Abnormal Pre-mRNA Splicing in Exonic Fabry Disease-Causing GLA Mutations
Abstract
:1. Introduction
2. Results
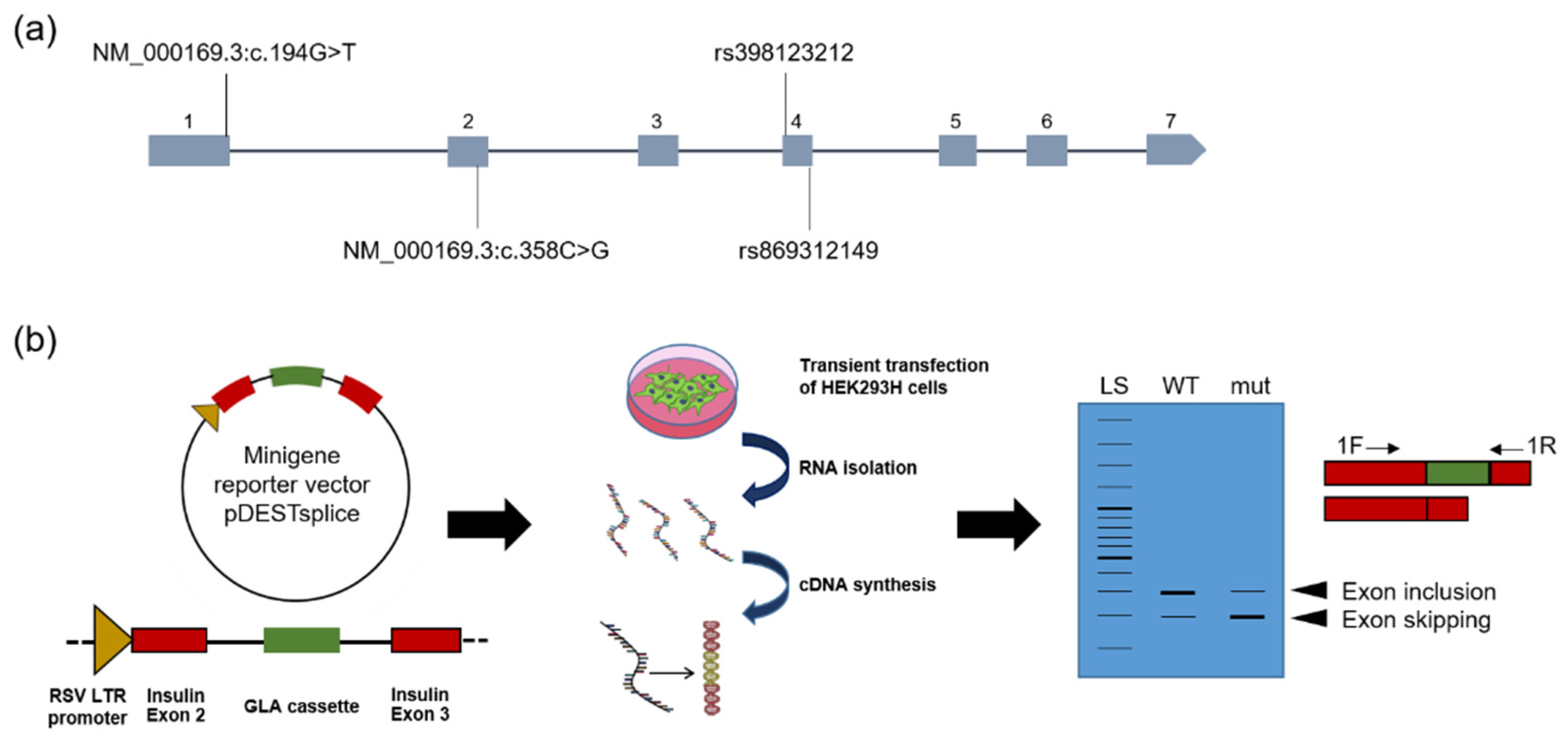
2.1. In Silico Splicing Prediction
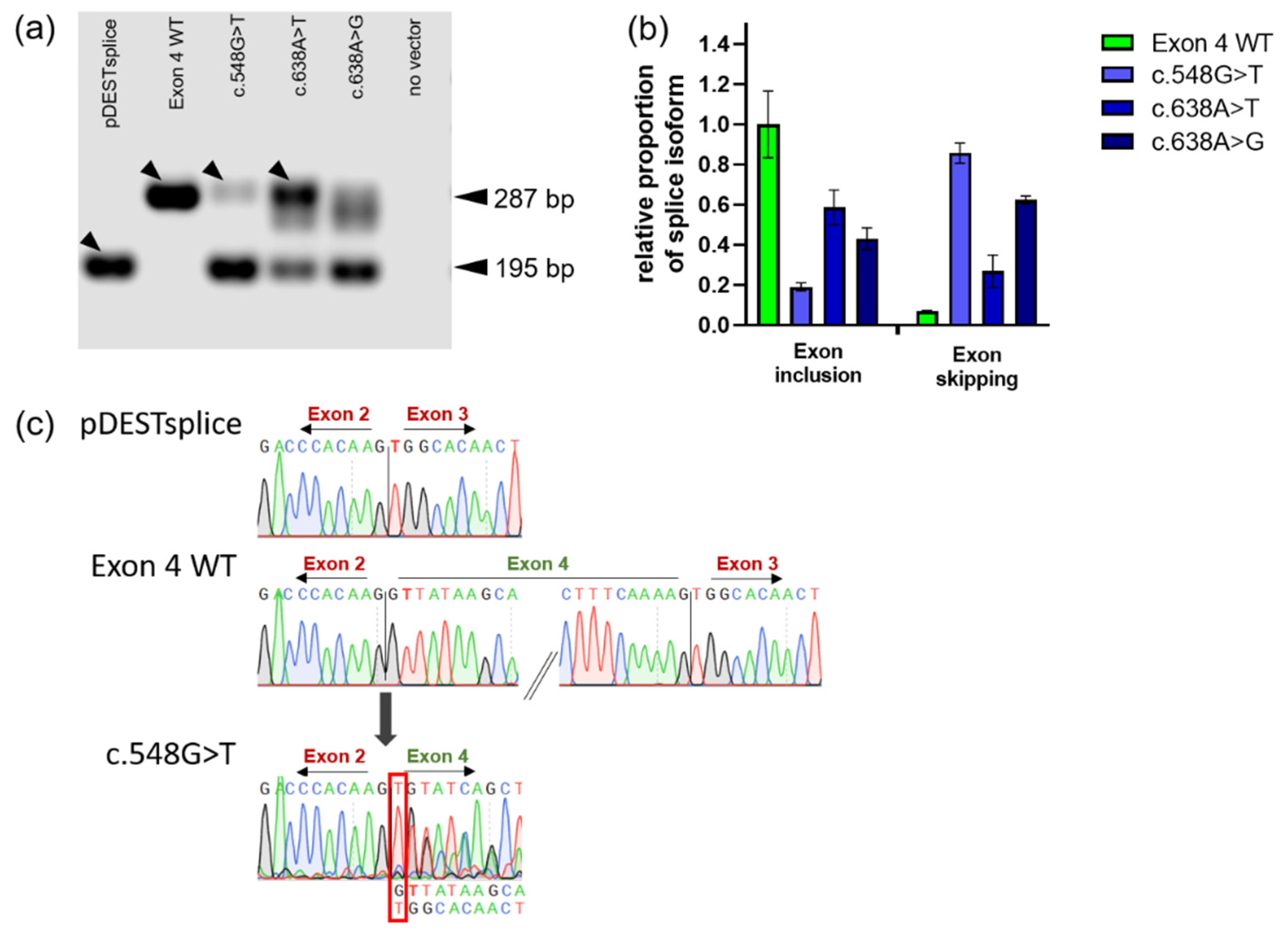
2.2. Corroboration of Splicing Abnormalities Due to GLA Exon 4 Mutations
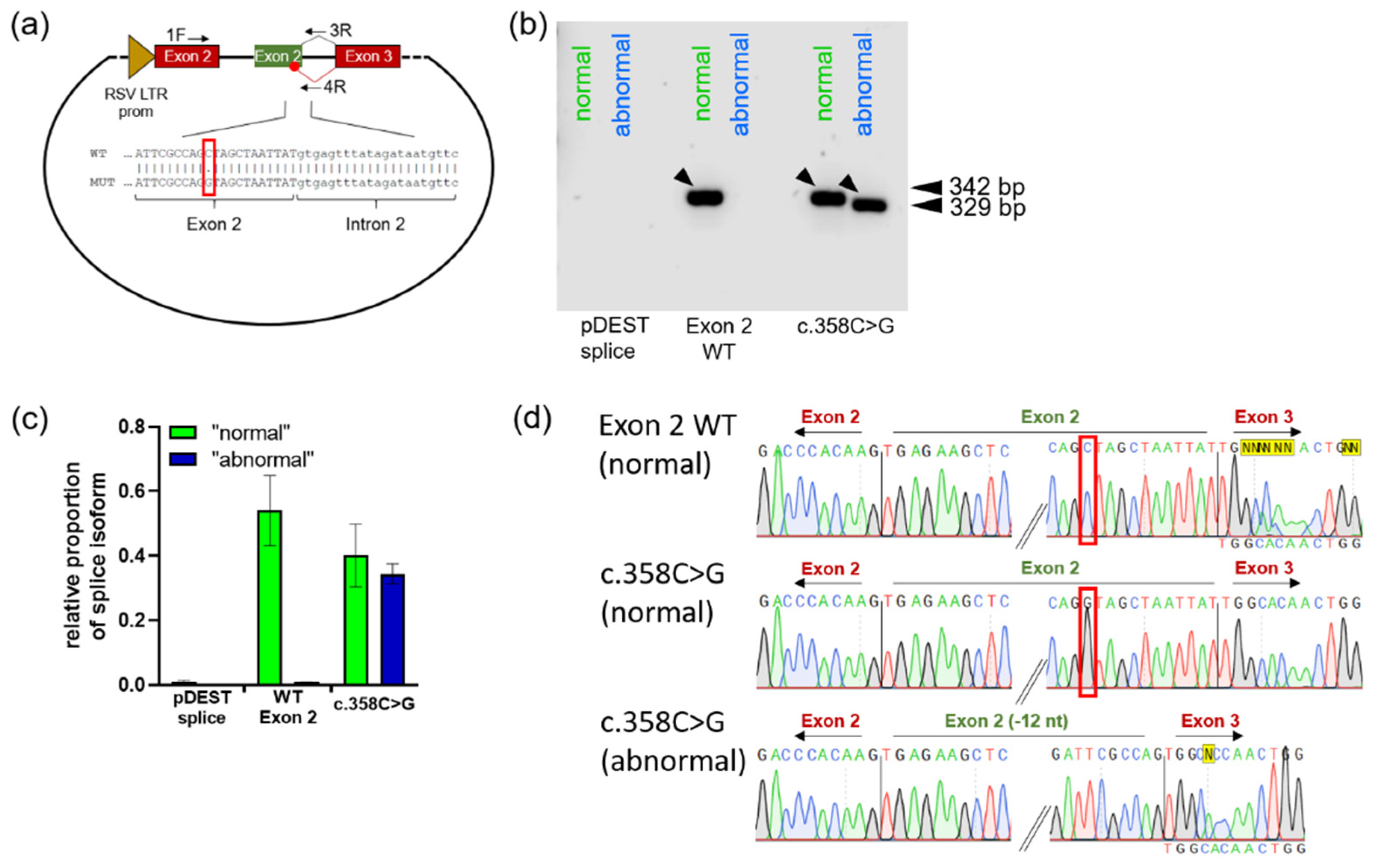
2.3. Utilization of Alternative Splicing Sites as a Result of Mutations in Exons 1 and 2
3. Discussion
4. Materials and Methods
4.1. Splicing Prediction
4.2. Cell Culture
4.3. AGAL Enzyme Activity Measurement
4.4. Construction of the Minigen Reporter Vectors
4.5. Isoform-Specific PCR Assay
4.6. Agarose Gel DNA Band Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mehta, A.; Beck, M.; Eyskens, F.; Feliciani, C.; Kantola, I.; Ramaswami, U.; Rolfs, A.; Rivera, A.; Waldek, S.; Germain, D.P. Fabry disease: A review of current management strategies. QJM 2010, 103, 641–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wendrich, K.; Whybra, C.; Ries, M.; Gal, A.; Beck, M. Neurological manifestation of Fabry disease in females. Contrib. Nephrol. 2001, 241–244. [Google Scholar] [CrossRef]
- MacDermot, K.D.; Holmes, A.; Miners, A.H. Anderson-Fabry disease: Clinical manifestations and impact of disease in a cohort of 60 obligate carrier females. J. Med. Genet. 2001, 38, 769–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arends, M.; Wanner, C.; Hughes, D.; Mehta, A.; Oder, D.; Watkinson, O.T.; Elliott, P.M.; Linthorst, G.E.; Wijburg, F.A.; Biegstraaten, M.; et al. Characterization of Classical and Nonclassical Fabry Disease: A Multicenter Study. J. Am. Soc. Nephrol. 2017, 28, 1631–1641. [Google Scholar] [CrossRef] [Green Version]
- Ortiz, A.; Germain, D.P.; Desnick, R.J.; Politei, J.; Mauer, M.; Burlina, A.; Eng, C.; Hopkin, R.J.; Laney, D.; Linhart, A.; et al. Fabry disease revisited: Management and treatment recommendations for adult patients. Mol. Genet. Metab. 2018, 123, 416–427. [Google Scholar] [CrossRef]
- GALAFOLD® GLA Mutation Search. Available online: https://www.galafoldamenabilitytable.com/ (accessed on 12 October 2022).
- Ishii, S.; Nakao, S.; Minamikawa-Tachino, R.; Desnick, R.J.; Fan, J.-Q. Alternative splicing in the alpha-galactosidase A gene: Increased exon inclusion results in the Fabry cardiac phenotype. Am. J. Hum. Genet. 2002, 70, 994–1002. [Google Scholar] [CrossRef] [Green Version]
- Dardis, A.; Buratti, E. Impact, Characterization, and Rescue of Pre-mRNA Splicing Mutations in Lysosomal Storage Disorders. Genes 2018, 9, 73. [Google Scholar] [CrossRef] [Green Version]
- Filoni, C.; Caciotti, A.; Carraresi, L.; Donati, M.A.; Mignani, R.; Parini, R.; Filocamo, M.; Soliani, F.; Simi, L.; Guerrini, R.; et al. Unbalanced GLA mRNAs ratio quantified by real-time PCR in Fabry patients’ fibroblasts results in Fabry disease. Eur. J. Hum. Genet. 2008, 16, 1311–1317. [Google Scholar] [CrossRef] [Green Version]
- Yokoi, T.; Shinoda, K.; Ohno, I.; Kato, K.; Miyawaki, T.; Taniguchi, N. A 3′ splice site consensus sequence mutation in the intron 3 of the alpha-galactosidase A gene in a patient with Fabry disease. Jinrui Idengaku Zasshi 1991, 36, 245–250. [Google Scholar] [CrossRef]
- Matsumura, T.; Osaka, H.; Sugiyama, N.; Kawanishi, C.; Maruyama, Y.; Suzuki, K.; Onishi, H.; Yamada, Y.; Morita, M.; Aoki, M.; et al. Novel acceptor splice site mutation in the invariant AG of intron 6 of α-galactosidase A gene, causing Fabry disease. Hum. Mutat. 1998, 11, 483. [Google Scholar] [CrossRef]
- Li, P.; Zhang, L.; Zhao, N.; Xiong, Q.; Zhou, Y.-A.; Wu, C.; Xiao, H. A Novel α-Galactosidase A Splicing Mutation Predisposes to Fabry Disease. Front. Genet. 2019, 10, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varela, P.; Caldas, M.M.; Pesquero, J.B. Novel GLA Mutation Promotes Intron Inclusion Leading to Fabry Disease. Front. Genet. 2019, 10, 783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soukarieh, O.; Gaildrat, P.; Hamieh, M.; Drouet, A.; Baert-Desurmont, S.; Frébourg, T.; Tosi, M.; Martins, A. Exonic Splicing Mutations Are More Prevalent than Currently Estimated and Can Be Predicted by Using In Silico Tools. PLoS Genet. 2016, 12, e1005756. [Google Scholar] [CrossRef] [Green Version]
- Lukas, J.; Giese, A.-K.; Markoff, A.; Grittner, U.; Kolodny, E.; Mascher, H.; Lackner, K.J.; Meyer, W.; Wree, P.; Saviouk, V.; et al. Functional characterisation of alpha-galactosidase a mutations as a basis for a new classification system in fabry disease. PLoS Genet. 2013, 9, e1003632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukas, J.; Scalia, S.; Eichler, S.; Pockrandt, A.-M.; Dehn, N.; Cozma, C.; Giese, A.-K.; Rolfs, A. Functional and Clinical Consequences of Novel α-Galactosidase A Mutations in Fabry Disease. Hum. Mutat. 2016, 37, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Citro, V.; Cammisa, M.; Liguori, L.; Cimmaruta, C.; Lukas, J.; Cubellis, M.V.; Andreotti, G. The Large Phenotypic Spectrum of Fabry Disease Requires Graduated Diagnosis and Personalized Therapy: A Meta-Analysis Can Help to Differentiate Missense Mutations. Int. J. Mol. Sci. 2016, 17, 2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukas, J.; Knospe, A.-M.; Seemann, S.; Citro, V.; Cubellis, M.V.; Rolfs, A. In Vitro Enzyme Measurement to Test Pharmacological Chaperone Responsiveness in Fabry and Pompe Disease. J. Vis. Exp. 2017, 130, 56550. [Google Scholar] [CrossRef]
- Sanchez-Niño, M.D.; Ortiz, A. Is it or is it not a pathogenic mutation? Is it or is it not the podocyte? J. Nephropathol. 2012, 1, 152–154. [Google Scholar] [CrossRef] [Green Version]
- Putscher, E.; Hecker, M.; Fitzner, B.; Lorenz, P.; Zettl, U.K. Principles and Practical Considerations for the Analysis of Disease-Associated Alternative Splicing Events Using the Gateway Cloning-Based Minigene Vectors pDESTsplice and pSpliceExpress. Int. J. Mol. Sci. 2021, 22, 5154. [Google Scholar] [CrossRef]
- Germain, D.P.; Shabbeer, J.; Cotigny, S.; Desnick, R.J. Fabry disease: Twenty novel alpha-galactosidase A mutations and genotype-phenotype correlations in classical and variant phenotypes. Mol. Med. 2002, 8, 306–312. [Google Scholar] [CrossRef]
- Filoni, C.; Caciotti, A.; Carraresi, L.; Cavicchi, C.; Parini, R.; Antuzzi, D.; Zampetti, A.; Feriozzi, S.; Poisetti, P.; Garman, S.C.; et al. Functional studies of new GLA gene mutations leading to conformational Fabry disease. Biochim. Biophys. Acta 2010, 1802, 247–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamin, E.R.; Della Valle, M.C.; Wu, X.; Katz, E.; Pruthi, F.; Bond, S.; Bronfin, B.; Williams, H.; Yu, J.; Bichet, D.G.; et al. The validation of pharmacogenetics for the identification of Fabry patients to be treated with migalastat. Genet. Med. 2017, 19, 430–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef] [PubMed]
- Hwu, W.-L.; Chien, Y.-H.; Lee, N.-C.; Chiang, S.-C.; Dobrovolny, R.; Huang, A.-C.; Yeh, H.-Y.; Chao, M.-C.; Lin, S.-J.; Kitagawa, T.; et al. Newborn screening for Fabry disease in Taiwan reveals a high incidence of the later-onset GLA mutation c.936+919GA (IVS4+919GA). Hum. Mutat. 2009, 30, 1397–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishii, S.; Suzuki, Y.; Fan, J.Q. Role of Ser-65 in the activity of alpha-galactosidase A: Characterization of a point mutation (S65T) detected in a patient with Fabry disease. Arch. Biochem. Biophys. 2000, 377, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Andreotti, G.; Citro, V.; de Crescenzo, A.; Orlando, P.; Cammisa, M.; Correra, A.; Cubellis, M.V. Therapy of Fabry disease with pharmacological chaperones: From in silico predictions to in vitro tests. Orphanet J. Rare Dis. 2011, 6, 66. [Google Scholar] [CrossRef] [Green Version]
- Auray-Blais, C.; Boutin, M.; Gagnon, R.; Dupont, F.O.; Lavoie, P.; Clarke, J.T.R. Urinary globotriaosylsphingosine-related biomarkers for Fabry disease targeted by metabolomics. Anal. Chem. 2012, 84, 2745–2753. [Google Scholar] [CrossRef]
- Lukas, J.; Torras, J.; Navarro, I.; Giese, A.-K.; Böttcher, T.; Mascher, H.; Lackner, K.J.; Fauler, G.; Paschke, E.; Cruzado, J.M.; et al. Broad spectrum of Fabry disease manifestation in an extended Spanish family with a new deletion in the GLA gene. Clin. Kidney J. 2012, 5, 395–400. [Google Scholar] [CrossRef] [Green Version]
- Sueoka, H.; Ichihara, J.; Tsukimura, T.; Togawa, T.; Sakuraba, H. Nano-LC-MS/MS for Quantification of Lyso-Gb3 and Its Analogues Reveals a Useful Biomarker for Fabry Disease. PLoS ONE 2015, 10, e0127048. [Google Scholar] [CrossRef]
- Center for Drug Evaluation and Research. Definitions; Qeios: London, UK, 2018. [Google Scholar]
- Lai, L.-W.; Whitehair, O.; Wu, M.-J.; O’Meara, M.; Lien, Y.-H.H. Analysis of splice-site mutations of the alpha-galactosidase A gene in Fabry disease. Clin. Genet. 2003, 63, 476–482. [Google Scholar] [CrossRef]
- Markham, A. Migalastat: First Global Approval. Drugs 2016, 76, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Parenti, G.; Pignata, C.; Vajro, P.; Salerno, M. New strategies for the treatment of lysosomal storage diseases (review). Int. J. Mol. Med. 2013, 31, 11–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukas, J.; Cimmaruta, C.; Liguori, L.; Pantoom, S.; Iwanov, K.; Petters, J.; Hund, C.; Bunschkowski, M.; Hermann, A.; Cubellis, M.V.; et al. Assessment of Gene Variant Amenability for Pharmacological Chaperone Therapy with 1-Deoxygalactonojirimycin in Fabry Disease. Int. J. Mol. Sci. 2020, 21, 956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferri, L.; Covello, G.; Caciotti, A.; Guerrini, R.; Denti, M.A.; Morrone, A. Double-target Antisense U1snRNAs Correct Mis-splicing Due to c.639+861CT and c.639+919GA GLA Deep Intronic Mutations. Mol. Ther. Nucleic Acids 2016, 5, e380. [Google Scholar] [CrossRef]
- Genomnis_HSF_Pro. Genomnis_HSF_Pro. Available online: https://hsf.genomnis.com/login (accessed on 12 October 2022).


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alfen, F.; Putscher, E.; Hecker, M.; Zettl, U.K.; Hermann, A.; Lukas, J. Abnormal Pre-mRNA Splicing in Exonic Fabry Disease-Causing GLA Mutations. Int. J. Mol. Sci. 2022, 23, 15261. https://doi.org/10.3390/ijms232315261
Alfen F, Putscher E, Hecker M, Zettl UK, Hermann A, Lukas J. Abnormal Pre-mRNA Splicing in Exonic Fabry Disease-Causing GLA Mutations. International Journal of Molecular Sciences. 2022; 23(23):15261. https://doi.org/10.3390/ijms232315261
Chicago/Turabian StyleAlfen, Franziska, Elena Putscher, Michael Hecker, Uwe Klaus Zettl, Andreas Hermann, and Jan Lukas. 2022. "Abnormal Pre-mRNA Splicing in Exonic Fabry Disease-Causing GLA Mutations" International Journal of Molecular Sciences 23, no. 23: 15261. https://doi.org/10.3390/ijms232315261





