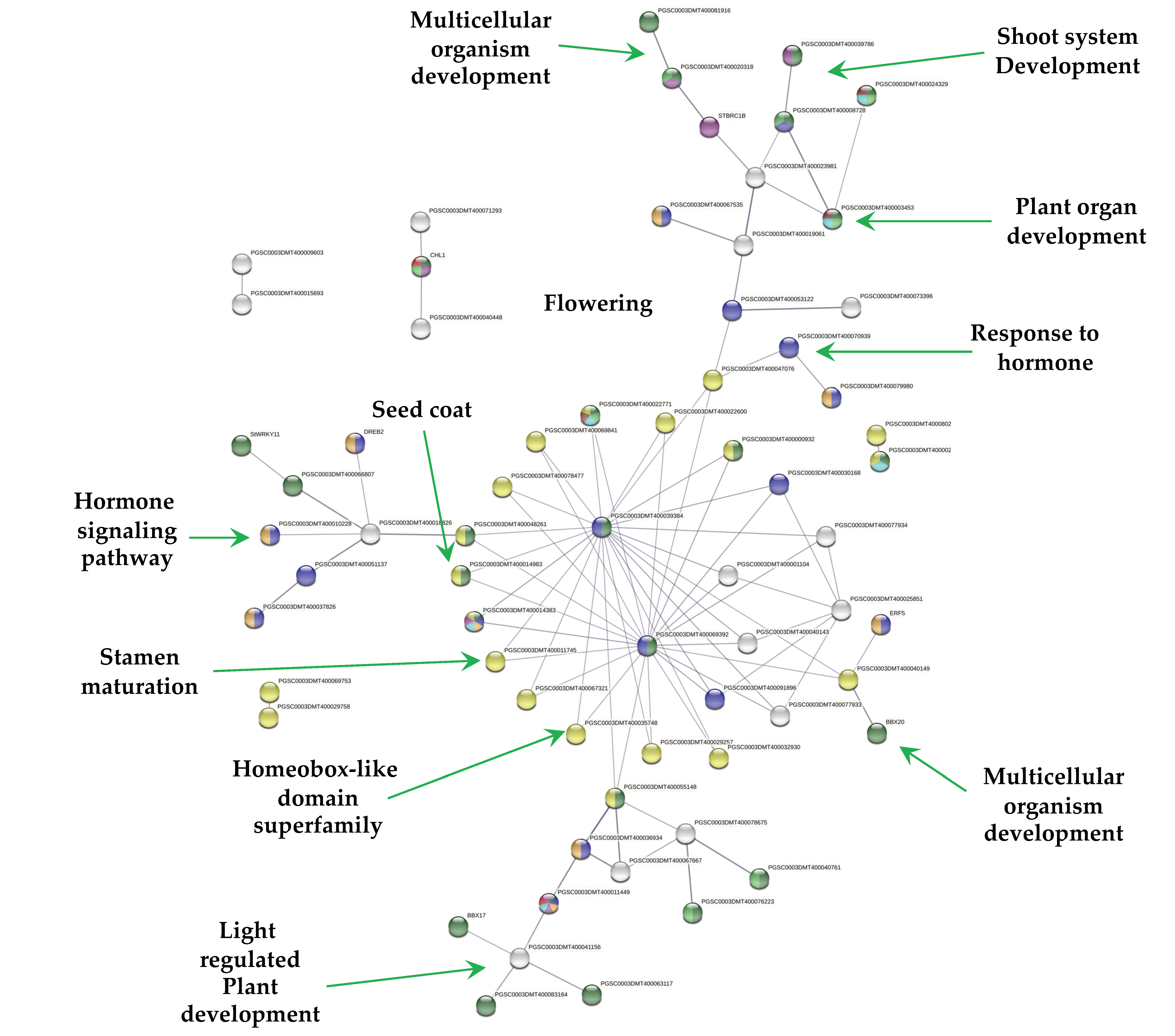
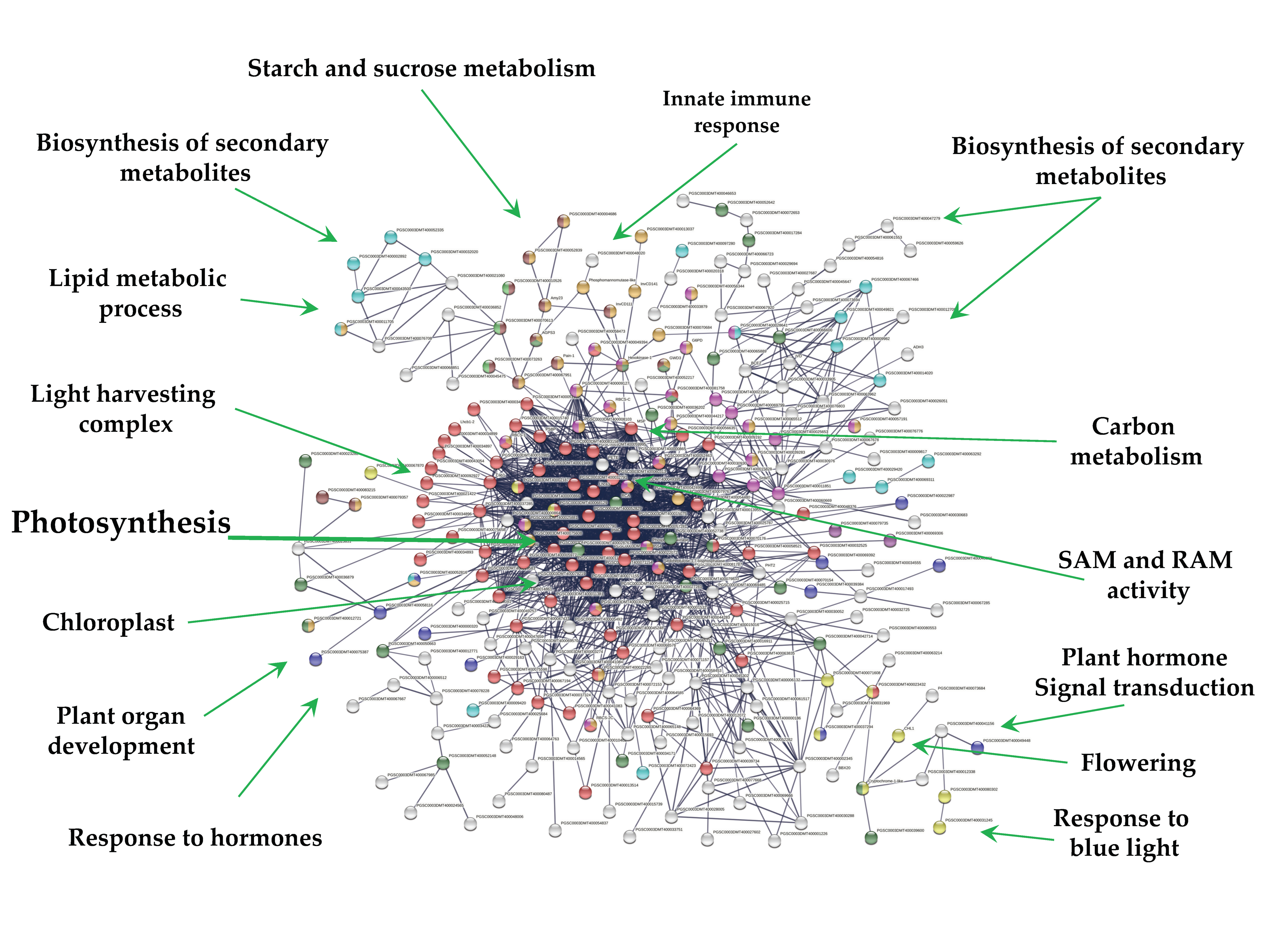
2.4. Down-Regulated Genes
Overall, 271 out of 1616 down-regulated genes were present in the PPI network with high confidence (0.700). Since MTs were induced in darkness, the network consists in one cluster related to photosynthesis (69) tightly interacting with several functions (Figure 7). These functions include; light harvesting complex (26), chloroplast (134), biosynthesis of secondary metabolites (65), carbohydrate derivative binding (27), cellular lipid metabolic process (17), starch and sucrose metabolism and glycosyl hydrolases (14), carbohydrate metabolic process (38), carbon metabolism (28), cellular lipid metabolic process (17), homeobox and calmodulin binding motif (2), response to hormone (11), flowering (1), plant hormone signal transduction (2), and response to blue light (10) (Figure 7).
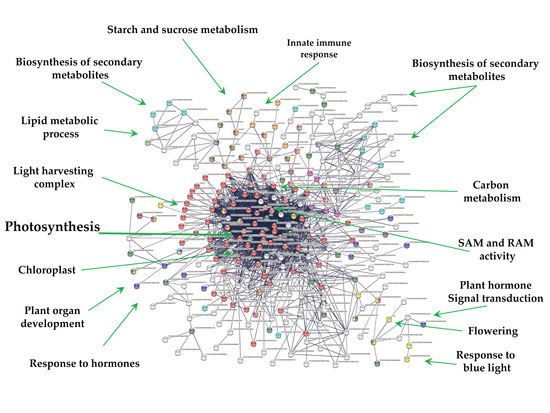
Figure 7.
PPI network of down-regulated genes derived from the STRING database of potato S. tuberosum from the transcriptomic-wide analysis with high confidence (0.700). Cluster is highlighted with the name of the function. The figure represents a full network, the edges indicate both functional and physical protein associations.
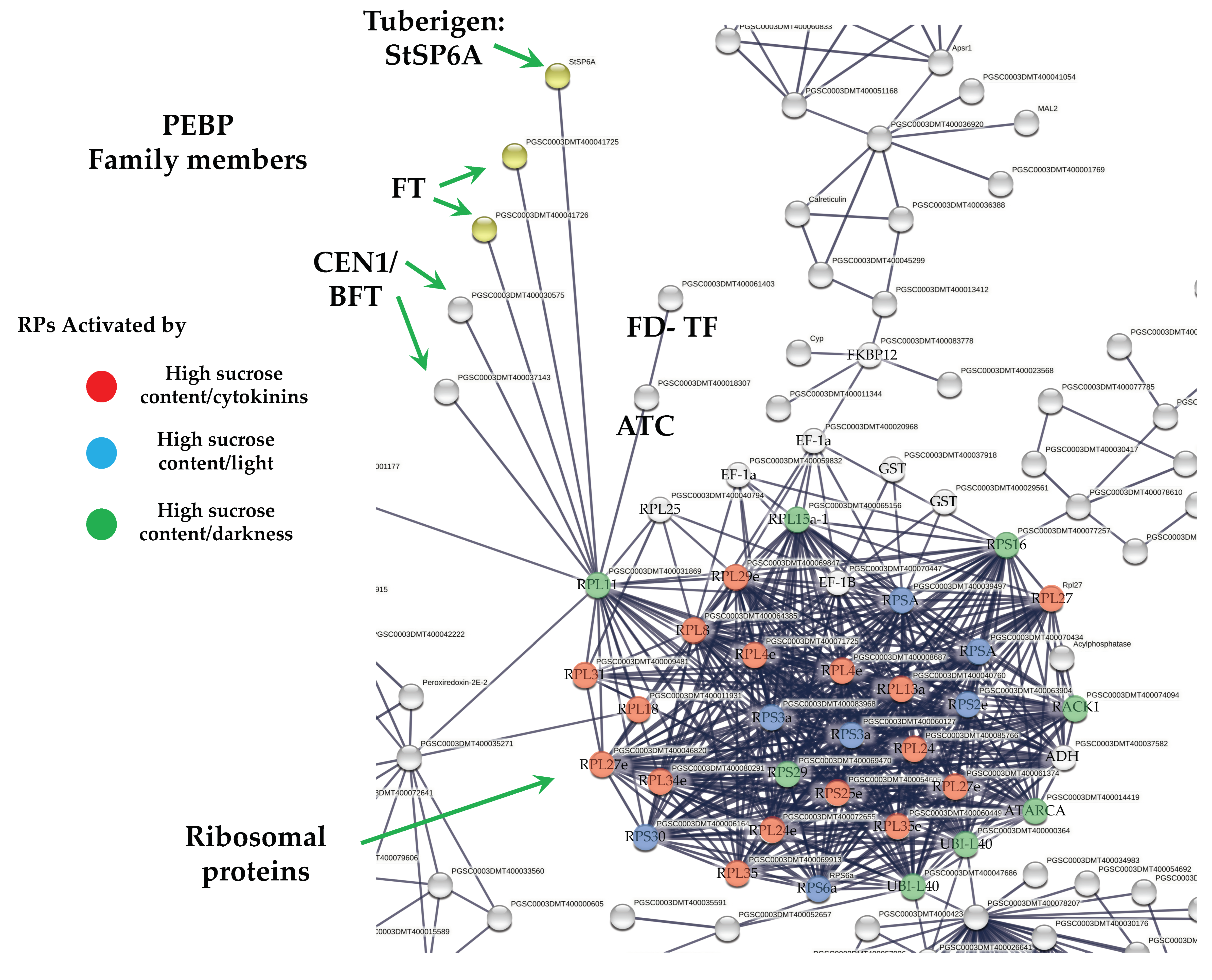
2.5. DEG Related to PEBP Family Members
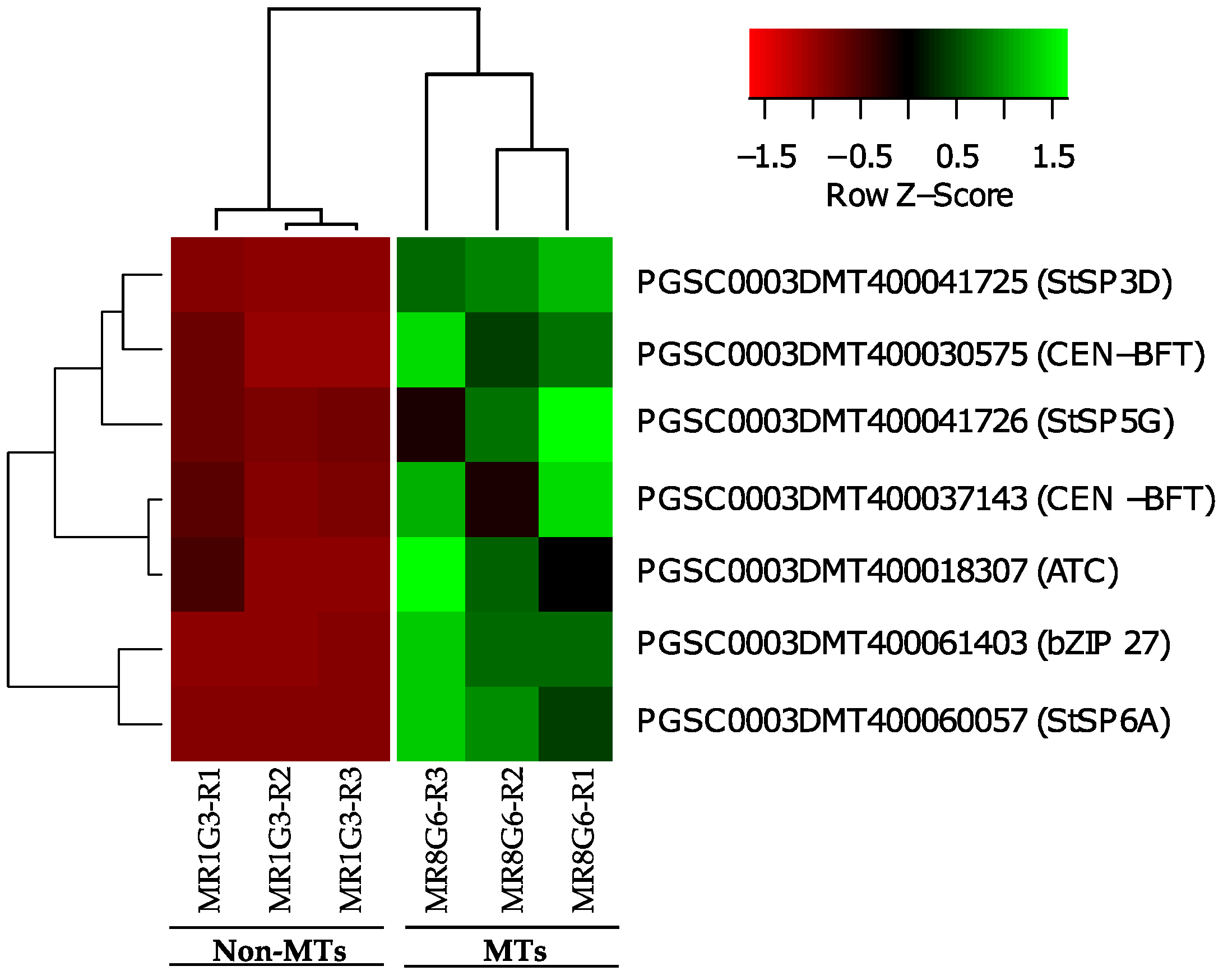
Overall, six PEBP family members were present in MTs development, three FT orthologs of PEBP family are present in potato, StSP6A, StSP5G (PGSC0003DMT400041726), and StSP3D (PGSC0003DMT400041725), two CEN1-BFT (PGSC0003DMT400030575), CEN1-BFT (PGSC0003DMT400037143), and a self-pruning ATC (PGSC0003DMT400018307). The TF FD (PGSC0003DMT400061403) a bZIP transcription factor-27 self-pruning was found interacting with ATC (Figure 8).
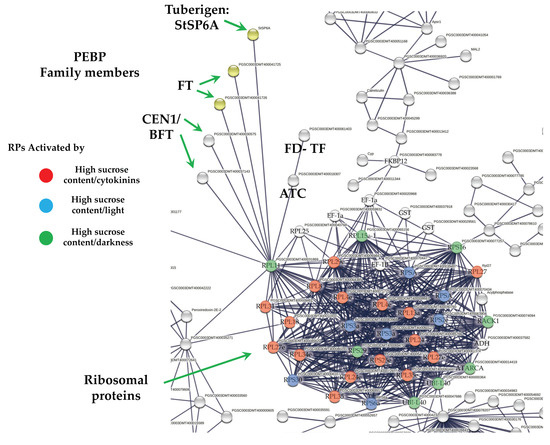
Figure 8.
PPI network of up-regulated PEBP family members and RPs cluster of potato S. tuberosum from the transcriptomic-wide analysis with high confidence (0.860). Selected proteins are highlighted with the name of the gene. RPs highlighted in red, blue, and green correspond to activated proteins by sucrose and CKs. The list of RPs and levels of gene regulation are shown in the heat map (Figure 9).
Gene ontology (GO) from functional annotation revealed a relationship between florigen genes present in the PPI network and RPs. bZIP27 FD (PGSC0003DMT400061403) was associated in 35 out of 59 GO-terms in which RPs were present (GO:0009889, GO:0009891, GO:0009893, GO:0010468, GO:0010556, GO:0010557, GO:0010604, GO:0010628, GO:0019222, GO:0031323, GO:0031325, GO:0031326, GO:0031328, GO:0048522, GO:0050789, GO:0050794, GO:0051171, GO:0051173, GO:0060255, GO:0065007, GO:0065009, GO:0080090, GO:2000112, GO:0005488, GO:0097159, GO:0098772, GO:0140110, GO:1901363, GO:0005622, GO:0005634, GO:0043226, GO:0043227, GO:0043229, GO:0043231, GO:0110165).
In the case of StSP6A, it was associated in 12 out of 51 GO-terms in which RPs were present (GO:0009987, GO:0050789, GO:0065007, GO:0005488, GO:0005622, GO:0005634, GO:0005737, GO:0043226, GO:0043227, GO:0043229, GO:0043231, GO:0110165).
StSP3D (PGSC0003DMT400041725 and PGSC0003DMT40004172a) were associated in 12 out of 51 GO-terms in which RPs were present (GO:0009987, GO:0050789, GO:0065007, GO:0005488, GO:0005622, GO:0005634, GO:0005737, GO:0043226, GO:0043227, GO:0043229, GO:0043231, GO:0110165).
CEN1/BFT/TFL1C genes (PGSC0003DMT400030575 and PGSC0003DMT400037143) were associated in 3 out of 17 GO-terms in which RPs were present (GO:0005622, GO:0005737, GO:0110165). The list of PEBP family members and levels of gene regulation are shown in the heat map (Figure 9).
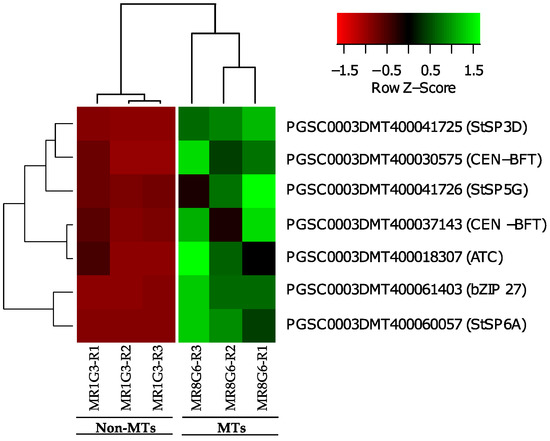
Figure 9.
Hierarchical clustering analyses (HCA) and heat map of PEBP family members during MTs development under darks conditions; levels of up-regulation are present in Log2.
2.6. DEG Related to RPs
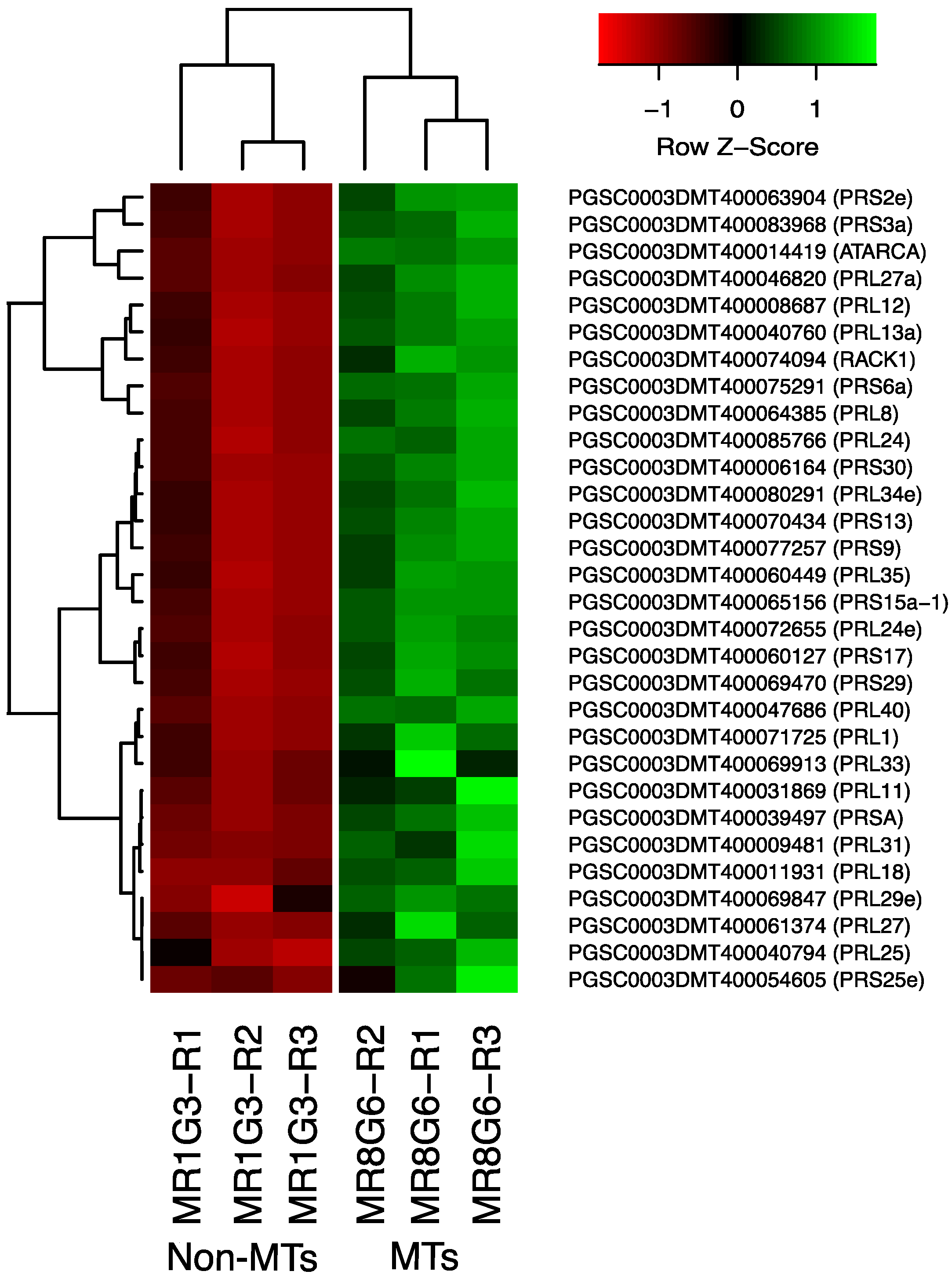
The PPI network analysis for the up-regulated transcripts corresponding to RPs in the established MTs developmental conditions yielded a cluster of 31 RPs interacting proteins, from which 11 proteins corresponded for the small subunit and 18 for the large subunit, three for eukaryotic elongation factors, two for WD40, RACK1, and ATARCA, and two for glutathione S-transferase genes (Figure 10). The architecture of the RPs cluster yielded in this analysis is quite similar to the evolutionary traits previously determined in PPI network analyses present in the Eukarya domain [19]. Moreover, in the PPI network analysis we were able to disentangle the protein interactions in four main categories. The RPs transcripts that were activated under high sucrose content conditions were highlighted in yellow. Those RPs transcripts that were activated under high sucrose content/darkness conditions, according to Gamm et al. [20], were highlighted in green. The RPs transcripts that were activated under high sucrose content/light conditions were highlighted in blue [20] as well as the RPs transcripts that were activated under high sucrose content/CKs according to Brenner and Schmülling [21]. We also found that among the RPs transcripts was present RPL29e (PGSC0003DMT400069847) that directly interacts with FKBP12 (PGSC0003DMT400083778) and RPL11 (PGSC0003DMT400031869), which in turn interacts with florigens and the thioredoxin-dependent protein (PGSC0003DMT400035271), the ubiquitin fusion protein, and the proliferating cell nuclear antigen (PGSC0003DMT400078207).
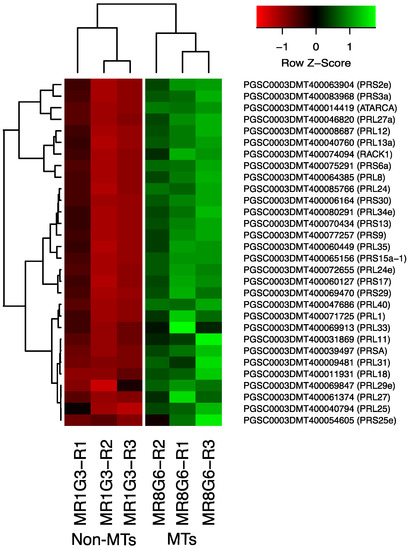
Figure 10.
Hierarchical clustering analyses (HCA) and heat map of RPs during MTs development under darkness conditions from levels of up-regulation are present in Log2.
2.7. DEG Related to CC
We found 117 up-regulated transcript genes directly related to the CC of potato MTs under darkness condition, from which 36 transcripts were directly related with mitotic regulation, 31 transcripts corresponded to E2F factors, 25 transcripts were related with cytoskeleton polymerization, 5 transcripts were related with motor proteins, 13 with epigenetic regulation through DNA/Histone methylation, and 5 with histone/nucleosome binding (Figure 11 and Figure S1, Table S1).
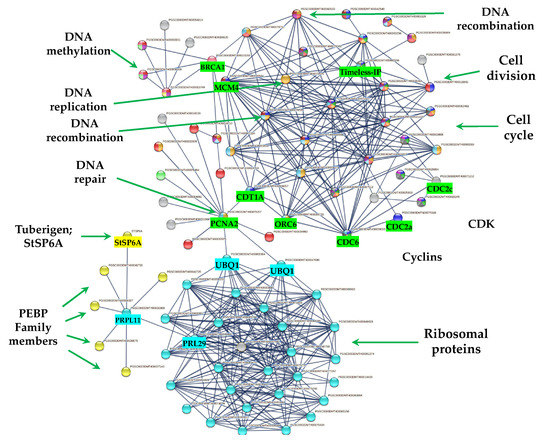
Figure 11.
PPI physical network of up-regulated transcripts genes of potato S. tuberosum from the transcriptomic-wide analysis with high confidence (0.860). Selected proteins are highlighted with the name of the gene.
2.7.1. Cyclins/CDKs
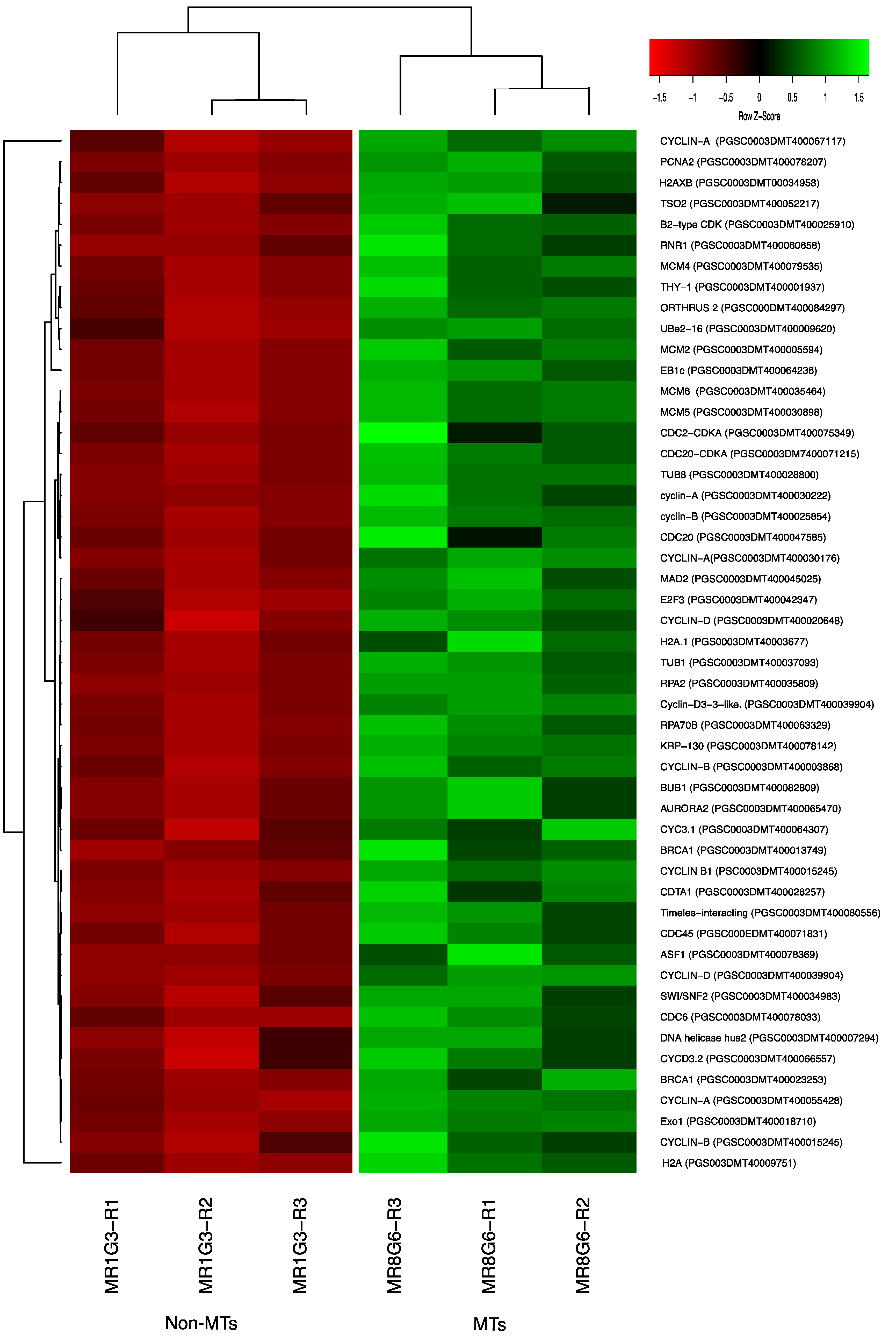
Two CDKs: CDC2-CDKA (PGSC0003DMT400075349) and CDC20-CDKA (PGSC0003DMT400071215) were found among the up-regulated transcripts in this work. The plant CDC2 is homologous to CDKA and in mammals to CDK1 and CDK2. CDKA function has been characterized as a regulatory protein at the G1/S and G2/M checkpoints. In addition, among the up-regulated transcripts related to Cyclins, we found 10 cyclins, which correspond to cyclin-A (four transcripts), cyclin-B (three transcripts), and cyclin-D (three transcripts). Furthermore, in the PPI network analysis the CDKA (PGSC0003DMT400071215) protein interacts with all the cyclins A and B in the network (Figure 11 and Figure S1, Table S1). The list of CC genes and levels of gene regulation are shown in the heat map (Figure 12).
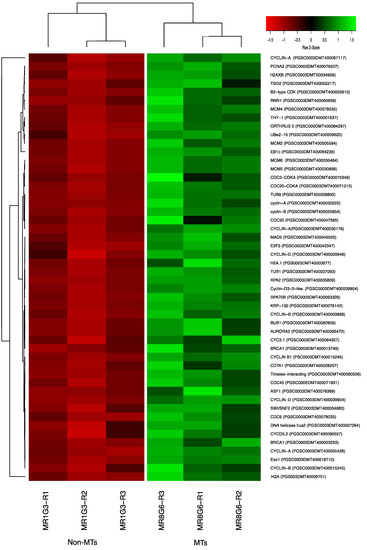
Figure 12.
Heat map of CC regulation proteins during MTs development under dark conditions; levels of up-regulation are present in Log2.
2.7.2. E2F Factors
In this work, we found up-regulated transcripts belonging to the E2F TFs family, which are directly involved in the regulation of the DNA replication in the onset of S phase. Among those transcripts, the PCNA2 (PGSC0003DMT400078207) interacts with E2F3 (PGSC0003DMT400042347), which in turn interacts with THY-1 (PGSC0003DMT400001937). The THY-1 interacts with LOG10, PGSC0003DMT400057413) which their transcriptional activation is CK-dependent. Further, PCNA2 interacts with SWI/SNF2 (PGSC0003DMT400034983), RNR1 (PGSC0003DMT400060658), and TSO2 (PGSC0003DMT400052217). The E2F3 transcription factor also interacts with BRCA1 (PGSC0003DMT400023253) (Figure S1, Table S1).
2.7.3. Histone Binding Proteins
Among the proteins present in the CC cluster, we also found three histone proteins: H2A (PGS003DMT40009751), H2A.1 (PGS0003DMT40003677), and H2AXB (PGSC0003DMT00034958). Those three proteins interact with the plant protein BRCA1 that in turn interacts with ORTHRUS 2 (PGSC000DMT400084297) and PCNA2 (PGSC0003DMT400078207). Moreover, ASF1 (PGSC0003DMT400078369) interacts with PCNA2 and with THY-1.
2.7.4. PPI Network Interaction of CC with RPs and Florigens
The PPI network analyses shed light on to protein interactions and allow a glimpse as to their biological and functional relationship. In this regard, we found that PEBP family members including the tuberigen StSP6A interact with RPs via RPL11 (PGSC0003DMT400031869), then with RPL29e (GSC0003DMT400069847) and UBQ1 (PGSC0003DMT400000364) within the cluster of RPs. UBQ1 interacts with the first protein of CC, PCNA2 (PGSC0003DMT400078207) (Figure 11). PCNA2 interacts with CDT1A (PGSC0003DMT00028257). CDT1A interacts with CDC6 (PGSC0003DMT400078033), then with CDC2a (PGSC0003DMT400075349), followed with CDC45 (PGSC000EDMT400071831) and with B1-type cyclin-dependent kinase (PGSC0003DM7400071215) interacting with CYCLIN B1 (PSC0003DMT400015245) and with B2-type cyclin-dependent kinase (PGSC0003DMT400025910) (Figure 11). In addition, CDT1A interacts with Exonuclease 1 (PGSC0003DMT400018710), with HUS2 (PGSC0003DMT400007294), then with BRCT domain BRCA1 (PGSC0003DMT400023253), with BRCA1 (PGSC0003DMT400013749), with Ubiquitin-conjugating enzyme e2-16 (PGSC0003DMT400009620), MCM4 (PGSC0003DMT400079535), then with MCM2 (PGSC0003DMT400005594), and with Timeless-interacting (PGSC0003DMT400080556) and two replication proteins RPA2 (PGSC0003DMT400035809) and RPA70B (PGSC0003DMT400063329).
2.8. DEG Transcripts with Biological Functions
The list of the transcripts genes found in the DEG analysis under MTs developmental darkness condition were directly related to cytokinin response, osmotic stress, one-carbon, carbon metabolism, glycolysis/gluconeogenesis, citrate cycle, fatty acid metabolism, thylakoids, acyl-carrier proteins, redoxins, response to oxidative stress, immunophilins, ribosomal proteins, sulfur metabolism, mitotic cell cycle, E2F activation, cytoskeleton, motor proteins, methylation, histone binding, sterol biosynthetic process and terpenoid backbone biosynthesis, amino sugar and nucleotide sugar metabolism, disulfide isomerase activity, LRR; leucine rich repeats, and sucrose metabolism, which are available in Table S1.
2.9. Validation of the Transcriptome-Wide Analysis
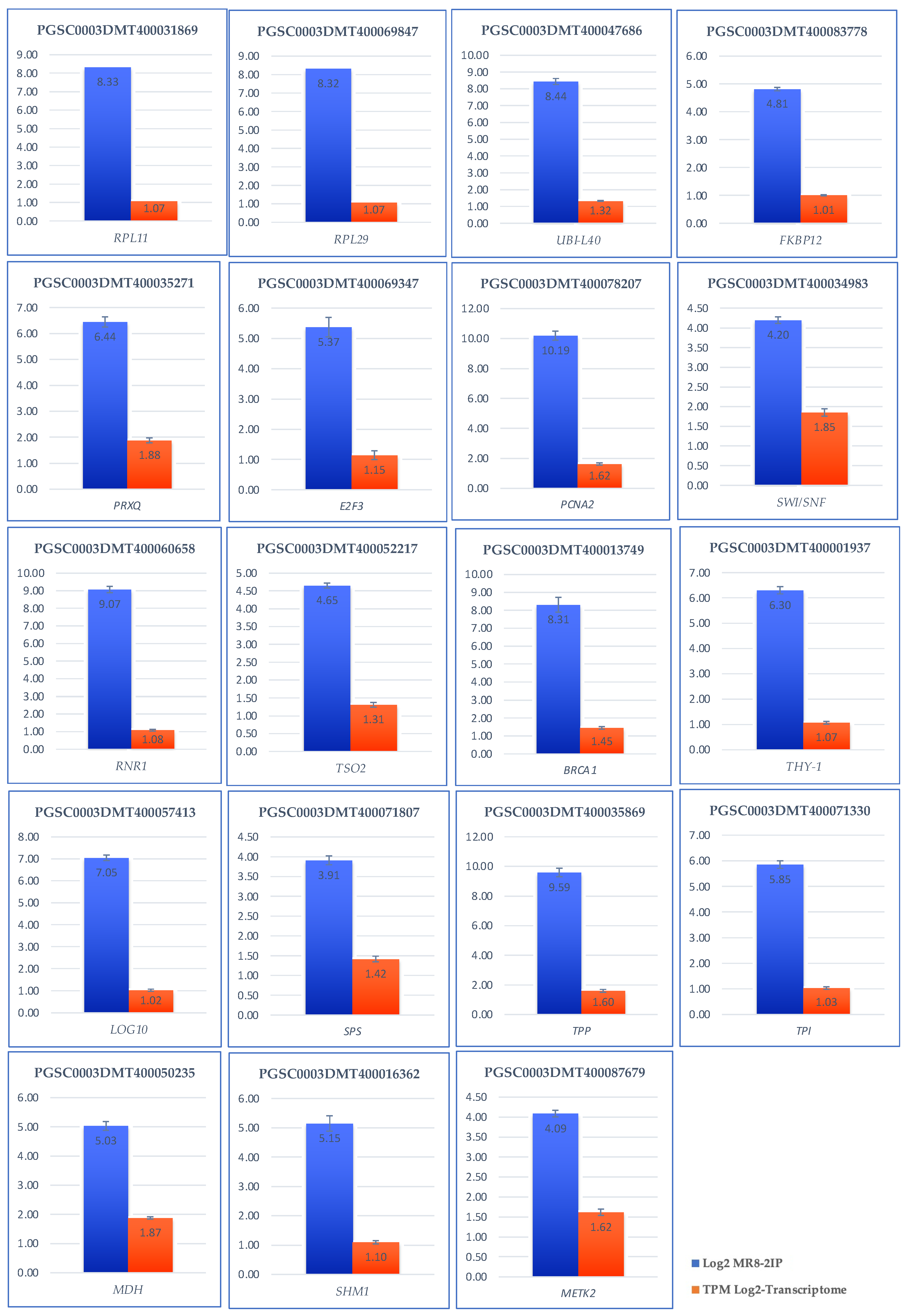
Validation of the transcriptomic-wide analysis was made by selecting 19 DEG and analyzing their regulation by quantitative reverse transcription PCR (qRT-PCR), using the primers described in Supplementary Table S1. The results are shown in Figure 13, indicating that the values are consistent with those obtained in the transcriptomic-wide analysis.
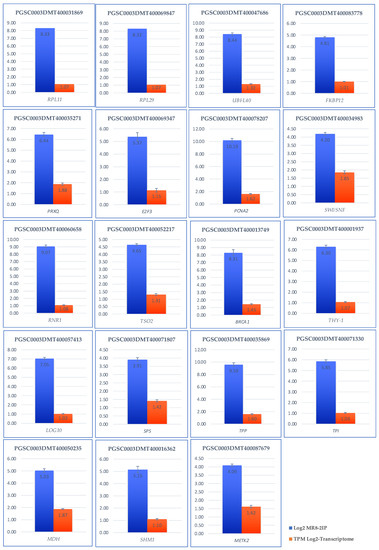
Figure 13.
Validation of the transcriptomic-wide analysis by quantitative reverse transcription PCR (qRT-PCR) of 19 DEG up-regulated genes involved in RPs, PEBP, CC, carbon metabolism of potato S. tuberosum. Blue columns correspond to absolute gene expression derived from the genome-wide analysis. The orange bars represent the expression of the transcriptome expressing the number of transcripts per million.
The genes selected to validate de transcriptomic-wide analysis of MTs development were: StSP6, ATC, UBQ1, FKBP12, RPL29, RPL11, PRXQ, MDH1, SHM1, METK2, TPI, TPP, SPP, BRCA1, RNR1, TSO2, THY-1, LOG10, SWI/SNF, PCNA2, and E2F3. EFa1, SEC3 were used as endogens.
3. Discussion
MTs development were induced from a stolon in medium containing sucrose 8%, gelrite 6 g/L, a CK 2iP 10 mg/L under darkness. In this environment, light modulating/repressing signals from leaves to underground were absent.
The molecular mechanisms of tuber development depend on genes related to light perception, temperature, and nutrient supply. These mechanisms have been associated mainly with phytochromes (PHYB and PHYF), homeobox TFs (BEL5 and POTH1), florigens belonging to the PEBP gene family (FT, StSP3D, StSP6A, StSP5G, TFL1), St14-3-3, StFDL1, TAC complex, StABI5 (StABL1), and several molecular regulators controlled under length day conditions (for review see [3,4,5]).
3.1. PPI Network of Up-Regulated TFs
TFs are proteins that control the transcription of genes by binding to a defined region of the genome. TFs are essential for the regulation of biological process, such as body planning, development, differentiation, and responses to various environmental signals [22].
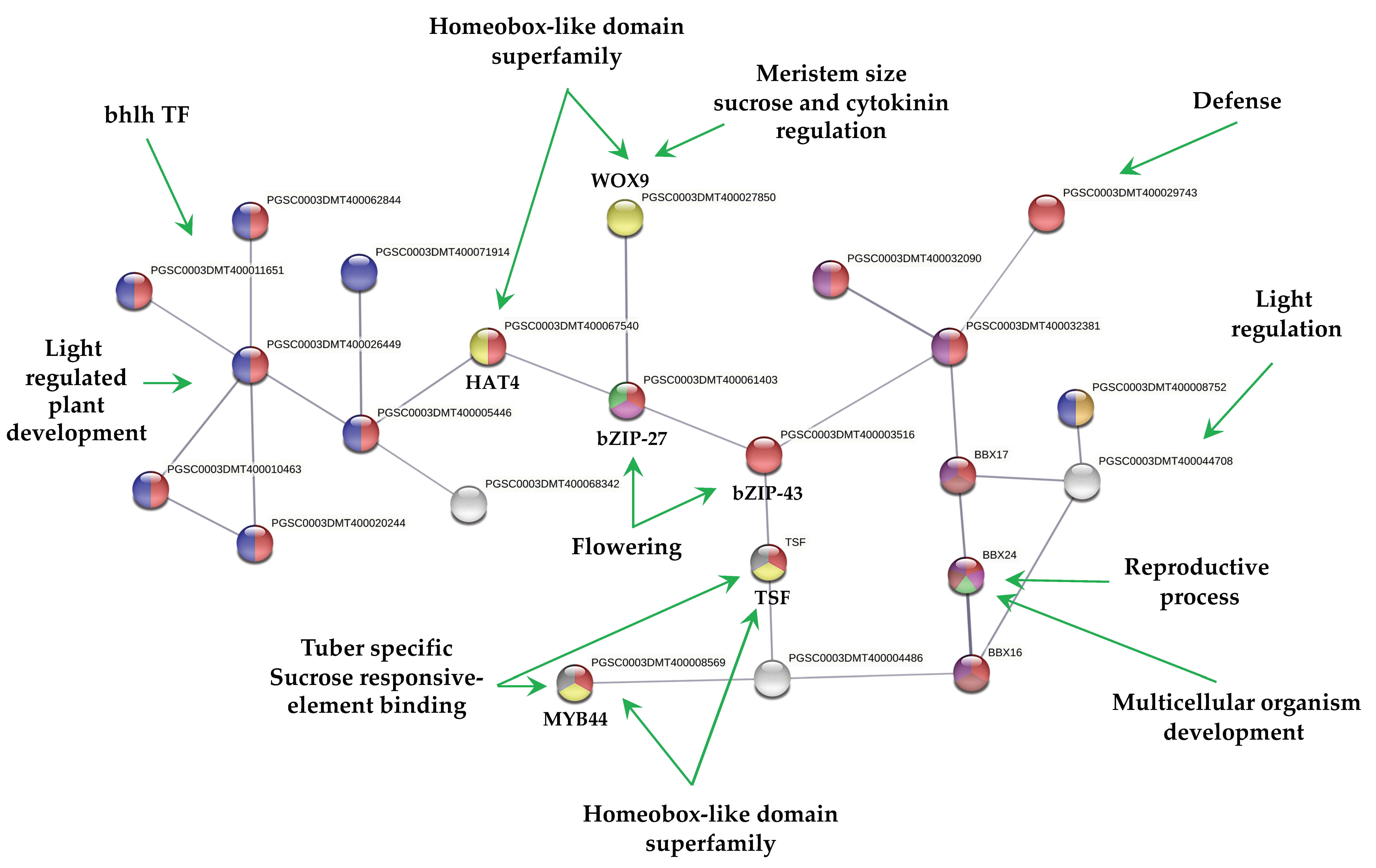
MTs development showed 23 up-regulated TFs interacting in a PPI network (Figure 3). The interpretation of the molecular mechanisms from a stolon to a tuber development in high content of sucrose and gelrite, 2iP as CK under darkness is as follows:
The MYB44 TF (PGSC000DMT400008569) a tuber-specific and sucrose-responsive element binding factor. MYB44 confers resistance to abiotic stresses dependent on ABA. Enrichment analysis with 50 interactors revealed that MYB44 is involved in auxin signaling pathways and tissue vascularization. MYB44 interacts with an AFR protein (PGSC0003DMT400004486), F-box/kelch-repeat skip25-like. AFR protein interacts with another tuber-specific and sucrose-responsive element binding factor TSF (PGSC0003DMT000794169). Enrichment analysis with 50 interactors revealed that TSF is involved in cell fate specification and commitment, multicellular organism development, cell cycle regulation, spliceosomal and ribonucleoprotein complex. TSF interacts with bZIP43 (PGSC0003DMT400003516); it is expressed in inflorescence meristem during petal differentiation and expansion stage. An enrichment analysis with 50 interactors revealed that bZIP43 is involved in histone modification, h3-k9 methylation, ribonucleoprotein complex, 14-3-3 interaction, and amino acid biosynthesis. bZIP43 interacts with a bZIP27-like, self-pruning TF (PGSC0003DMT400061403), required for positive regulation of flowering. Enrichment analysis with 50 interactors revealed that bZIP27 is involved in structural component of ribosomes, cell cycle regulation, cyrcadian rhythm, protein transport, methylation. bZIP43 interacts with a wuschel-related homeobox 9-like (WOX9), COMPOUND INFLORESCENCE (PGSC0003DMT400027850). WOX9 is required for meristem growth and development, and acts through a positive regulation of wuschel. Mutant phenotypes include embryo lethality, reduced root development, and smaller meristems. Phenotypes can be rescued by the addition of sucrose in the growth media. Overexpression can partially rescue the triple mutants of CK receptor phenotype, as a downstream effector of CK signaling. Enrichment analysis with 50 interactors revealed that WOX9 is involved in ribosome, cell cycle regulation, cell fate commitment, meristem growth and maintenance, translation initiation factor 4f, and flower development.
bZIP43 also interacts with a homeobox-leucine zipper protein HAT4 (PGSC0003DMT400067540). HAT4 acts as mediator of the red/far-red light effects on leaf cell expansion in the shading response. It is also involved in the negative regulation of cell elongation and proliferation processes, such as root formation and secondary growth of the vascular system. HAT4 TF interacts with 6 bhlh TFs. Enrichment analysis with 50 interactors revealed that HAT4 is involved in flower development, meristem maintenance, photoreception, sugar signaling, and abiotic stimulus.
The above information about PPI network of up-regulated TFs during MTs development may explain partially some molecular events required to develop MTs: vascularization (MYB44), proliferation process (HAT4), cell cycle, ribosomes (bZIP27), cell fate commitment and meristems (WOX9), light regulation (HAT4), and epigenetic regulation (bZIP43) present during MTs differentiation. The rest of TFs in the PPI network may participate during MTs development (Figure 3). Further analysis of each component is required to elucidate it.
3.2. PPI Network of Down-Regulated TFs
MTs development revealed that 57 down-regulated TFs interact in a PPI network (Figure 5). The morphological transformation from a stolon explant (modified stem) to a tuber must involve the down-regulation of TFs involved in stem, leaf, root, and flower development. From 57 down-regulated TFs, 14 TFs were related to shoot system development, 7 to flower and gametophyte development, 6 to phyllome development, 32 in response to hormone, and 23 in hormone signaling pathways.
The PPI network revealed a cluster of 18 homedomain TF superfamily/16 MYB interacting with two BTI TF (PGSC0003DMT400039384) and (PGSC0003DMT400069392) involved in gametophyte development. The MYB TF are involved in multicellular organism development, stamen maturation and dehiscence, seed coat, lateral organ boundaries, and root and xylem differentiation (Figure 5). The gametophyte cluster interacts with a light regulated plant development consisting in BBX17, PIL6 (phytochrome interacting factor), two FAMA TFs, involved in differentiation of stomatal guard cells, derepresses stem cell gene expression, and 10 TF required during fertilization of ovules by pollen.
Another cluster interacting with the gametophyte is composed by TCP TFs. These TFs are involved in leaf development, branching, circadian rhythm, embryonic growth, floral organ morphogenesis and hormone signaling. Recently, Nicolas et al. [23], demonstrated that the TCP TF BRC1b is a tuber repressor in aerial axillary buds, limits sucrose accumulation, promotes dormancy, abscisic acid response, and a reduced number of plasmodesmata. In addition, it interacts with StSP6A, and inhibits its tuber-inducing activity in aerial nodes. In our work, StBRC1b (PGSC0003DMT400010380) was found to be down-regulated and interacting within the network (Figure 5). StBRC1b interacts with TCP23-like and F-box/lrr-repeat MAX2 homolog a-like. A combined up- and down-regulated TFs networks revealed that StBRC1b interacts close to one up-regulated TF, the tuber-specific and sucrose-responsive element TSF (PGSC0003DMT000794169). STBRC1b interacts with MAX2 (PGSC0003DMT400020318) and KUF1 (PGSC0003DMT400004486), a karrikin protein that interacts with TSF TF, up-regulated in this work (Supplementary Figure S2).
Tang et al. [24], reported that the gene Soltu.DM.06GO25210, named by authors IT1, is identical to StBRC1b. When silenced by CRISPR/Cas, stolons were converted to branches instead of swelling with the capability of developing small tubers within time. StSP6A interacts with IT1 (StBRC1b), and authors suggest that it forms a complex during tuber initiation. They also found that there was an impaired interaction between the StSP6A gene from Etuberosum, a non-tuber producer Solanum plant species, due to the deleted fourth exon in Etuberosum. MAX2 is required for responses to strigolactones and karrikins as well as the repression of axillary shoots outgrowth. MEE1 is required during endosperm development in embryos.
3.3. Up-Regulated DEG PPI Network
3.3.1. PEBP Family Members and FD TF Interacting with RPs
The PEBP gene family is well-conserved across species from animals to plants, which play central roles in several biological processes. In Homo sapiens, PEBP1 (RKIP) has been implicated in several cancers, probably acting as a metastasis suppressor [25]. In addition, it has been proposed to be an active regulator for growth, transformation, differentiation [26], and a cell cycle modulator [27]. In flowering plants, PEBP gene family members have undergone duplication. PEBP are involved in determination of plant architecture. PEBP family consists of three groups: the FLOWERING LOCUS (FT)-like, the TERMINAL FLOWER1 (TFL1)-like, and CENTRORADIALS (CEN)-like. The MOTHER OF FT AND TFL1-like (MFT), and the BROTHER OF FT AND TFL1 (BFT)-like are present as a separate subclade within the CEN group [28]. In potato, multiple FT paralogues are related to both sexual and vegetative reproduction. SP3D, an FT orthologue, controls flowering, while SP5G and SP6A, two other PEBP genes, act as a repressor and inducer of tuber formation, respectively [29]. The balance between FT/TFL1 defines the plant growth habit as indeterminate or determinate by modulating the pattern of formation of vegetative and reproductive structures in the apical and axillary meristems [30]. In our study, six PEBP family members were transcriptionally active in MTs development in darkness conditions, such as three orthologs SP6A, StSP5G (PGSC0003DMT400041726), SP3D (PGSC0003DMT400041725), and two orthologs of CEN1-BFT (PGSC0003DMT400030575). In the PPI network analysis, we found that all PEBP protein family members interact with the universal plastid RPL11 (PGSC0003DMT400031869). PEBPs lack a DNA-binding domain but can exert the function as transcriptional co-regulators through complexing with TFs, regulating the expression of downstream target genes [31]. The TF FD (PGSC0003DMT400061403), a bZIP transcription factor-27 self-pruning was also found interacting with ATC (Figure 8).
Purwestri et al. [32] and Wang et al. [33], through a yeast two-hybrid screening approach, found that SP6A can interact with RPs and other proteins involved in protein synthesis, RNA and DNA binding proteins, histones, initiation factors, signaling and carbon metabolism, and regulate flowering. In addition, the PPI network analysis unveiled 77 interactors, widely distributed throughout the cells in nearly all subcellular components, including plastids, ribosomes, thylakoids, cytosol, and chloroplasts. SP6A functions as positive regulator of tuber development under high contents of sucrose [30,34] and represses bud flower formation under short day conditions [35]. It is involved in several pathways during tuberization, including temperature, microRNA, and photoperiod [36,37,38]. Several transcriptomic analysis of potato S. tuberosum have revealed the presence of RPs during the tuberization process [12,15,16,39,40]. In Sharma et al. [15], we found through PPI network analysis with the highest confidence (0.900) that RPs proteins interacted with SP6A, RP40SA, RP60S, RPS8, RPS4A, RPL10, BEL5, and RPL14.
The ABI5-like 1 (StABL1) bind to SP6A in a 14-3-3 manner forming an alternative tuberigen activation complex (aTAC), thereby resulting in early tuberization [41]. In our study, the transcript for abscisic acid-insensitive 5-like protein (PGSC0003DMT400072260) was up-regulated; it did not interact directly with SP6A in the PPI network analysis, even using the lower confidence (0.150) value. This suggest that a 14-3-3 protein is essential to establish a protein interacting complex with SP6A. In our study, the 14-3-3, (34G; PGSC0003DMT400079172) transcript was up-regulated, and its protein interacts with SP6A, using a high confidence (0.780) value. It is very likely that SP6A and 14-3-3 might be forming a complex under MTs darkness conditions. 34G (14-3-3) is involved in cell cycle regulation, mitotic cycle, ribosome biogenesis, response to stress, starch and sucrose metabolism, reproductive process, and basal transcription factor [42,43].
CENTRORADIALS (CEN) represses the initiation of floral meristems in several species. The DHvCEN5 barley plants developed spikelet initiation in both photoperiods; GO enrichment analysis demonstrate that HvCEN had a negative impact on to chromatin modification, cell cycle regulation, CK signaling, and cell growth through RPs [44]. A member of the TERMINAL FLOWER-1/CENTRORADIALS gene family (CEN) has shown to function as a negative regulator of tuberization. At the protein level, CEN competes with SP6A for the interaction with other members of the TAC complex [45]. Moreover, in potato plants, CEN overexpression induced a lower rate of sprout growth, lower content of CKs, and higher content of abscisic acid (ABA) [46].
Another transcript for a FT was found in this work–the SP5G (PGSC0003DMT400041726) which was up-regulated. The function of this FT is to blockade the tuberization process. In the absence of light/darkness, SP5G might not repress SP6A. In addition, it has been shown that PHYB and PHYF co-stabilized CONSTANS-like 1 protein (COL1), binds SP5G, and represses SP6A and tuberization [47]. We also found that the transcript for SP3D was up-regulated and that this protein is involved in flowering, and it has been shown that florigens can promote tuberization; therefore, the SP3D might be able to promote this process [29,48].
3.3.2. RPs Cluster
Several transcriptomic analyses in S. tuberosum have revealed the transcriptional activation of the RPs coding genes in the tuberization process [12,15,16,20,39,40]. Specifically, the work of Taylor et al. [49] found a 15-20-fold increase in transcript levels of two RPs, S19 and L7, during the early stages of tuberization in stolon tips. Moreover, the RP proteins have been directly related with growth, development, embryo viability, flowering, and leaf development [50,51,52,53,54,55,56].
Sucrose Activation of RPs
The up-regulation of RPs in Arabidopsis is activated by different biotic and abiotic factors [57,58,59,60,61,62,63] such as sucrose [20,64,65,66,67] and CKs [21].
In our study, a high content of sucrose (8%), a CK 2iP 10 mg/L, and osmotic stress conditions were applied under darkness conditions. Hummel et al. [67] found that in response to 6% sucrose feeding, 166 out of 204 RPs were up-regulated. In our study, 31 RPs were detected, and according to the data obtained by Gamm et al. [20] who applied 5% of sucrose, we obtained 14 RPs activated in dark conditions and 12 with similar in light and dark RPs activated by sucrose.
CK Activation of RPs
In our study, 12 transcripts for RPs were directly related to CKs activation [21], 12 transcripts for RPs were transcriptionally activated by both sucrose and CK treatment. In Arabidopsis, 87 transcripts for RP of the 474 identified responded specifically to CKs; CK-responsive RPs related genes encode proteins for both plastidic and cytoplasmatic localization [21].
CKs binds directly to ribosomes in higher plants with high affinity [68,69]. CKs alter the polyribosome formation, ribosomal protein synthesis, number of ribosomes, and the phosphorylation status of ribosomal proteins [70,71,72,73,74,75,76,77]. In Arabidopsis, Karunadasa et al. [74] demonstrated that protein synthesis is induced by CKs and CK signaling and requires isoforms of the RP L4. The loss of function of RPL4 increases the tolerance to osmotic stress and decreases sensitivity to CK-induced growth. In our study, 12 RPs were activated by CKs [21], and 12 RPs were activated by both sucrose and CKs treatment. In Arabidopsis, 87 out of 474 respond to CKs [21]. The CK-responsive genes encode proteins of both plastidic and cytoplasmatic localization.
Stress Activation of RPs
The expression levels of RPs are altered under stress conditions [57,58,61,62,63,75], suggesting that it participates in signal detection and stress response. In rice, RPs large subunits are water and salt stress responsive proteins [61,62,63]. In maize, ribosome footprints from chloroplast and cytosol have been analyzed [76] and provided insights into the dynamic translation mechanisms during drought [77] and viral infection [78]. In cotton, RPL14-2 interact with several RPs to activate drought and salt tolerance [79]. In tea (Camellia sinensis L.), RPL32 is up-regulated in drought stress [80].
RPs and Their Relationship with CC
In Saccharomyces cerevisiae, the repression of nine RPs 60S genes (L4, L7, L18, L3, L17, L28, L35, L37, and L40) resulted in G2/M phase arrest. The repression of nine other 60S RPs (L1, L3, L9, L16, L19, L21, L25, L30, and L43) and 22 40S genes (S0, S1, S2, S3, S4, S5, S6, S9, S10, S13, S14, S15, S17, S19, S20, S21, S22, S26, S27, S29, and S30) caused arrest in the G1 phase [81].
Overall, 14 of 38 proteins analyzed in yeast were present in the potato RPs network: RPL1, RPL18, RPL25, RPL35, RPL40, RPS2, RPS3, RPS6, RPS9, RPS13, RPS15, RPS17, RPS29, and RPS30. Accordingly, they could contribute to the activation of CC during potato MTs development.
Maitra et al. [82] demonstrated a relationship between RPs, CC, and one-carbon metabolism. Yeasts regulate the one-carbon flow balance between donors by controlling the activity of cytoplasmic serine hydroxymethyl transferase (SHM2P) [83]. In cells lacking certain ribosomal proteins, SHM2 translation efficiency and Gly:Ser ratios were found to be low. These observations are consistent with low flow through the 1C pathways.
We proposed that RPs present in our analysis, in addition to being essential in the progression of CC in MTs development, could also participate in the one-carbon pathways that ultimately comprise a series of interrelated metabolic pathways that include the methionine and folate cycles–critical for cell function–providing 1C units (methyl groups) for the synthesis of DNA, polyamines, amino acids, creatine, and phospholipids [84].
3.3.3. CC Cluster
Tuber development relies on the temporal and spatial control of cell proliferation and cell growth to correctly develop an organ that will help the potato plant to survive under adverse conditions. Potato tubers develop from stolons, from which swelling growth occurs in the subapical region of the meristem. Initial expansion and radial cell division of cells in the pith and cortex, in combination with restricted longitudinal growth is followed by cell division and enlargement in random orientations in the perimedullary region once the swelling has reached a diameter of 2 to 4 mm, continuing until the tuber reaches its full size. In this study, genes involved in CC regulation were present, cyclins/CDKs, E2F factors and histone binding proteins.
Cyclins/CDKs
Overall, 2 cyclin-dependent kinase (CDKs); CDC2 (PGSC0003DMT400075349) and CDKB1;2 (PGSC0003DMT400071215), 10 cyclins: 4 cyclin-A, 3 cyclins-B and 3 cyclin-D were found. CDKB1;2 (PGSC0003DMT400071215) interacts with cyclins A and B present in our network. CKs in the presence of sucrose activate the expression of cyclins-D that have been shown to be essential in CC regulation. Yang et al. [85] demonstrated that CKs (Zeatin, BAP, and 2iP) regulate cell cycle by promoting the nuclear shuttling of the MYB-domain protein 3R4. MYB3R4 binds 5′-AACGG-3′ motifs of promoters of G2/M-specific genes and E2F targets. In our analysis, a gene homologous of MYB3R4 was found (PGSC0003DMT00047825). In accordance, it is very feasible that the potato MYB3R4 activate the cell cycle regulation genes during MTs development.
Rhee et al. [86] analyzed cyclins-D gene expression in potato exposed to different light regimes, hormones (CKs and auxins), and carbohydrates (SUC and glucose) and found that StCYCD3;1 and StCYCD3;3 were more expressed in above-ground organs whereas StCYCD3;2 was expressed more abundant in undergrounds organs. The above expression levels were when cytokinins like zeatin and BAP combined with sucrose induced higher expression compared to 2,4-D. In our work, StCYCD3;2 was found interacting within the PPI network with a cyclin-dependent kinase B2 (CDKB) (PGSC0003DMT400025910). The levels of expression found were similar; StCYCD3;2 was 3.5, while StCYCD3;1 was 3.03, and StCYCD3;3 was a 2.85 fold change.
In H. sapiens, PEBP1 (RKIP) is a metastasis suppressor in aggressive cancers [25] and cell cycle modulator [27]. Silenced-RKIP accelerate DNA synthesis and G1/S transition entry by inducing the expression of CDC6, MCM2,4,6,7, cyclinD2, D1, and E2. It also enhances G2/M transition and down-regulates G2/M checkpoints like AURORA-B, cyclin G1, and slow G2/M transition time. This led to a higher proliferation rate compared to control cells [27].
In our work, the components of G2/M checkpoints were present, showing similarities with the function of PEBP1(RKIP) in H. sapiens.
The CC regulation of mitosis is summarized as follows: The CYCB-CDKA complex (PGSC0003DMT400071215) interacts with MAD2 (PGSC0003DMT400045025) (mitotic arrest deficient), MAD2 interacts with CDC20 protein (PGSC0003DMT400047585) (APC metaphase/anaphase complex). MAD2 and CDC20 are checkpoint proteins locally catalyzed by kinetochores, the central player in the distribution of genetic material to the daughter cells during mitosis [87].
MAD2 interacts with EB1c (PGSC0003DMT400064236) (microtubule stability). MAD2 interacts with TUB1 (PGSC0003DMT400037093) (tubulin formation). In Arabidopsis, TUB1 expression is linked to light induction [88].
In hypocotyl explants, the TUB1 highest expression levels are found in dark conditions. The TUB1 transcriptional behavior in previous reports is in accordance with our data, since our work was carried out in dark conditions [88].
MAD2 interacts with TUB8 (PGSC0003DMT400028800). During the initiation of tuber formation, there are changes in the expression of tubulins at the tip of the stolons [87].
MAD2 interacts with AURORA2 (PGSC0003DMT400065470), a serine/threonine protein kinase. It is abundant in tissues with high division, such as roots and flowers, but low in leaves and stems. In mitosis it is intimately linked with the cytoskeleton (pre-prophase band, phragmoplast, metaphase plate) required for cytokinesis [89].
MAD2 interacts with KRP-130 (PGSC0003DMT400078142). KRP-130 are involved in microtubule dynamics, morphogenesis, chromosome segregation, organelle transport, and vesicles [90].
KRP-130 interacts with BUB1 (PGSC0003DMT400082809); BUB1 binds to the kinetochore and has an important role in establishing the mitotic spindle checkpoint and chromosome aggregation [91].
BUB1 interacts with the CDKA-CYCA complex (PGSC0003DMT400071215). BUB1 interacts in the same way with the complex of CDKA-CYCA. CDKA (PGSC0003DMT400071215) interacts with four B cyclins. CDKA has been identified at the G1/S and G2/M checkpoints [92] and is modulated by cyclins A and B [93].
E2F Factors
The E2F proteins form a family of TFs that regulate the transition from the G1 to the S phase in the cell cycle. We found 31 E2F factors in this analysis. The activation of genes such as PCNA2, RNR1, TSO2, THY-1, SWI/SNF2, ORC6, ORC4, ORC5, SPT16, MCM6, SMC3, MCM2, CHR11, POLA2, and PSF2 demonstrates that DNA replication was carried out correctly, validating that the conditions evaluated are optimal for the development of MTs. Abnormal induction of the S phase re-entry in mature leaf cells is observed in overexpression of E2F in Arabidopsis [94]. Unregulated activity of this family of TFs has been characterized broadly in human cancers [95].
Abiotic stress is perceived through SWI/SNF2 and its interaction with PCNA2. CKs are perceived through the activation of LOG10 to the synthesis of dihydrofolate reductase. Key proteins were found during mitosis, such as those of the cytoskeleton, microtubules, tubulins, motor proteins, and chromosome stability, as well as the proteins of the synthesis phase. Like PCNA2, RNR1, TSO2, THY-1, SWI/SNF2.
Histone Binding Proteins
Three histone proteins are present in the CC cluster; histone H2A (PGS003DMT40009751), histone H2A.1 (PGS0003DMT40003677), and histone H2AXB (PGSC0003DMT00034958).
Chromatin structure is essential for normal cell cycle progression, chromosome segregation, and centromere function [96].
Loss of function mutations in H2A cause an increase in ploidy phenotype, and an increase rate of chromosome loss and defects in completing the G2/M phase of the cell cycle. Altered centromeric chromatin structure and mutations in genes encoding kinetochore components revealed the role for H2A in the proper centromere-kinetochore function [97].
Histones also regulate a DNA damage response by epigenetic modifications. The expression of a loss of function mutant of H2AX Ser139Ala, that partially disrupts the phosphorylation site, suppressed the normal G2/M cell cycle arrest following DNA damage by ionizing radiation. The G2/M cell cycle arrest by DNA damage is promoted by checkpoints such as BRCA1 [98]. Taken together, H2AXB (PGSC0003DMT00034958) regulates the DNA damage response via BRCA1 (PGSC0003DMT400023253) to arrest mitotic cell cycle and ensure an appropriate cell division during MTs development.
These three proteins interact with BRCA1, a heterodimeric ubiquitin E3 ligase, required for the accumulation of ubiquitin conjugates at sites of DNA damage and for silencing at DNA satellite repeat regions. BRCA1 interact with E3 ubiquitin-protein ligase ORTHRUS 2 (PGSC000DMT400084297) and PCNA2.
Histone chaperone ASF1 (PGSC0003DMT400078369) interacts with PCNA2 with a combined score of 0.970 and with THY-1 with a score (0.860). ASF1 play an important role in chromatin replication, maintenance of genome integrity, and cell proliferation during plant development. Histones prevent DNA from becoming tanged, protect it from DNA damage, and play important roles in gene regulation and DNA replication.
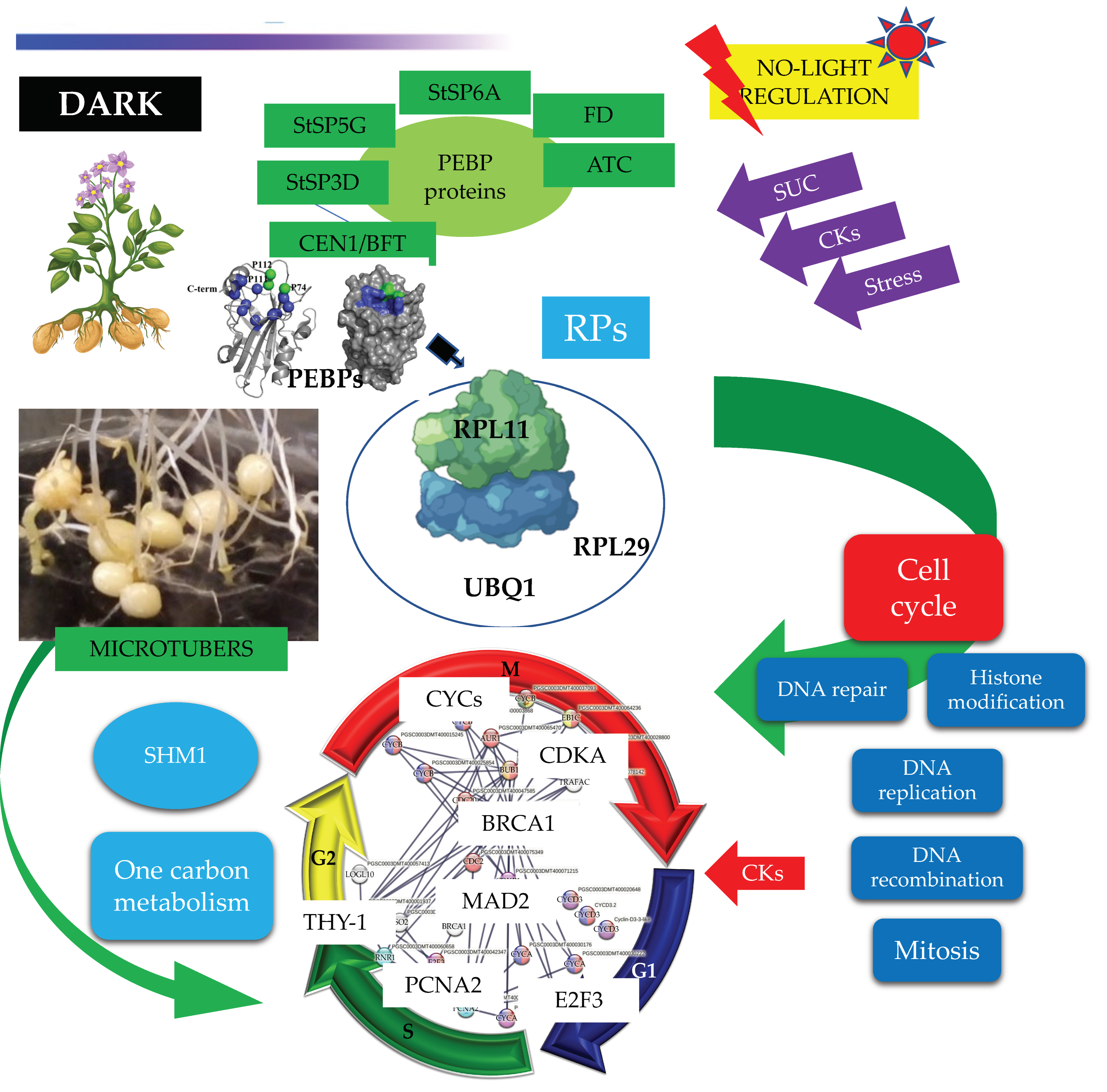
MTs development under high content of sucrose/gelrite, 2iP and darkness is activated by PEBP members (StSP6A, StSP5G, StSP3D, CEN1/BFT, ATC) and the TF FD. PEBP members interact with RPs to modulate CC regulation (DNA synthesis and mitosis) (Figure 14).
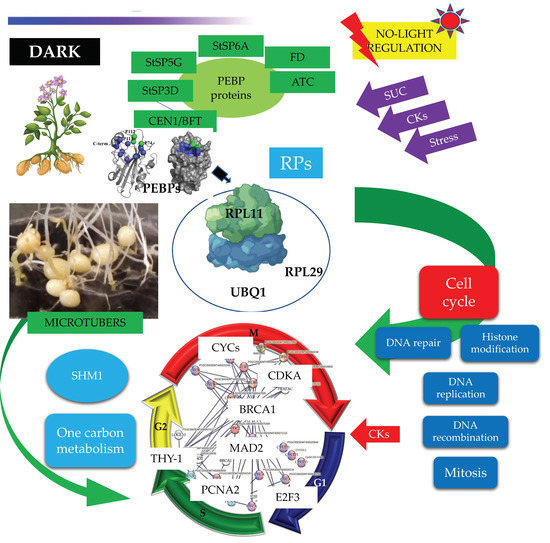
Figure 14.
PPI network of up-regulated CC proteins involved in mitosis (M), S, G2, G1 phase of potato S. tuberosum from transcriptomic-wide analysis with high confidence (0.860).
3.4. Validation of Transcriptomic-Wide Analysis by RT-qPCR
To validate the results of the transcriptomic-wide analysis and the conclusions reached, we performed an analysis by RT-qPCR of 21 genes interacting within the PPI network.
Figure 13 shows the results of validation by RT-qPCR. For all cases, the correct validation is observed as they are up-regulated both in the transcriptome and in the relative quantification.
4. Materials and Methods
4.1. Plant Material
4.1.1. Potato Shoot Micropropagation
Potato cv. Alpha plantlets were propagated by shoot proliferation under in vitro conditions in MS medium [99], supplemented with sucrose 30 g/L (CAT 57-50-1 Sigma-Aldrich, St. Louis, MO, USA), activated charcoal 3 g/L (CAT: 242276 Sigma-Aldrich, St. Louis, MO, USA), pH 5.8 and solidified with gelrite 3 g/L (GELZAN CAT. G1910 Sigma-Aldrich, St. Louis, MO, USA). Shoots were incubated at 25/17 °C under fluorescent light at 25 μmol/m2/s of irradiance. Stolon explants derived from propagated shoots were used for MTs induction.
4.1.2. Potato MTs Induction
Potato MTs induction protocol was made according to Herrera-Isidron et al. [10]. Stolon explants were cultured in flasks containing MR8-G6-2iP medium: MS medium, supplemented with 2iP 10 mg/L, 8% sucrose, 6 g/L gelrite, 3 g/L activated charcoal, pH 5.8, osmotic potential of 2.02 mPa. As control, stolon explants were cultured in MR1-G3-2iP: MS medium supplemented with 10 mg/2iP, 1% sucrose, 3 g/L gelrite, 3 g/L activated charcoal, pH 5.8, osmotic potential of −0.88 Mpa.). Flasks were sealed with plastic and incubated in dark at 25/17 °C for 15 days of incubation.
4.1.3. Transcriptome Sequencing and Assembly
To isolate RNA from MTs, tissue samples were collected after 15 days of incubation; at this time, MTs initiate to develop [10].
Total RNA was isolated by using Trizol (Invitrogen, Carlsbad, CA, USA), RNA concentration was measured by its absorbance at 260 nm, ratio 260 nm/280 nm was assessed, and its integrity confirmed by electrophoresis in agarose 1% (w/v) gels.
Samples of cDNA were amplified by PCR using SYBRTM Green (ThermoFisher CAT: 4312704, Waltham, MA, USA) in Real-Time PCR Systems (CFX96 BioRad, Herules, CA, USA).
Final product cDNA samples for sequencing were sent to GENEWIZ, Plainfield, NJ, USA. Illumina HiSeq 2500 (Illumina, San Diego, CA, USA) was used for sequencing. Sequenced reads were tested for quality using the FastQC software package (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and preprocessed to remove sequence adapters and low-quality bases using the software Trimmomatics [100].
4.1.4. Analysis of DEG
Adapter removal was performed using the Trimmomatic v0.3.6 program [100]. RNA-seq reads were aligned to the Solanum tuberosum reference genome available in Phytozome v12.1. (https://phytozome.jgi.doe.gov/pz/portal.html) with the STAR aligner v.2.5.2b. [101]. In this step, the BAM (Binary Alignment/Map) files were generated. Subsequently, a count and set of transcripts were made using the featureCounts program of the Subread v.1.5.2 package [102]. A quantification and differential analysis of the transcripts was performed using the DESeq2 v1.12.4 program. Finally, an ontology analysis was performed using Blast2GO.
4.1.5. PPI Analysis of Microtuberization
A gene network with high confidence (0.860) was performed with the STRING database v11.5 [103], based on S. tuberosum, homologous genes present in the S. tuberosum genome in Sol genomics network [104]. Gene identifier (Id) was made according to the UNIPROT [105], NCBI [106] database. Homologous in S. tuberosum greater than 60% in protein sequence with A. thaliana were considered. Oligonucleotides were designed to qPCR (2−∆∆CT method analysis) [107] gene expression or transcriptional analysis (Table S1).
5. Conclusions
- A PPI network of up-regulated TFs revealed that at least six TFs–MYB43, TSF, bZIP27, bZIP43, HAT4, WOX9–may be involved during MTs development.
- Two fundamental biological process essential for life and highly conserved through organisms were found interacting tightly: RPs comprising 29 and CC 117 proteins.
- PEBP members interact with RPs and CC process to activate MTs development under high content of sucrose and gelrite, 2iP under darkness.
- Further experiments by yeast two-hybrid screening approach of genome edited up- and down-regulated PEBP members with RPL11 are required to demonstrate our model of MTs development under darkness.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232213835/s1. References [55,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142] are cited in Supplementary Materials Table S1.
Author Contributions
E.V.-L. and L.H.-I.: Conceptualization, methodology, validation, formal analysis, investigation, data curation, writing original draft preparation, review and editing, funding acquisition; J.A.F.-L. and O.S.R.-M.: Methodology, software, validation, investigation; A.B.: Software, data curation, formal analysis, supervision, writing, review and editing; J.L.C.-P.: Conceptualization, methodology, validation, formal analysis, investigation, data curation, writing original draft preparation, review and editing, funding acquisition, writing, review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded, by SIP 20222061 IPN, proyectos de desarrollo tecnológico o inovación para alumnos del IPN # 120.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Rosalia Vuelvas-Nolasco for her excellent technical assistance of plant tissue culture of potato and medium preparation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Food and Agriculture Organization Corporate Statistical Database. FAOSTAT. 2022. Available online: https://www.fao.org/faostat/es/#data/QCL/visualize (accessed on 26 July 2022).
- Khosa, J.; Bellinazzo, F.; Kamenetsky, R.; Macknight, R.; Immink, R.G.H. PHOSPHATIDYLETHANOLAMONE-BINDING PROTEINS: The conductors of dual reproduction in plants with vegetative storage organs. J. Exp. Bot. 2021, 72, 2845–2856. [Google Scholar] [CrossRef] [PubMed]
- Dutt, S.; Manjul, A.S.; Raigond, P.; Singh, B.; Siddappa, S.; Bhardwaj, V.; Kawar, P.G.; Patil, V.U.; Kardile, H.B. Key Players Associated with Tuberization in Potato: Potential Candidates for Genetic Engineering. Crit. Rev. Biotechnol. 2017, 37, 942–957. [Google Scholar] [CrossRef] [PubMed]
- Kondhare, K.R.; Malankar, N.N.; Devani, R.S.; Banerjee, A.K. Genome-Wide Transcriptome Analysis Reveals Small RNA Profiles Involved in Early Stages of Stolon-to-Tuber Transitions in Potato under Photoperiodic Conditions. BMC Plant Biol. 2018, 18, 284. [Google Scholar] [CrossRef] [PubMed]
- Zierer, W.; Rüscher, D.; Sonnewald, U.; Sonnewald, S. Tuber and Tuberous Root Development. Annu. Rev. Plant Biol. 2021, 72, 551–580. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, D.J.; Coleman, W.K.; Coleman, S.E. Potato Microtuber Production and Performance: A Review. Am. J. Pot. Res. 2003, 80, 103–115. [Google Scholar] [CrossRef]
- Hannapel, D.J. Signalling the Induction of Tuber Formation. In Potato Biology and Biotechnology; Elsevier: Amsterdam, The Netherlands, 2007; pp. 237–256. [Google Scholar] [CrossRef]
- Vinterhalter, D.; Dragicevic, I.; Vinterhalter, B. Potato in vitro culture techniques and biotechnology. Fruit Veg. Cereal Sci. Biotechnol. (Spec. Issue 1) 2008, 2, 16–45. [Google Scholar]
- Coleman, W.K.; Donnelly, D.J.; Coleman, S.E. Potato Microtubers as Research Tools: A Review. Am. J. Pot. Res. 2001, 78, 47–55. [Google Scholar] [CrossRef]
- Herrera-Isidron, L.; Valencia-Lozano, E.; Rosiles-Loeza, P.Y.; Robles-Hernández, M.G.; Napsuciale-Heredia, A.; Cabrera-Ponce, J.L. Gene Expression Analysis of Microtubers of Potato Solanum Tuberosum L. Induced in Cytokinin Containing Medium and Osmotic Stress. Plants 2021, 10, 876. [Google Scholar] [CrossRef]
- Crookshanks, M.; Emmersen, J.; Welinder, K.G.; Lehmann-Nielsen, K. The potato tuber transcriptome analysis of 6067 expressed sequence tags. FEBS Lett. 2001, 506, 123–126. [Google Scholar] [CrossRef]
- Shan, J.; Song, W.; Zhou, J.; Wang, X.; Xie, C.; Gao, X.; Xie, T.; Liu, J. Transcriptome analysis reveals novel genes potentially involved in photoperiodic tuberization in potato. Genomics 2013, 102, 388–396. [Google Scholar] [CrossRef]
- Kumar, J.; Sapna, T.; Sundaresha, D.S.; Chandel, P.; Ali, N.; Singh, B.; Bhardwaj, V.; Singh, B.P. Microarray analysis of gene expression patterns in the leaf during potato tuberization in the potato somatic hybrid Solanum tuberosum and Solanum etuberosum. Genome 2014, 58, 305–313. [Google Scholar] [CrossRef]
- Gong, L.; Zhang, H.; Gan, X.; Zhang, L.; Chen, Y.; Nie, F.; Shi, L.; Li, M.; Guo, Z.; Zhang, G.; et al. Transcriptome Profiling of the Potato (Solanum tuberosum L.) Plant under Drought Stress and Water-Stimulus Conditions. PLoS ONE 2015, 10, e0128041. [Google Scholar] [CrossRef]
- Sharma, P.; Lin, T.; Hannapel, D.J. Targets of the StBEL5 Transcription Factor Include the FT Ortholog StSP6A. Plant Physiol. 2016, 170, 310–324. [Google Scholar] [CrossRef]
- Vulavala, V.K.R.; Fogelman, E.; Faigenboim, A.; Shoseyov, O.; Ginzberg, I. The Transcriptome of Potato Tuber Phellogen Reveals Cellular Functions of Cork Cambium and Genes Involved in Periderm Formation and Maturation. Sci. Rep. 2019, 9, 10216. [Google Scholar] [CrossRef]
- Wilson, D.N.; Doudna Cate, J.H. The Structure and Function of the Eukaryotic Ribosome. Cold Spring Harb. Perspect. Biol. 2012, 4, a011536. [Google Scholar] [CrossRef]
- Inzé, D.; De-Veylder, L. Cell Cycle Regulation in Plant Development. Annu. Rev. Genet. 2006, 40, 77–105. [Google Scholar] [CrossRef]
- Timsit, Y.; Sergeant-Perthuis, G.; Bennequin, D. Evolution of ribosomal protein network architectures. Sci. Rep. 2021, 11, 625. [Google Scholar] [CrossRef]
- Gamm, M.; Peviani, A.; Honsel, A.; Snel, B.; Smeekens, S.; Hanson, J. Increased Sucrose Levels Mediate Selective mRNA Translation in Arabidopsis. BMC Plant Biol. 2014, 14, 306. [Google Scholar] [CrossRef]
- Brenner, W.G.; Schmülling, T. Transcript Profiling of Cytokinin Action in Arabidopsis Roots and Shoots Dcovers Largely Similar but Also Organ-Specific Responses. BMC Plant Biol. 2012, 12, 112. [Google Scholar] [CrossRef]
- Latchman, D.S. Transcription factors: An overview. Int. J. Biochem. Cell Biol. 1997, 29, 1305–1312. [Google Scholar] [CrossRef]
- Nicolas, M.; Torres-Pérez, R.; Wahl, V.; Cruz-Oró, E.; Rodríguez-Buey, M.L.; Zamarreño, A.M.; Martín-Jouve, B.; García-Mina, J.M.; Oliveros, J.C.; Prat, S.; et al. Spatial control of potato tuberization by the TCP transcription factor BRANCHED1b. Nat. Plants 2022, 8, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Jia, Y.; Zhang, J.; Li, H.; Cheng, L.; Wang, P.; Bao, Z.; Liu, Z.; Feng, S.; Zhu, X.; et al. Genome evolution and diversity of wild and cultivated potatoes. Nature 2022, 606, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Lai, T.H.; Kim, D.R. A functional Network Model of the Metastasis Supressor PEBP1/RKIP and its Regulators in Breast Cancer Cells. Cancers 2021, 13, 6098. [Google Scholar] [CrossRef] [PubMed]
- Yeung, K.; Seitz, T.; Li, S.; Janosch, P.; McFerran, B.; Kaiser, C.; Fee, F.; Katsanakis, K.D.; Rose, D.W.; Mischak, H.; et al. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 1999, 401, 173–177. [Google Scholar] [CrossRef]
- Al-Mulla, F.; Bitar, M.S.; Taqi, Z.; Rath, O.; Kolch, W. RAF kinase activity signalling by (RKIP) modulates cell cycle kinetics and motility. Mol. Biosyst. 2011, 7, 928–941. [Google Scholar] [CrossRef]
- Voogd, C.; Brian, L.A.; Wang, T.; Allan, A.C.; Varkony-Gasic, E. Three FT and multiple CEN and BFT genes regulate maturity, flowering, and vegetative phenology in kiwifruit. J. Exp. Bot. 2017, 68, 1539–1553. [Google Scholar] [CrossRef]
- Navarro, C.; Abelenda, J.A.; Cruz-Oró, E.; Cuéllar, C.A.; Tamaki, S.; Silva, J.; Shimamoto, K.; Prat, S. Control of Flowering and Storage Organ Formation in Potato by FLOWERING LOCUS T. Nature 2011, 478, 119–122. [Google Scholar] [CrossRef]
- Moraes, T.S.; Dornelas, M.C.; Martinelli, A.P. FT/TFL1: Calibrating Plant Architecture. Front. Plant Sci. 2019, 10, 97. [Google Scholar] [CrossRef]
- Collani, S.; Neumann, M.; Yant, L.; Schmid, M. FT Modulates Genome-wide DNA-Binding of the bZIP Transcription Factor FD. Plant Physiol. 2019, 180, 367–380. [Google Scholar] [CrossRef]
- Purwestri, Y.A.; Susanto, F.A.; Tsuji, H. Hd3a florigen recruits different proteins to reveal its function in plant growth and development. Intech 2017, 4, 49–67. [Google Scholar] [CrossRef][Green Version]
- Wang, E.; Liu, T.; Sun, X.; Jing, S.; Zhou, T.; Liu, T.; Song, B. Profiling of the Candidate Interacting Proteins of SELF-PRUNING 6A (SP6A) in Solanum tuberosum. Int J. Mol. Sci. 2022, 15, 9126. [Google Scholar] [CrossRef] [PubMed]
- Abelenda, J.A.; Bergonzi, S.; Oortwijn, M.; Sonnewald, S.; Du, M.; Visser, R.G.; Bachem, C.W. Source-sink regulation is mediated by interaction of an FT homolog with a SWEET protein in potato. Curr. Biol. 2019, 29, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Plantenga, F.D.M.; Bergonzi, S.; Abelenda, J.A.; Bachem, C.W.B.; Visser, R.G.F.; Heuvelink, E.; Marcelis, L.F.M. The tuberization signal StSP6A represses flower bud development in potato. J. Exp. Bot. 2019, 70, 937–948. [Google Scholar] [CrossRef]
- Navarro, C.; Cruz-Oró, E.; Pratt, S. Conserved function of flowering (FT) homologues as signals for storage organ differentiation. Curr. Opin. Plant Biol. 2015, 23, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Hastilestari, B.R.; Lorenz, J.; Reid, S.; Hofman, J.; Pscheidt, D.; Sonnewald, U.; Sonnewald, S. Deciphering source and sink responses of potato plants (Solanum tuberosum L.) to elevated temperatures. Plant Cell Envirom. 2018, 41, 2600–2616. [Google Scholar] [CrossRef] [PubMed]
- Lehretz, G.G.; Sonnewald, S.; Hornyik, C.; Corral, J.M.; Sonnewald, U. Post-transcriptional Regulation of FLOWERING LOCUS T Modulates Heat-Dependent Source-Sink Development in Potato. Curr. Biol. 2019, 29, 1614–1624.e3. [Google Scholar] [CrossRef] [PubMed]
- Salvato, F.; Havelund, J.F.; Chen, M.; Rao, R.S.P.; Rogowska-Wrzesinska, A.; Jensen, O.N.; Gang, D.R.; Thelen, J.J.; Møller, I.M. The Potato Tuber Mitochondrial Proteome. Plant Physiol. 2014, 16, 637–653. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, J.P.; Cankar, K.; Scheffer, S.J.; Beenen, H.G.; Shepherd, L.V.T.; Stewart, D.; Davies, H.V.; Wilkockson, S.J.; Leifert, C.; Gruden, K.; et al. Transcriptome Analysis of Potato Tubers Effects of Different Agricultural Practices. J. Agric. Food Chem. 2009, 57, 1612–1623. [Google Scholar] [CrossRef] [PubMed]
- Jing, S.; Sun, X.; Yu, L.; Wang, E.; Cheng, Z.; Liu, H.; Jiang, P.; Qin, J.; Begum, S.; Song, B. Transcription factor StABI5-like 1 binding to the FLOWERING LOCUS T homologs promotes early maturity in potato. Plant Physiol. 2022, 18, 1677–1693. [Google Scholar] [CrossRef]
- Mhawech, P. 14-3-3 proteins-an update. Cell Res. 2005, 15, 228–236. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, W.; Yu, H.; Peng, J.; Hu, Z.; Chen, L. The role of 14-3-3 proteins in plant growth and response to abiotic stress. Plant Cell Rep. 2022, 41, 833–852. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; van Esse, W.; Mulki, M.A.; Kirschner, G.; Zhong, J.; Simon, R.; von Korff, M. CENTRORADIALIS Interacts with FLOWERING LOCUS T-Like Genes to Control Floret Development and Grain Number. Plant Physiol. 2019, 180, 1013–1030. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Campbell, R.; Ducreux, L.J.M.; Morris, J.; Hedley, P.E.; Mellado-Ortega, E.; Roberts, A.G.; Stephens, J.; Bryan, G.J.; Torrance, L.; et al. TERMINAL FLOWER-1/CENTRORADIALIS inhibits tuberization via protein interaction with the tuberigen activation complex. Plant J. 2020, 103, 2263–2278. [Google Scholar] [CrossRef] [PubMed]
- Morris, W.L.; Alamar, M.C.; Lopez-Cobollo, R.M.; Castillo Cañete, J.; Bennett, M.; Van der Kaay, J.; Stevens, J.; Kumar Sharma, S.; McLean, K.; Thompson, A.J.; et al. A member of the TERMINAL FLOWER 1/CENTRORADIALIS gene family controls sprout growth in potato tubers. J. Exp. Bot. 2019, 70, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Abelenda, J.A.; Cruz-Oró, E.; Franco-Zorrilla, J.M.; Prat, S. Potato StCONSTANS-Like1 Suppresses Storage Organ Formation by Directly Activating the FT-like StSP5G Repressor. Curr. Biol. 2016, 26, 872–881. [Google Scholar] [CrossRef]
- Teo, C.J.; Takahashi, K.; Shimizu, K.; Shimamoto, K.; Taoka, K.I. Potato Tuber Induction is Regulated by Interactions Between Components of a Tuberigen Complex. Plant Cell Physiol. 2017, 58, 365–374. [Google Scholar] [CrossRef]
- Taylor, M.A.; Arif, S.A.M.; Pearce, S.R.; Davies, H.V.; Kumar, A.; George, L.A. Differential Expression and Sequence Analysis of Ribosomal Protein Genes Induced in Stolon Tips of Potato (Solanum Tuberosum L.) during the Early Stages of Tuberization. Plant Physiol. 1992, 100, 1171–1176. [Google Scholar] [CrossRef]
- Van Lijsebettens, M.; Vanderhaeghen, R.; De Block, M.; Bauw, G.; Villarroel, R.; Van Montagu, M. An S18 Ribosomal Protein Gene Copy at the Arabidopsis PFL Locus Affects Plant Development by Its Specific Expression in Meristems. EMBO J. 1994, 13, 3378–3388. [Google Scholar] [CrossRef]
- Tsugeki, R.; Kochieva, E.Z.; Fedoroff, N.V. A Transposon Insertion in the Arabidopsis SSR16 Gene Causes an Embryo-Defective Lethal Mutation. Plant J. 1996, 10, 479–489. [Google Scholar] [CrossRef]
- Zinn, A.R.; Ross, J.L. Turner Syndrome and Haploinsufficiency. Curr. Opin. Genet. Dev. 1998, 8, 322–327. [Google Scholar] [CrossRef]
- Lambertsson, A. The Minute Genes in Drosophila and Their Molecular Functions. Adv. Genet. 1998, 38, 69–134. [Google Scholar] [CrossRef] [PubMed]
- Volarević, S.; Stewart, M.J.; Ledermann, B.; Zilberman, F.; Terracciano, L.; Montini, E.; Grompe, M.; Kozma, S.C.; Thomas, G. Proliferation, But Not Growth, Blocked by Conditional Deletion of 40 S Ribosomal Protein S6. Science 2000, 288, 2045–2047. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Kim, G.-T.; Shinozaki, K. Disruption of an Arabidopsis Cytoplasmic Ribosomal Protein S13-Homologous Gene by Transposon-Mediated Mutagenesis Causes Aberrant Growth and Development. Plant J. 2000, 22, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Seidel, F.; Beine-Golovchuk, O.; Hsieh, Y.-C.; Kopka, J. Systematic Review of Plant Ribosome Heterogeneity and Specialization. Front. Plant Sci. 2020, 11, 948. [Google Scholar] [CrossRef]
- Ferreyra, M.L.F.; Pezza, A.; Biarc, J.; Burlingame, A.L.; Casati, P. Plant L10 Ribosomal Proteins Have Different Roles during Development and Translation under Ultraviolet-B Stress. Plant Physiol. 2010, 153, 1878–1894. [Google Scholar] [CrossRef]
- Wang, J.; Lan, P.; Gao, H.; Zheng, L.; Li, W.; Schmidt, W. Expression Changes of Ribosomal Proteins in Phosphate- and Iron-Deficient Arabidopsis Roots Predict Stress-Specific Alterations in Ribosome Composition. BMC Genom. 2013, 14, 783. [Google Scholar] [CrossRef]
- Cheong, B.E.; Beine-Golovchuk, O.; Gorka, M.; Ho, W.W.H.; Martinez-Seidel, F.; Firmino, A.A.P.; Skirycz, A.; Roessner, U.; Kopka, J. Arabidopsis REI-LIKE Proteins Activate Ribosome Biogenesis during Cold Acclimation. Sci. Rep. 2021, 11, 2410. [Google Scholar] [CrossRef]
- Zhang, C.; Li, H.; Yuan, C.; Liu, S.; Li, M.; Zhu, J.; Lin, X.; Lu, Y.; Guo, X. CKB 1 Regulates Expression of Ribosomal Protein L10 Family Gene and Plays a Role in UV -B Response. Plant Biol. J. 2020, 22, 143–152. [Google Scholar] [CrossRef]
- Moin, M.; Bakshi, A.; Saha, A.; Dutta, M.; Madhav, S.M.; Kirti, P.B. Rice Ribosomal Protein Large Subunit Genes and Their Spatio-Temporal and Stress Regulation. Front. Plant Sci. 2016, 7, 1284. [Google Scholar] [CrossRef]
- Moin, M.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Expression Profiling of Ribosomal Protein Gene Family in Dehydration Stress Responses and Characterization of Transgenic Rice Plants Overexpressing RPL23A for Water-Use Efficiency and Tolerance to Drought and Salt Stresses. Front. Chem. 2017, 5, 97. [Google Scholar] [CrossRef]
- Moin, M.; Saha, A.; Bakshi, A.; Divya, D.; Madhav, M.S.; Kirti, P.B. Study on Transcriptional Responses and Identification of Ribosomal Protein Genes for Potential Resistance against Brown Planthopper and Gall Midge Pests in Rice. Curr. Genom. 2021, 22, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Gonzali, S.; Loreti, E.; Solfanelli, C.; Novi, G.; Alpi, A.; Perata, P. Identification of Sugar-Modulated Genes and Evidence for in Vivo Sugar Sensing in Arabidopsis. J. Plant Res. 2006, 119, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A Central Integrator of Transcription Networks in Plant Stress and Energy Signalling. Nature 2007, 44, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Bläsing, O.E.; Gibon, Y.; Retzlaff, K.; Höhne, M.; Günther, M.; Stitt, M. Global Transcript Levels Respond to Small Changes of the Carbon Status during Progressive Exhaustion of Carbohydrates in Arabidopsis Rosettes. Plant Physiol. 2008, 146, 1834–1861. [Google Scholar] [CrossRef]
- Hummel, M.; Cordewener, J.H.G.; de Groot, J.C.M.; Smeekens, S.; America, A.H.P.; Hanson, J. Dynamic Protein Composition of Arabidopsis Thaliana Cytosolic Ribosomes in Response to Sucrose Feeding as Revealed by Label Free MSE Proteomics. Proteomics 2012, 12, 1024–1038. [Google Scholar] [CrossRef]
- Berridge, M.V.; Ralph, R.K.; Letham, D.S. The Binding of Kinetin to Plant Ribosomes. Biochem. J. 1970, 119, 75–84. [Google Scholar] [CrossRef]
- Fox, J.E.; Erion, J.L.A. Cytokinin Binding Protein from Higher Plant Ribosomes. Biochem. Biophys. Res. Commun. 1975, 64, 694–700. [Google Scholar] [CrossRef]
- Tepfer, D.A.; Fosket, D.E. Hormone-Mediated Translational Control of Protein Synthesis in Cultured Cells of Glycine Max. Dev. Biol. 1978, 62, 486–497. [Google Scholar] [CrossRef]
- Feierabend, J.; de Boer, J. Comparative Analysis of the Action of Cytokinin and Light on the Formation of Ribulosebisphosphate Carboxylase and Plastid Biogenesis. Planta 1978, 142, 75–82. [Google Scholar] [CrossRef]
- Yakovleva, L.; Kulaeva, O. The effect of phytohormones on phosphorylation of ribosomal proteins in detached pumpkin cotyledons. Biochem. Physiol. Pflanz. 1987, 182, 359–365. [Google Scholar] [CrossRef]
- Cherepneva, G.N.; Schmidt, K.-H.; Kulaeva, O.N.; Oelmüller, R.; Kusnetsov, V.V. Expression of the Ribosomal Proteins S14, S16, L13a and L30 Is Regulated by Cytokinin and Abscisic Acid. Plant Sci. 2003, 165, 925–932. [Google Scholar] [CrossRef]
- Karunadasa, S.S.; Kurepa, J.; Shull, T.E.; Smalle, J.A. Cytokinin-induced Protein Synthesis Suppresses Growth and Osmotic Stress Tolerance. New Phytol. 2020, 227, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Hulm, J.L.; McIntosh, K.B.; Bonham-Smith, P.C. Variation in Transcript Abundance among the Four Members of the Arabidopsis thaliana RIBOSOMAL PROTEIN S15a Gene Family. Plant Science 2005, 169, 267–278. [Google Scholar] [CrossRef]
- Chotewutmontri, P.; Stiffler, N.; Watkins, K.P.; Barkan, A. Ribosome Profiling in Maize. In Maize; Methods in Molecular, Biology; Lagrimini, L.M., Ed.; Springer: New York, NY, USA, 2018; Volume 1676, pp. 165–183. [Google Scholar] [CrossRef]
- Lei, L.; Shi, J.; Chen, J.; Zhang, M.; Sun, S.; Xie, S.; Li, X.; Zeng, B.; Peng, L.; Hauck, A.; et al. Ribosome Profiling Reveals Dynamic Translational Landscape in Maize Seedlings under Drought Stress. Plant J. 2015, 84, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Lei, L.; Shi, J.; Wang, X.; Chen, J.; Xue, M.; Sun, S.; Zhan, B.; Xia, Z.; Jiang, N.; et al. Characterization of Maize Translational Responses to Sugarcane Mosaic Virus Infection. Virus Res. 2019, 259, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Shiraku, M.L.; Magwanga, R.O.; Cai, X.; Kirungu, J.N.; Xu, Y.; Mehari, T.G.; Hou, Y.; Wang, Y.; Agong, S.G.; Peng, R.; et al. Functional Characterization of GhACX3 Gene Reveals Its Significant Role in Enhancing Drought and Salt Stress Tolerance in Cotton. Front. Plant Sci. 2021, 12, 658755. [Google Scholar] [CrossRef]
- Mehdi, R.; Mojtaba, K.; Mojtaba, M.; Sanam, S.C. Identification of Drought-Responsive Proteins of Sensitive and Tolerant Tea (Camellia sinensis L.) Clones under Normal and Drought Stress Conditions. Curr. Prot. 2020, 17, 227–240. [Google Scholar] [CrossRef]
- Thapa, M.; Bommakanti, A.; Shamsuzzaman, M.; Gregory, B.; Samsel, L.; Zengel, J.M.; Lindahl, L. Repressed Synthesis of Ribosomal Proteins Generates Protein-Specific Cell Cycle and Morphological Phenotypes. MBoC. 2013, 24, 3620–3633. [Google Scholar] [CrossRef]
- Maitra, N.; He, C.; Blank, H.M.; Tsuchiya, M.; Schilling, B.; Kaeberlein, M.; Aramayo, R.; Kennedy, B.K.; Polymenis, M. Translational Control of One-Carbon Metabolism Underpins Ribosomal Protein Phenotypes in Cell Division and Longevity. eLife 2020, 9, e53127. [Google Scholar] [CrossRef]
- Piper, M.D.; Hong, S.-P.; Ball, G.E.; Dawes, I.W. Regulation of the Balance of One-Carbon Metabolism in Saccharomyces Cerevisiae. J. Biol. Chem. 2000, 275, 30987–30995. [Google Scholar] [CrossRef]
- Clare, C.E.; Brassington, A.H.; Kwong, W.Y.; Sinclair, K.D. One-Carbon Metabolism: Linking Nutritional Biochemistry to Epigenetic Programming of Long-Term Development. Annu. Rev. Anim. Biosci. 2019, 7, 263–287. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Cortijo, S.; Korsbo, N.; Roszak, P.; Schiessl, K.; Gurzadyan, A.; Wightman, R.; Jönsson, H.; Meyerowitz, E. Molecular mechanism of cytokinin-activated cell division in Arabidopsis. Science 2021, 26, 1350–1355. [Google Scholar] [CrossRef] [PubMed]
- Rhee, Y.; Hwang, K.; Cho, S.; Lee, M.; Kil, E.J.; Choi, S.; Hahn, B.S.; Kim, D.; Auh, C.K.; Lee, S. Expression analysis of D-type cyclin in potato (Solanum tuberosum L.) under different culture conditions. Acta Physiol. Plant. 2016, 38, 36. [Google Scholar] [CrossRef]
- Lara-Gonzalez, P.; Kim, T.; Oegema, K.; Corbett, K.; Desai, A. A tripartite mechanism catalyzes Mad2-Cdc20 assembly at unattached kinetochores. Science 2021, 374, 64–67. [Google Scholar] [CrossRef]
- Leu, W.M.; Cao, X.L.; Wilson, T.J.; Snustad, D.P.; Chua, N.H. Phytochrome A and phytochrome B mediate the hypocotyl-specific down-regulation of TUB1 by light in arabidopsis. Plant Cell 1995, 7, 2187–2196. [Google Scholar] [CrossRef]
- Demidov, D.; Van Damme, D.; Geelen, D.; Blattner, F.R.; Houben, A. Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell 2005, 17, 836–848. [Google Scholar] [CrossRef]
- Cai, G.; Cresti, M. Are kinesins required for organelle trafficking in plant cells? Front. in Plant Sci. 2012, 3, 170. [Google Scholar] [CrossRef]
- Weaver, R.L.; Limzerwala, J.F.; Naylor, R.M.; Jeganathan, K.B.; Baker, D.J.; van Deursen, J.M. BubR1 alterations that reinforce mitotic surveillance act against aneuploidy and cancer. Elife 2016, 5, e16620. [Google Scholar] [CrossRef]
- Hemerly, A.; Engler, J.D.A.; Bergounioux, C.; Van Montagu, M.; Engler, G.; Inzé, D.; Ferreira, P. Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 1995, 14, 3925–3936. [Google Scholar] [CrossRef]
- Boruc, J.; Van den Daele, H.; Hollunder, J.; Rombauts, S.; Mylle, E.; Hilson, P.; Russinova, E. Functional modules in the Arabidopsis core cell cycle binary protein–protein interaction network. Plant Cell 2010, 22, 1264–1280. [Google Scholar] [CrossRef]
- Rossignol, P.; Stevens, R.; Perennes, C.; Jasinski, S.; Cella, R.; Tremousaygue, D.; Bergounioux, C. AtE2F-a and AtDP-a, members of the E2F family of transcription factors, induce Arabidopsis leaf cells to re-enter S phase. Mol. Genet. Genom. 2002, 266, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Z.; Tsai, S.Y.; Leone, G. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat. Rev. Cancer 2009, 11, 785–797. [Google Scholar] [CrossRef]
- Schulman, I.; Bloom, K.S. Centromeres: An integrated protein/DNA complex required for chromosome movement. Annu. Rev. of Cell Biol. 1991, 7, 311–336. [Google Scholar] [CrossRef] [PubMed]
- Pinto, I.; Winston, F. Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J. 2002, 19, 1598–1612. [Google Scholar] [CrossRef] [PubMed]
- Rios-Doria, J.; Velkova, A.; Dapic, V.; Galán-Caridad, J.M.; Dapic, V.; Carvalho, M.A.; Melendez, J.; Monteiro, A.N.A. Ectopic expression of histone H2AX mutants reveals a role for its post-translational modifications. Cancer Biol. Ther. 2009, 8, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Von Mering, C. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Bombarely, A.; Menda, N.; Tecle, I.Y.; Buels, R.M.; Strickler, S.; Fischer-York, T.; Mueller, L.A. The Sol Genomics Network (solgenomics. net): Growing tomatoes using Perl. Nucleic Acids Res. 2010, 39, D1149–D1155. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, G.-H.; Liu, M.-J.; Xiong, Y.; Sheen, J.; Wu, S.-H. TOR and RPS6 Transmit Light Signals to Enhance Protein Translation in Deetiolating Arabidopsis Seedlings. Proc. Natl. Acad. Sci. USA 2018, 115, 12823–12828. [Google Scholar] [CrossRef]
- Kim, Y.-K.; Kim, S.; Shin, Y.; Hur, Y.-S.; Kim, W.-Y.; Lee, M.-S.; Cheon, C.-I.; Verma, D.P.S. Ribosomal Protein S6, a Target of Rapamycin, Is Involved in the Regulation of RRNA Genes by Possible Epigenetic Changes in Arabidopsis. J. Biol. Chem. 2014, 289, 3901–3912. [Google Scholar] [CrossRef]
- Tzafrir, I. The Arabidopsis SeedGenes Project. Nucleic Acids Res. 2003, 31, 90–93. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, J.; Jia, M.; Xu, N.; Liang, S.; Shao, J.; Qi, Y.; Liu, X.; An, L.; Yu, F. Balance between Cytosolic and Chloroplast Translation Affects Leaf Variegation. Plant Physiol. 2018, 176, 804–818. [Google Scholar] [CrossRef]
- Creff, A.; Sormani, R.; Desnos, T. The Two Arabidopsis RPS6 Genes, Encoding for Cytoplasmic Ribosomal Proteins S6, Are Functionally Equivalent. Plant Mol. Biol. 2010, 73, 533–546. [Google Scholar] [CrossRef]
- Sun, L.; Yu, Y.; Hu, W.; Min, Q.; Kang, H.; Li, Y.; Hong, Y.; Wang, X.; Hong, Y. Ribosomal Protein S6 Kinase1 Coordinates with TOR-Raptor2 to Regulate Thylakoid Membrane Biosynthesis in Rice. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2016, 1861, 639–649. [Google Scholar] [CrossRef]
- Kim, K.-Y. Molecular Cloning of Low-Temperature-Inducible Ribosomal Proteins from Soybean. J. Exp. Bot. 2004, 55, 1153–1155. [Google Scholar] [CrossRef] [PubMed]
- Romani, I.; Tadini, L.; Rossi, F.; Masiero, S.; Pribil, M.; Jahns, P.; Kater, M.; Leister, D.; Pesaresi, P. Versatile Roles of Arabidopsis Plastid Ribosomal Proteins in Plant Growth and Development: Different Functions of Plastid Ribosome Subunits. Plant J. 2012, 72, 922–934. [Google Scholar] [CrossRef] [PubMed]
- Szick-Miranda, K.; Zanial, A.S.; Zanial, A.S.; Abidayo, S.; Slater, K.L.C. Analysis of RPS15aE, an Isoform of a Plant-Specific Evolutionarily Distinct Ribosomal Protein in Arabidopsis Thaliana, Reveals Its Potential Role as a Growth Regulator. Plant Mol. Biol. Rep. 2010, 28, 239–252. [Google Scholar] [CrossRef]
- Lu, C.; Xie, Z.; Yu, F.; Tian, L.; Hao, X.; Wang, X.; Chen, L.; Li, D. Mitochondrial Ribosomal Protein S9M Is Involved in Male Gametogenesis and Seed Development in Arabidopsis. Plant Biol. J. 2020, 22, 655–667. [Google Scholar] [CrossRef]
- Ma, Z.; Dooner, H.K. A Mutation in the Nuclear-Encoded Plastid Ribosomal Protein S9 Leads to Early Embryo Lethality in Maize. Plant J. 2004, 37, 92–103. [Google Scholar] [CrossRef]
- Qiu, Z.; Chen, D.; He, L.; Zhang, S.; Yang, Z.; Zhang, Y.; Wang, Z.; Ren, D.; Qian, Q.; Guo, L.; et al. The Rice White Green Leaf 2 Gene Causes Defects in Chloroplast Development and Affects the Plastid Ribosomal Protein S9. Rice 2018, 11, 39. [Google Scholar] [CrossRef]
- Tzafrir, I.; Pena-Muralla, R.; Dickerman, A.; Berg, M.; Rogers, R.; Hutchens, S.; Sweeney, T.C.; McElver, J.; Aux, G.; Patton, D.; et al. Identification of Genes Required for Embryo Development in Arabidopsis. Plant Physiol. 2004, 135, 1206–1220. [Google Scholar] [CrossRef]
- Schultes, N.P.; Sawers, R.J.H.; Brutnell, T.P.; Krueger, R.W. Maize High Chlorophyll Fluorescent 60 Mutation Is Caused by an Ac Disruption of the Gene Encoding the Chloroplast Ribosomal Small Subunit Protein 17. Plant J. 2000, 21, 317–327. [Google Scholar] [CrossRef]
- Chan, Y.L.; Paz, V.; Olvera, J.; Wool, I.G. The Primary Structure of L37—A Rat Ribosomal Protein with a Zinc Finger-like Motif. Biochem. Biophys. Res. Commun. 1993, 192, 590–596. [Google Scholar] [CrossRef]
- Finkelshtein, A.; Khamesa, H.; Tuan, L.A.; Rabanim, M.; Chamovitz, D.A. Overexpression of the Ribosomal S30 Subunit Leads to Indole-3-carbinol Tolerance in Arabidopsis Thaliana. Plant J. 2021, 105, 668–677. [Google Scholar] [CrossRef]
- Nagaraj, S.; Senthil-Kumar, M.; Ramu, V.S.; Wang, K.; Mysore, K.S. Plant Ribosomal Proteins, RPL12 and RPL19, Play a Role in Nonhost Disease Resistance against Bacterial Pathogens. Front. Plant Sci. 2016, 6, 1192. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Zhang, C.; Li, Q.; Yang, Q.; Gu, M.; Liu, Q. A Residue Substitution in the Plastid Ribosomal Protein L12/AL1 Produces Defective Plastid Ribosome and Causes Early Seedling Lethality in Rice. Plant Mol. Biol. 2016, 91, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xie, C.; Li, W.; Zhang, R.; Jue, D.; Yang, Q. Expression of a Wild Eggplant Ribosomal Protein L13a in Potato Enhances Resistance to Verticillium Dahliae. Plant Cell Tissue Organ Cult 2013, 115, 329–340. [Google Scholar] [CrossRef]
- Song, J.; Wei, X.; Shao, G.; Sheng, Z.; Chen, D.; Liu, C.; Jiao, G.; Xie, L.; Tang, S.; Hu, P. The Rice Nuclear Gene WLP1 Encoding a Chloroplast Ribosome L13 Protein Is Needed for Chloroplast Development in Rice Grown under Low Temperature Conditions. Plant Mol. Biol. 2014, 84, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jang, S.; Ryu, S.; Lee, S.; Park, J.; Lee, S.; An, G.; Park, S.K. Mutation of Plastid Ribosomal Protein L13 Results in an Albino Seedling-Lethal Phenotype in Rice. Plant Breed. Biotechnol. 2019, 7, 395–404. [Google Scholar] [CrossRef]
- Zhang, H.; Luo, M.; Day, R.C.; Talbot, M.J.; Ivanova, A.; Ashton, A.R.; Chaudhury, A.M.; Macknight, R.C.; Hrmova, M.; Koltunow, A.M. Developmentally Regulated HEART STOPPER, a Mitochondrially Targeted L18 Ribosomal Protein Gene, Is Required for Cell Division, Differentiation, and Seed Development in Arabidopsis. J. Exp. Bot. 2015, 66, 5867–5880. [Google Scholar] [CrossRef]
- Yan, H.; Chen, D.; Wang, Y.; Sun, Y.; Zhao, J.; Sun, M.; Peng, X. Ribosomal Protein L18aB Is Required for Both Male Gametophyte Function and Embryo Development in Arabidopsis. Sci. Rep. 2016, 6, 31195. [Google Scholar] [CrossRef]
- Nishimura, T.; Wada, T.; Yamamoto, K.T.; Okada, K. The Arabidopsis STV1 Protein, Responsible for Translation Reinitiation, Is Required for Auxin-Mediated Gynoecium Patterning. Plant Cell 2005, 17, 2940–2953. [Google Scholar] [CrossRef]
- Li, S.; Liu, K.; Zhang, S.; Wang, X.; Rogers, K.; Ren, G.; Zhang, C.; Yu, B. STV1, a Ribosomal Protein, Binds Primary MicroRNA Transcripts to Promote Their Interaction with the Processing Complex in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 1424–1429. [Google Scholar] [CrossRef]
- Park, S.-H.; Chung, M.-S.; Lee, S.; Lee, K.-H.; Kim, C.S. Loss of Ribosomal Protein L24A (RPL24A) Suppresses Proline Accumulation of Arabidopsis Thaliana Ring Zinc Finger 1 (Atrzf1) Mutant in Response to Osmotic Stress. Biochem. Biophys. Res. Commun. 2017, 494, 499–503. [Google Scholar] [CrossRef]
- Szakonyi, D.; Byrne, M.E. Ribosomal Protein L27a Is Required for Growth and Patterning in Arabidopsis Thaliana: Ribosomal Protein-Mediated Patterning. Plant J. 2011, 65, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Zsögön, A.; Szakonyi, D.; Shi, X.; Byrne, M.E. Ribosomal Protein RPL27a Promotes Female Gametophyte Development in a Dose-Dependent Manner. Plant Physiol. 2014, 165, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Oristian, D.S.; Sloofman, L.G.; Zhou, X.; Wang, L.; Farach-Carson, M.C.; Kirn-Safran, C.B. Ribosomal Protein L29/HIP Deficiency Delays Osteogenesis and Increases Fragility of Adult Bone in Mice. J. Orthop. Res. 2009, 27, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Aseev, L.V.; Boni, I.V. Extraribosomal Functions of Bacterial Ribosomal Proteins. Mol. Biol. 2011, 45, 739–750. [Google Scholar] [CrossRef]
- Finley, D.; Bartel, B.; Varshavsky, A. The Tails of Ubiquitin Precursors Are Ribosomal Proteins Whose Fusion to Ubiquitin Facilitates Ribosome Biogenesis. Nature 1989, 338, 394–401. [Google Scholar] [CrossRef]
- Chen, J.-G. RACK1 Mediates Multiple Hormone Responsiveness and Developmental Processes in Arabidopsis. J. Exp. Bot. 2006, 57, 2697–2708. [Google Scholar] [CrossRef]
- Guo, J.; Chen, J.-G. RACK1 Genes Regulate Plant Development with Unequal Genetic Redundancy in Arabidopsis. BMC Plant Biol. 2008, 8, 108. [Google Scholar] [CrossRef]
- Cheng, Z.; Li, J.-F.; Niu, Y.; Zhang, X.-C.; Woody, O.Z.; Xiong, Y.; Djonović, S.; Millet, Y.; Bush, J.; McConkey, B.J.; et al. Pathogen-Secreted Proteases Activate a Novel Plant Immune Pathway. Nature 2015, 521, 213–216. [Google Scholar] [CrossRef]
- Kakehi, J.-I.; Kawano, E.; Yoshimoto, K.; Cai, Q.; Imai, A.; Takahashi, T. Mutations in Ribosomal Proteins, RPL4 and RACK1, Suppress the Phenotype of a Thermospermine-Deficient Mutant of Arabidopsis Thaliana. PLoS ONE 2015, 10, e0117309. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).