Evolution of Transcriptomes in Early-Generation Hybrids of the Apomictic Ranunculus auricomus Complex (Ranunculaceae)
Abstract
1. Introduction
2. Results
2.1. Transcriptome Data
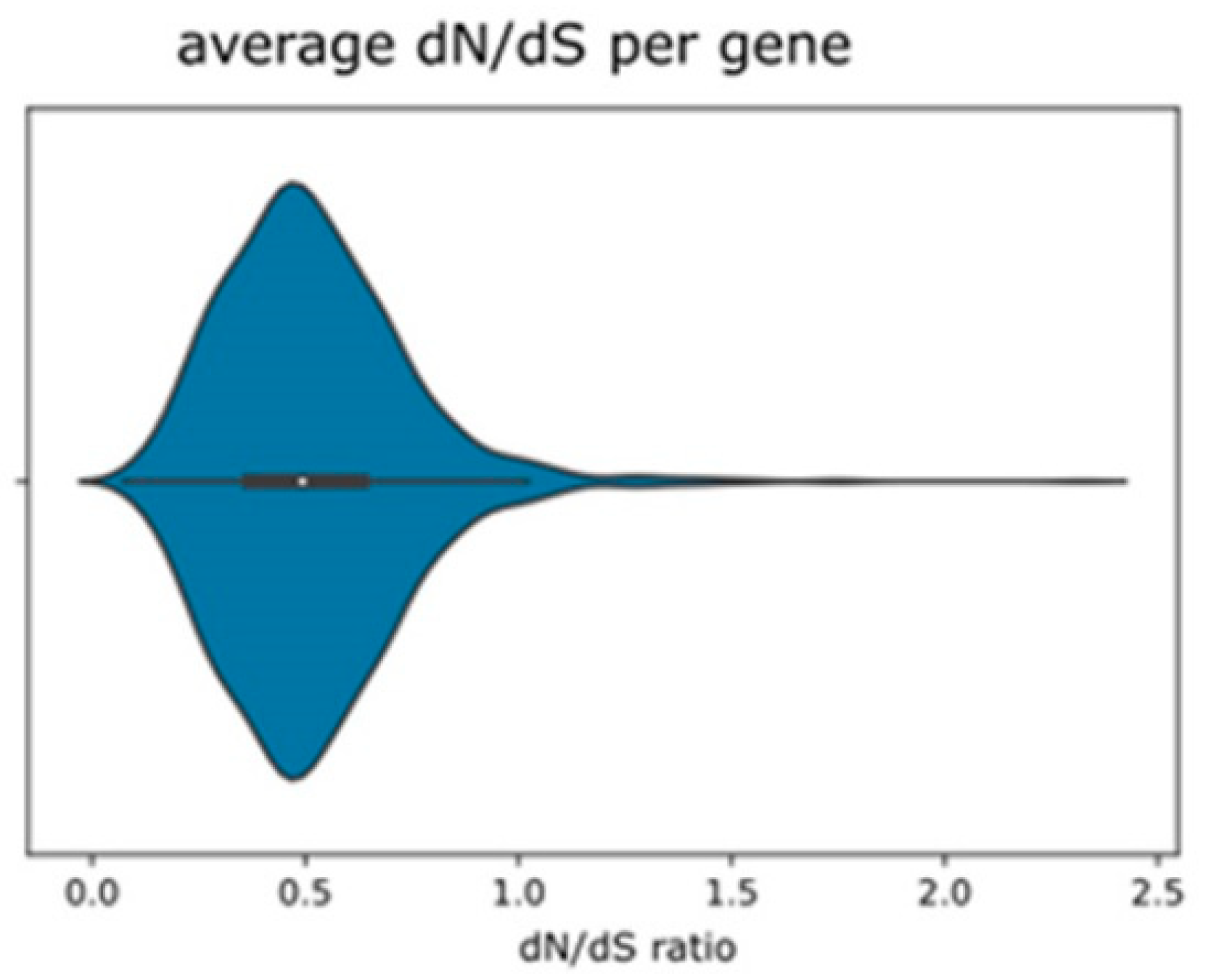
2.2. dN/dS Ratios and Genes under Diversifying Selection in Parent–Hybrid Comparisons
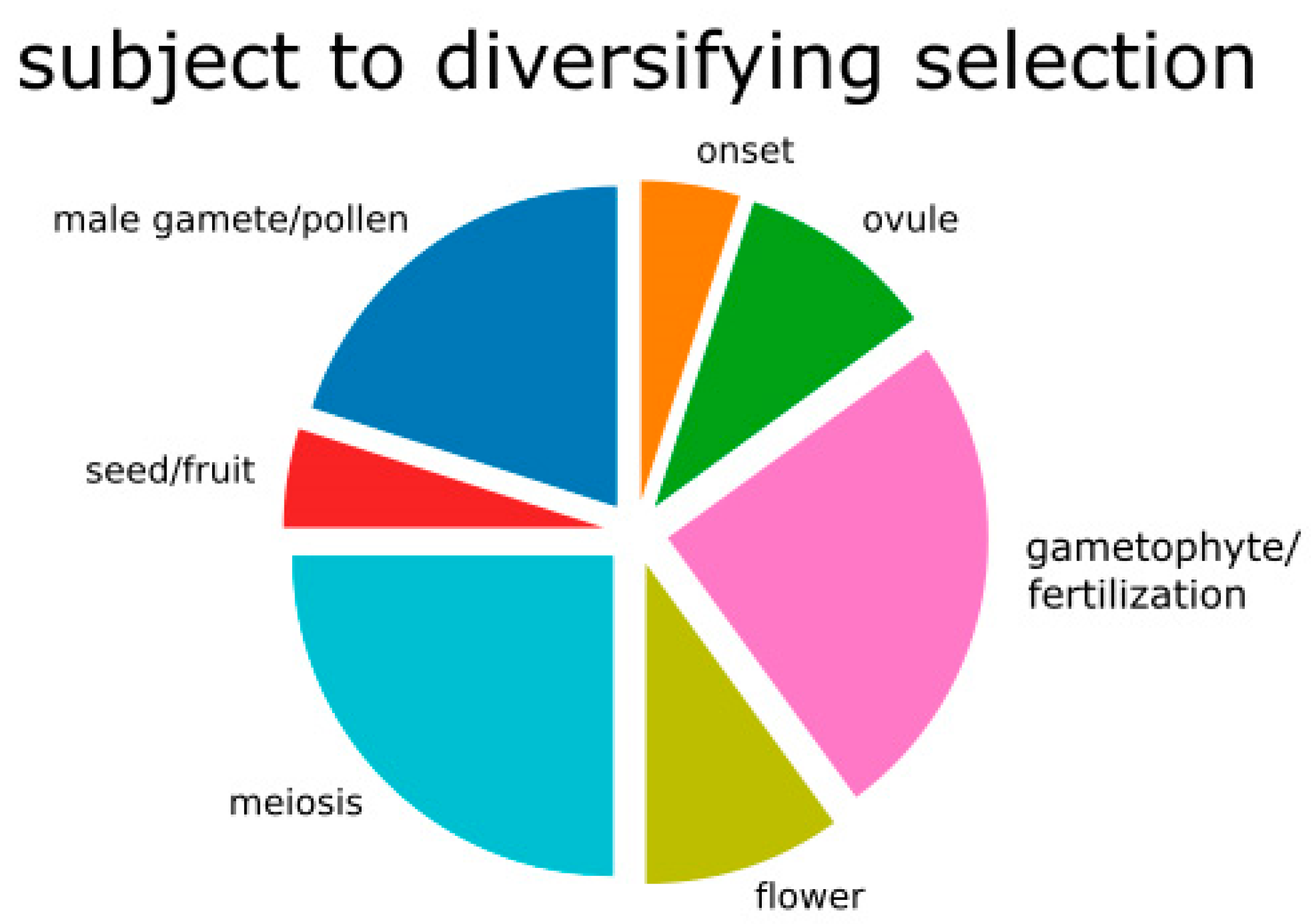
2.3. Gene Annotation Related to Reproduction
3. Discussion
3.1. Factors Influencing Selection Regimes and dN/dS Values
3.2. Transcriptome Evolution between Hybrids and Parents Related to Apospory
3.3. Scenarios for the De Novo Evolution of Apomixis in Diploid Hybrids
4. Materials and Methods
4.1. Plant Material
4.2. RNA Extraction, Library Preparation, and Sequencing
4.3. Read Processing, De Novo Assembly, and Data Processing
4.4. Hybridisation Networks
4.5. Analyses of Loci under Selection (dN/dS Ratio Analyses)
4.6. Filtering dN/dS Ratios
4.7. Gene Ontology
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abbott, R.; Albach, D.; Ansell, S.; Arntzen, J.W.; Baird, S.J.E.; Bierne, N.; Boughman, J.W.; Brelsford, A.; Buerkle, C.A.; Buggs, R.; et al. Hybridization and speciation. J. Evol. Biol. 2013, 26, 229–246. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Soltis, D.E. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 2009, 60, 561–588. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.L. Natural Hybridization and Evolution; Oxford University Press: New York, NY, USA, 1997. [Google Scholar]
- Rieseberg, L.H.; Raymond, O.; Rosenthal, D.M.; Lai, Z.; Livingstone, K.; Nakazato, T.; Durphy, J.L.; Schwarzbach, A.E.; Donovan, L.A.; Lexer, C. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 2003, 301, 1211–1216. [Google Scholar] [CrossRef] [PubMed]
- Karrenberg, S.; Lexer, C.; Rieseberg, L. Reconstructing the History of Selection during Homoploid Hybrid Speciation. Am. Nat. 2007, 169, 725–737. [Google Scholar] [CrossRef]
- Asker, S.; Jerling, L. Apomixis in Plants; CRC press: Boca Raton, FL, USA, 1992. [Google Scholar]
- Carman, J.G. Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn. Soc. 1997, 61, 51–94. [Google Scholar] [CrossRef]
- Hojsgaard, D.; Hörandl, E. The Rise of Apomixis in Natural Plant Populations. Front. Plant Sci. 2019, 10, 358. [Google Scholar] [CrossRef]
- Mogie, M.; Britton, N.F.; Stewart-Cox, A.J. Asexuality, polyploidy and the male function. In Apomixis: Evolution, Mechanisms and Perspectives; Hörandl, E., Grossniklaus, U., Van Dijk, P., Sharbel, T.F., Eds.; Gantner: Rugell, Liechtenstein, 2007; pp. 169–194. [Google Scholar]
- Ozias-Akins, P.; van Dijk, P.J. Mendelian genetics of apomixis in plants. Annu. Rev. Genet. 2007, 41, 509–537. [Google Scholar] [CrossRef]
- Grimanelli, D. Epigenetic regulation of reproductive development and the emergence of apomixis in angiosperms. Curr. Opin. Plant Biol. 2012, 15, 57–62. [Google Scholar] [CrossRef]
- Schmidt, A. Controlling Apomixis: Shared Features and Distinct Characteristics of Gene Regulation. Genes 2020, 11, 29. [Google Scholar] [CrossRef]
- De Arias, M.M.; Gao, L.; Sherwood, D.A.; Dwivedi, K.; Price, B.J.; Jamison, M.; Kowallis, B.M.; Carman, J.G. Whether gametophytes are reduced or unreduced in angiosperms might be determined metabolically. Genes 2020, 11, 1449. [Google Scholar] [CrossRef]
- Underwood, C.J.; Mercier, R. Engineering Apomixis: Clonal Seeds Approaching the Fields. Annu. Rev. Plant Biol. 2022, 73, 201–225. [Google Scholar] [CrossRef] [PubMed]
- Sharbel, T.F.; Voigt, M.L.; Corral, J.M.; Galla, G.; Kumlehn, J.; Klukas, C.; Schreiber, F.; Vogel, H.; Rotter, B. Apomictic and sexual ovules of Boechera display heterochronic global gene expression patterns. Plant Cell 2010, 22, 655–671. [Google Scholar] [CrossRef] [PubMed]
- Sharbel, T.F.; Voigt, M.L.; Corral, J.M.; Thiel, T.; Varshney, A.; Kumlehn, J.; Vogel, H.; Rotter, B. Molecular signatures of apomictic and sexual ovules in the Boechera holboellii complex. Plant J. 2009, 58, 870–882. [Google Scholar] [CrossRef] [PubMed]
- Kantama, L.; Sharbel, T.F.; Schranz, M.E.; Mitchell-Olds, T.; de Vries, S.; de Jong, H. Diploid apomicts of the Boechera holboellii complex display large-scale chromosome substitutions and aberrant chromosomes. Proc. Natl. Acad. Sci. USA 2007, 104, 14026–14031. [Google Scholar] [CrossRef]
- Stein, J.; Quarin, C.L.; Martinez, E.J.; Pessino, S.C.; Ortiz, J.P.A. Tetraploid races of Paspalum notatum show polysomic inheritance and preferential chromosome pairing around the apospory-controlling locus. Theor. Appl. Genet. 2004, 109, 186–191. [Google Scholar] [CrossRef]
- Akiyama, Y.; Conner, J.A.; Goel, S.; Morishige, D.T.; Mullet, J.E.; Hanna, W.W.; Ozias-Akins, P. High-resolution physical mapping in Pennisetum squamulatum reveals extensive chromosomal heteromorphism of the genomic region associated with apomixis. Plant Physiol. 2004, 134, 1733–1741. [Google Scholar] [CrossRef]
- Podio, M.; Siena, L.A.; Hojsgaard, D.; Stein, J.; Quarin, C.L.; Ortiz, J.P.A. Evaluation of meiotic abnormalities and pollen viability in aposporous and sexual tetraploid Paspalum notatum (Poaceae). Plant Syst. Evol. 2012, 298, 1625–1633. [Google Scholar] [CrossRef]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef]
- Siena, L.A.; Sartor, M.E.; Espinoza, F.; Quarin, C.L.; Ortiz, J.P.A. Genetic and embryological evidences of apomixis at the diploid level in Paspalum rufum support recurrent auto-polyploidization in the species. Sex. Plant Reprod. 2008, 21, 205–215. [Google Scholar] [CrossRef]
- Schinkel, C.C.F.; Kirchheimer, B.; Dellinger, A.S.; Klatt, S.; Winkler, M.; Dullinger, S.; Hörandl, E. Correlations of polyploidy and apomixis with elevation and associated environmental gradients in an alpine plant. Aob Plants 2016, 8, plw064. [Google Scholar] [CrossRef]
- Pellino, M.; Hojsgaard, D.; Hörandl, E.; Sharbel, T.E. Chasing the apomictic factors in the Ranunculus auricomus complex: Exploring gene expression patterns in microdissected sexual and apomictic ovules. Genes 2020, 11, 728. [Google Scholar] [CrossRef] [PubMed]
- Darlington, C.D. The Evolution of Genetic Systems; Cambridge University Press: Cambridge, UK, 1939. [Google Scholar]
- Barke, B.H.; Karbstein, K.; Daubert, M.; Hörandl, E. The relation of meiotic behaviour to hybridity, polyploidy and apomixis in the Ranunculus auricomus complex (Ranunculaceae). Bmc Plant Biol. 2020, 20, 1. [Google Scholar] [CrossRef] [PubMed]
- Rieseberg, L.H.; Willis, J.H. Plant speciation. Science 2007, 317, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Tiley, G.P.; Burleigh, G. The relationship of recombination rate, genome structure, and patterns of molecular evolution across angiosperms. Bmc Evol. Biol. 2015, 15, 194. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.J. The relation of recombination to mutational advance. Mutat. Res. 1964, 106, 2–9. [Google Scholar] [CrossRef]
- Pellino, M.; Hojsgaard, D.; Schmutzer, T.; Scholz, U.; Hörandl, E.; Vogel, H.; Sharbel, T.F. Asexual genome evolution in the apomictic Ranunculus auricomus complex: Examining the effects of hybridization and mutation accumulation. Mol. Ecol. 2013, 22, 5908–5921. [Google Scholar] [CrossRef]
- Hollister, J.D.; Greiner, S.; Wang, W.; Wang, J.; Zhang, Y.; Wong, G.K.-S.; Wright, S.I.; Johnson, M.T. Recurrent loss of sex is associated with accumulation of deleterious mutations in Oenothera. Mol. Biol. Evol. 2015, 32, 896–905. [Google Scholar] [CrossRef]
- Lovell, J.T.; Williamson, R.J.; Wright, S.I.; McKay, J.K.; Sharbel, T.F. Mutation Accumulation in an Asexual Relative of Arabidopsis. PLoS Genet. 2017, 13, e1006550. [Google Scholar] [CrossRef]
- Karbstein, K.; Tomasello, S.; Hodac, L.; Lorberg, E.; Daubert, M.; Hörandl, E. Moving beyond assumptions: Polyploidy and environmental effects explain a geographical parthenogenesis scenario in European plants. Mol. Ecol. 2021, 30, 2659–2675. [Google Scholar] [CrossRef]
- Karbstein, K.; Tomasello, S.; Hodac, L.; Wagner, N.; Marincek, P.; Barke, B.H.; Paetzold, C.; Hörandl, E. Untying Gordian knots: Unraveling reticulate polyploid plant evolution by genomic data using the large Ranunculus auricomus species complex. New Phytol. 2022, 235, 2081–2098. [Google Scholar] [CrossRef]
- Karbstein, K.; Tomasello, S.; Hodac, L.; Dunkel, F.G.; Daubert, M.; Hörandl, E. Phylogenomics supported by geometric morphometrics reveals delimitation of sexual species within the polyploid apomictic Ranunculus auricomus complex (Ranunculaceae). Taxon 2020, 69, 1191–1220. [Google Scholar] [CrossRef]
- Tomasello, S.; Karbstein, K.; Hodač, L.; Paetzold, C.; Hörandl, E. Phylogenomics unravels Quaternary vicariance and allopatric speciation patterns in temperate-montane plant species: A case study on the Ranunculus auricomus species complex. Mol. Ecol. 2020, 29, 2031–2049. [Google Scholar] [CrossRef] [PubMed]
- Nogler, G.A. Genetics of apospory in apomictic Ranunculus auricomus. 5. Conclusion. Bot. Helv. 1984, 94, 411–422. [Google Scholar]
- Hojsgaard, D.; Greilhuber, J.; Pellino, M.; Paun, O.; Sharbel, T.F.; Hörandl, E. Emergence of apospory and bypass of meiosis via apomixis after sexual hybridisation and polyploidisation. New Phytol. 2014, 204, 1000–1012. [Google Scholar] [CrossRef]
- Karbstein, K.; Rahmsdorf, E.; Tomasello, S.; Hodač, L.; Hörandl, E. Breeding system of diploid sexuals within the Ranunculus auricomus complex and its role in a geographical parthenogenesis scenario. Ecol. Evol. 2020, 10, 14435–14450. [Google Scholar] [CrossRef] [PubMed]
- Hörandl, E. Evolutionary implications of self-compatibility and reproductive fitness in the apomictic Ranunculus auricomus polyploid complex (Ranunculaceae). Int. J. Plant Sci. 2008, 169, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- Hodač, L.; Klatt, S.; Hojsgaard, D.; Sharbel, T.; Hörandl, E. A little bit of sex prevents mutation accumulation even in apomictic polyploid plants. BMC Evol. Biol. 2019, 19, 170. [Google Scholar] [CrossRef]
- Barke, B.H.; Daubert, M.; Hörandl, E. Establishment of apomixis in diploid F2 hybrids and inheritance of apospory from F1 to F2 hybrids of the Ranunculus auricomus complex. Front. Plant Sci. 2018, 9, 1111. [Google Scholar] [CrossRef]
- Del Amparo, R.; Branco, C.; Arenas, J.; Vicens, A.; Arenas, M. Analysis of selection in protein-coding sequences accounting for common biases. Brief. Bioinform. 2021, 22, bbaa431. [Google Scholar] [CrossRef]
- Hörandl, E.; Greilhuber, J. Diploid and autotetraploid sexuals and their relationships to apomicts in the Ranunculus cassubicus group: Insights from DNA content and isozyme variation. Plant Syst. Evol. 2002, 234, 85–100. [Google Scholar] [CrossRef]
- Leebens-Mack, J.H.; Barker, M.S.; Carpenter, E.J.; Deyholos, M.K.; Gitzendanner, M.A.; Graham, S.W.; Grosse, I.; Li, Z.; Melkonian, M.; Mirarab, S.; et al. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef]
- Koci, J.; Roslein, J.; Paces, J.; Kotusz, J.; Halacka, K.; Kosco, J.; Fedorcak, J.; Iakovenko, N.; Janko, K. No evidence for accumulation of deleterious mutations and fitness degradation in clonal fish hybrids: Abandoning sex without regrets. Mol. Ecol. 2020, 29, 3038–3055. [Google Scholar] [CrossRef] [PubMed]
- Hodač, L.; Barke, B.H.; Hörandl, E. Mendelian segregation of leaf phenotypes in experimental F-2 hybrids elucidates origin of morphological diversity of the apomictic Ranunculus auricomus complex. Taxon 2018, 67, 1082–1092. [Google Scholar] [CrossRef]
- Albrecht, B.; Scornavacca, C.; Cenci, A.; Huson, D.H. Fast computation of minimum hybridization networks. Bioinformatics 2012, 28, 191–197. [Google Scholar] [CrossRef]
- Degnan, J.H. Modeling Hybridization Under the Network Multispecies Coalescent. Syst. Biol. 2018, 67, 786–799. [Google Scholar] [CrossRef]
- Kubatko, L.S.; Chifman, J. An invariants-based method for efficient identification of hybrid species from large-scale genomic data. Bmc Evol. Biol. 2019, 19, 112. [Google Scholar] [CrossRef]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Pond, S.L.K. Detecting Individual Sites Subject to Episodic Diversifying Selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef] [PubMed]
- Welch, D.B.M.; Meselson, M.S. Rates of nucleotide substitution in sexual and anciently asexual rotifers. Proc. Natl. Acad. Sci. USA 2001, 98, 6720–6724. [Google Scholar] [CrossRef]
- Shah, J.N.; Kirioukhova, O.; Pawar, P.; Tayyab, M.; Mateo, J.L.; Johnston, A.J. Depletion of Key Meiotic Genes and Transcriptome-Wide Abiotic Stress Reprogramming Mark Early Preparatory Events Ahead of Apomeiotic Transition. Front. Plant Sci. 2016, 7, 1539. [Google Scholar] [CrossRef]
- Dean, P.J.; Siwiec, T.; Waterworth, W.M.; Schlogelhofer, P.; Armstrong, S.J.; West, C.E. A novel ATM-dependent X-ray-inducible gene is essential for both plant meiosis and gametogenesis. Plant J. 2009, 58, 791–802. [Google Scholar] [CrossRef]
- Wang, Y.B.; Hou, Y.N.; Gu, H.Y.; Kang, D.M.; Chen, Z.L.; Liu, J.J.; Qu, L.J. The Arabidopsis Anaphase-Promoting Complex/Cyclosome Subunit 1 is Critical for Both Female Gametogenesis and Embryogenesis. J. Integr. Plant Biol. 2013, 55, 64–74. [Google Scholar] [CrossRef]
- Zhou, W.; Li, Z.; Zhang, J.; Mou, B.; Zhou, W. The OsIME4 gene identified as a key to meiosis initiation by RNA in situ hybridization. Plant Biol. 2021, 23, 861–873. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.A.; de Palma, J.; Oane, R.; Gamuyao, R.; Luo, M.; Chaudhury, A.; Herve, P.; Xue, Q.; Bennett, J. OsTDL1A binds to the LRR domain of rice receptor kinase MSP1, and is required to limit sporocyte numbers. Plant J. 2008, 54, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Hojsgaard, D.H.; Martinez, E.J.; Quarin, C.L. Competition between meiotic and apomictic pathways during ovule and seed development results in clonality. New Phytol. 2013, 197, 336–347. [Google Scholar] [CrossRef]
- Zhang, J.Q.; Ma, H.L. The Female Gametophyte Characteristics and Gene Expression Analysis Involved in Apomixis of Wild Germplasm Materials of Kentucky Bluegrass in Gansu Province of China. J. Plant Growth Regul. 2022. [Google Scholar] [CrossRef]
- Selva, J.P.; Zappacosta, D.; Carballo, J.; Rodrigo, J.M.; Bellido, A.; Gallo, C.A.; Gallardo, J.; Echenique, V. Genes modulating the increase in sexuality in the facultative diplosporous grass Eragrostis curvula under water stress conditions. Genes 2020, 11, 969. [Google Scholar] [CrossRef]
- Mendes-Bonato, A.B.; Pagliarini, M.S.; Do Valle, C.B. Meiotic arrest compromises pollen fertility in an interspecific hybrid between Brachiaria ruziziensis x Brachiaria decumbens (Poaceae:Paniceae). Braz. Arch. Biol. Technol. 2007, 50, 831–837. [Google Scholar] [CrossRef]
- Otto, S.P.; Scott, M.F.; Immler, S. Evolution of haploid selection in predominantly diploid organisms. Proc. Natl. Acad. Sci. USA 2015, 112, 15952–15957. [Google Scholar] [CrossRef]
- Mau, M.; Liiving, T.; Fomenko, L.; Goertzen, R.; Paczesniak, D.; Bottner, L.; Sharbel, T.F. The spread of infectious asexuality through haploid pollen. New Phytol. 2021, 230, 804–820. [Google Scholar] [CrossRef]
- Zühl, L.; Volkert, C.; Ibberson, D.; Schmidt, A. Differential activity of F-box genes and E3 ligases distinguishes sexual versus apomictic germline specification in Boechera. J. Exp. Bot. 2019, 70, 5643–5657. [Google Scholar] [CrossRef]
- d’Erfurth, I.; Jolivet, S.; Froger, N.; Catrice, O.; Novatchkova, M.; Mercier, R. Turning meiosis into mitosis. PLoS. Biol. 2009, 7, e1000124. [Google Scholar] [CrossRef] [PubMed]
- Underwood, C.J.; Vijverberg, K.; Rigola, D.; Okamoto, S.; Oplaat, C.; Op den Camp, R.H.M.; Radoeva, T.; Schauer, S.E.; Fierens, J.; Jansen, K.; et al. A PARTHENOGENESIS allele from apomictic dandelion can induce egg cell division without fertilization in lettuce. Nat. Genet. 2022, 54, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.A.; Mookkan, M.; Huo, H.Q.; Chae, K.; Ozias-Akins, P. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc. Natl. Acad. Sci. USA 2015, 112, 11205–11210. [Google Scholar] [CrossRef]
- Grimanelli, D.; Garcia, M.; Kaszas, E.; Perotti, E.; Leblanc, O. Heterochronic expression of sexual reproductive programs during apomictic development in Tripsacum. Genetics 2003, 165, 1521–1531. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaumova, R.; Krahulcova, A.; Krahulec, F. The intriguing complexity of parthenogenesis inheritance in Pilosella rubra (Asteraceae, Lactuceae). Sex. Plant Reprod. 2012, 25, 185–196. [Google Scholar] [CrossRef]
- Curtis, M.D.; Grossniklaus, U. Molecular control of autonomous embryo and endosperm development. Sex. Plant Reprod. 2008, 21, 79–88. [Google Scholar] [CrossRef]
- Klatt, S.; Hadacek, F.; Hodač, L.; Brinkmann, G.; Eilerts, M.; Hojsgaard, D.; Hörandl, E. Photoperiod extension enhances sexual megaspore formation and triggers metabolic reprogramming in facultative apomictic Ranunculus auricomus. Front. Plant Sci. 2016, 7, 278. [Google Scholar] [CrossRef]
- Ulum, F.B.; Costa Castro, C.; Hörandl, E. Ploidy-dependent effects of light stress on the mode of reproduction in the Ranunculus auricomus complex (Ranunculaceae). Front. Plant Sci. 2020, 11, 104. [Google Scholar] [CrossRef]
- Hörandl, E.; Hadacek, F. The oxidative damage initiation hypothesis for meiosis. Plant Reprod. 2013, 26, 351–367. [Google Scholar] [CrossRef]
- Ulum, F.B.; Hadacek, F.; Hörandl, E. Polyploidy improves photosynthesis regulation within the Ranunculus auricomus complex (Ranunculaceae). Biology 2021, 10, 100191. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Prjibelski, A.D. rnaSPAdes: A de novo transcriptome assembler and its application to RNA-Seq data. Gigascience 2019, 8, giz100. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Huson, D.H.; Linz, S. Autumn Algorithm-Computation of Hybridization Networks for Realistic Phylogenetic Trees. Ieee-Acm Trans. Comput. Biol. Bioinform. 2018, 15, 398–410. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Scornavacca, C. Dendroscope 3: An Interactive Tool for Rooted Phylogenetic Trees and Networks. Syst. Biol. 2012, 61, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.; Findeiss, S.; Steiner, L.; Marz, M.; Stadler, P.F.; Prohaska, S.J. Proteinortho: Detection of (Co-)orthologs in large-scale analysis. BMC Bioinform. 2011, 12, 124. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Wernersson, R.; Pedersen, A.G. RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003, 31, 3537–3539. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Reichelt, N.; Wen, J.; Paetzold, C.; Appelhans, M.S. Target enrichment improves phylogenetic resolution in the genus Zanthoxylum (Rutaceae) and indicates both incomplete lineage sorting and hybridization events. Ann. Bot. 2021, 128, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Stajich, J.E.; Block, D.; Boulez, K.; Brenner, S.E.; Chervitz, S.A.; Dagdigian, C.; Fuellen, G.; Gilbert, J.G.R.; Korf, I.; Lapp, H.; et al. The bioperl toolkit: Perl modules for the life sciences. Genome Res 2002, 12, 1611–1618. [Google Scholar] [CrossRef] [PubMed]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramirez, F.; Vesztrocy, A.W.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef] [PubMed]
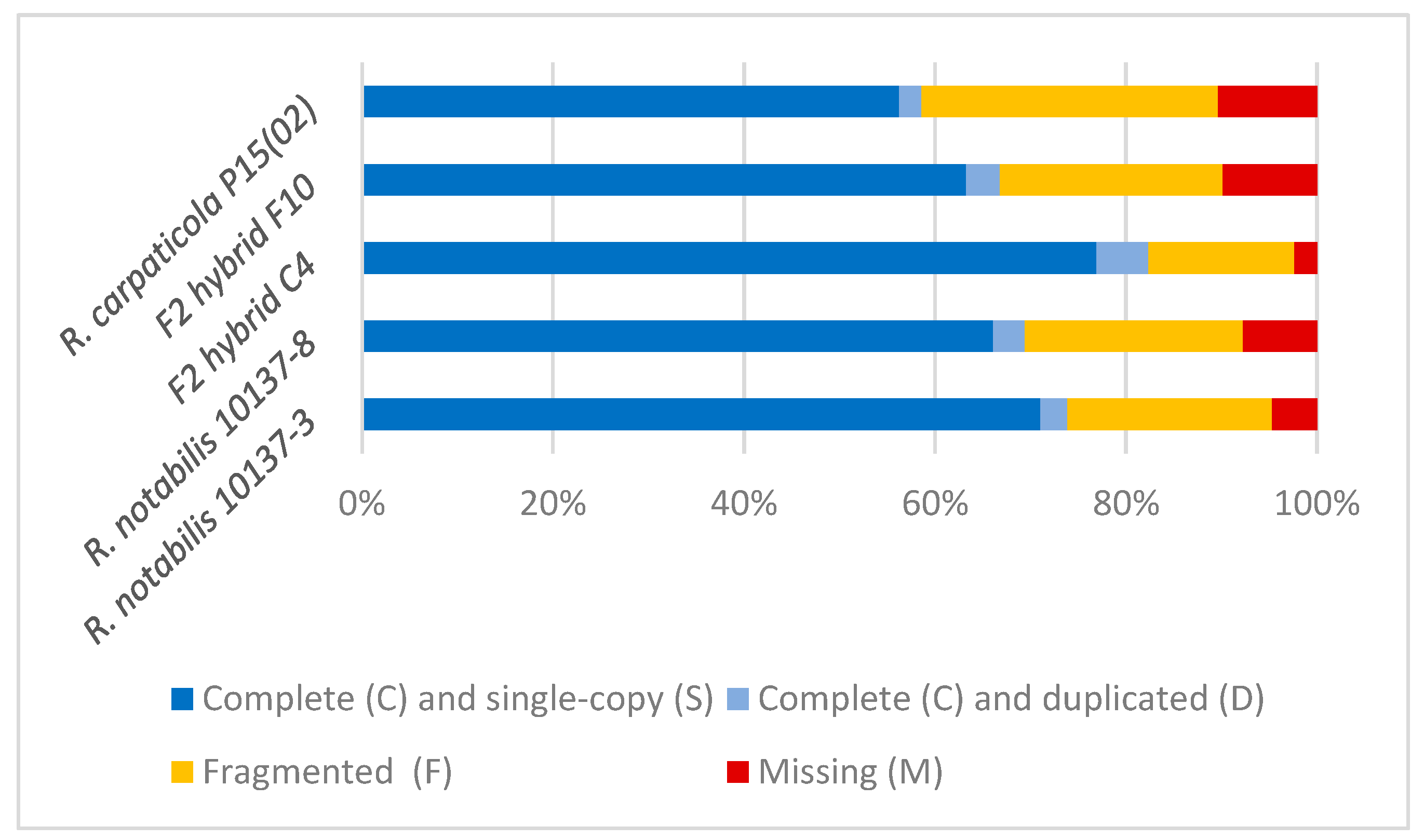
| R. notabilis 10137-3 | R. notabilis 10137-8 | R. carpaticola P15(02) | F2 Hybrid C4 | F2 Hybrid F10 | |
|---|---|---|---|---|---|
| Number of raw reads | 3,878,575 | 3,868,199 | 2,754,320 | 5,610,385 | 2,797,829 |
| Number of trimmed reads | 3,750,852 | 3,815,142 | 2,724,406 | 4,487,452 | 2,774,750 |
| Number of assembled contigs | 28,520 | 30,262 | 25,501 | 33,233 | 27,789 |
| Average contig length (bp) | 812.59 | 778.60 | 756.48 | 823.97 | 765.97 |
| Longest contig (bp) | 10,473 | 10,008 | 11,019 | 11,019 | 7131 |
| Shortest contig (bp) | 300 | 300 | 300 | 300 | 300 |
| Levels of dN/dS Values of 1514 Loci in Pairwise Comparisons | No. Loci Filtered | % |
|---|---|---|
| Parental divergence (average of 2 pairs, dN/dS > 1.0) | 139 | 9.18% |
| Intraspecific polymorphism in R. notabilis (1 pair, dN/dS > 1.0) | 81 | 5.35% |
| Hybrid–hybrid divergence (1 pair, dN/dS > 4.0) | 20 | 1.32% |
| Parent–hybrid comparison (average of 6 pairs, dN/dS < 1.0) | 1176 | 77.68% |
| Parent–hybrid comparison not annotated (average of 6 pairs, dN/dS > 1.0) | 19 | 1.25% |
| Parent–hybrid comparison annotated (average of 6 pairs, dN/dS > 1.0) | 79 | 5.22% |
| Total no. of loci | 1514 | 100.00% |
| Contig | Protein Coordinates | Symbol | Function | GO Association |
|---|---|---|---|---|
| contig_1127 | 1–978 [+] | THA8_ARATH | THYLAKOID ASSEMBLY 8, chloroplastic | |
| GO:0009793 embryo development ending in seed dormancy | ||||
| contig_1423 | 1–921 [+] | XRI1_ARATH | X-ray-induced transcript 1 | |
| GO:0007143 female meiotic nuclear division | ||||
| GO:0007140 male meiotic nuclear division | ||||
| GO:0009555 pollen development | ||||
| contig_145 | 1524–3197 [+] | APC1_ARATH | Anaphase-promoting complex subunit 1 | |
| GO:0009793 embryo development ending in seed dormancy | ||||
| GO:0009553 embryo sac development | ||||
| GO:0007091 metaphase/anaphase transition of mitotic cell cycle | ||||
| GO:0048481 plant ovule development | ||||
| contig_213 | 2–1033 [+] | ADT4_ARATH | ADP-ATP carrier protein ER-ANT1 | |
| GO:0048316 seed development | ||||
| contig_3193 | 242–916 [+] | DIV_ANTMA | Transcription factor DIVARICATA | |
| GO:0009908 flower development | ||||
| contig_3535 | 242–916 [+] | MSP1_ORYSJ | Leucine-rich repeat receptor protein kinase MSP1 | |
| GO:0048658 anther wall tapetum development | ||||
| GO:0009554 megasporogenesis | ||||
| GO:0009556 microsporogenesis | ||||
| contig_3826 | 1–384 [+] | NOC4_ARATH | Protein NUCLEOLAR COMPLEX ASSOCIATED 4 | |
| GO:0009793 embryo development ending in seed dormancy | ||||
| contig_4136 | 1–1857 [+] | ASY1_ARATH | Meiosis-specific protein ASY1 | |
| GO:0007129 homologous chromosome pairing at meiosis | ||||
| contig_52 | 919–3012 [+] | SYVM2_ARATH | Valine—tRNA ligase, chloroplastic/mitochondrial | |
| GO:0009793 embryo development ending in seed dormancy | ||||
| contig_583 | 1–1032 [+] | SC15B_ARATH | Exocyst complex component SEC15B | |
| GO:0060321 acceptance of pollen | ||||
| GO:0009846 pollen germination | ||||
| GO:0009860 pollen tube growth | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paetzold, C.; Barke, B.H.; Hörandl, E. Evolution of Transcriptomes in Early-Generation Hybrids of the Apomictic Ranunculus auricomus Complex (Ranunculaceae). Int. J. Mol. Sci. 2022, 23, 13881. https://doi.org/10.3390/ijms232213881
Paetzold C, Barke BH, Hörandl E. Evolution of Transcriptomes in Early-Generation Hybrids of the Apomictic Ranunculus auricomus Complex (Ranunculaceae). International Journal of Molecular Sciences. 2022; 23(22):13881. https://doi.org/10.3390/ijms232213881
Chicago/Turabian StylePaetzold, Claudia, Birthe H. Barke, and Elvira Hörandl. 2022. "Evolution of Transcriptomes in Early-Generation Hybrids of the Apomictic Ranunculus auricomus Complex (Ranunculaceae)" International Journal of Molecular Sciences 23, no. 22: 13881. https://doi.org/10.3390/ijms232213881
APA StylePaetzold, C., Barke, B. H., & Hörandl, E. (2022). Evolution of Transcriptomes in Early-Generation Hybrids of the Apomictic Ranunculus auricomus Complex (Ranunculaceae). International Journal of Molecular Sciences, 23(22), 13881. https://doi.org/10.3390/ijms232213881





