Comparison and Functional Analysis of Odorant-Binding Proteins and Chemosensory Proteins in Two Closely Related Thrips Species, Frankliniella occidentalis and Frankliniella intonsa (Thysanoptera: Thripidae) Based on Antennal Transcriptome Analysis
Abstract
1. Introduction
2. Results
2.1. Transcriptome Sequencing and Assembly
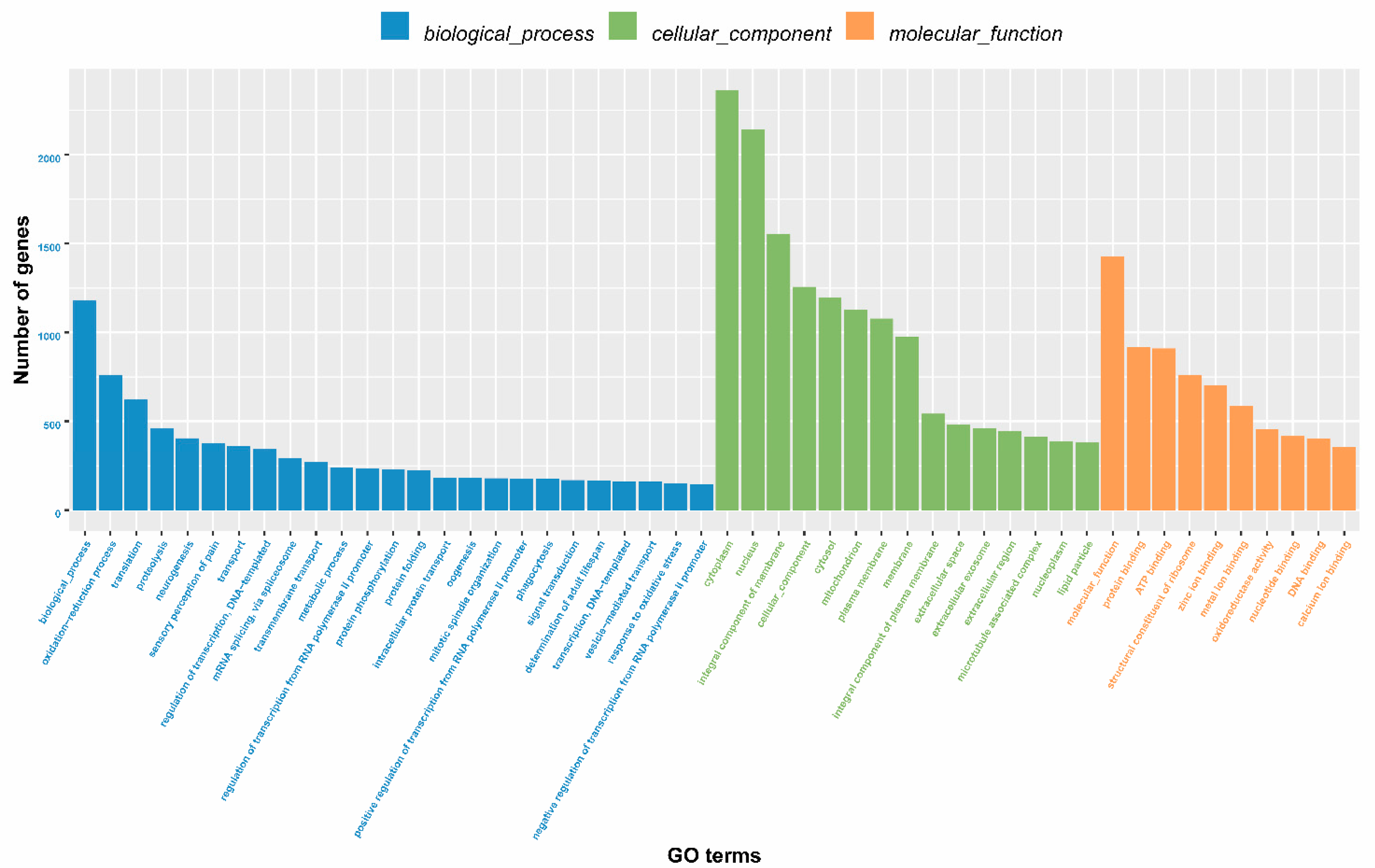
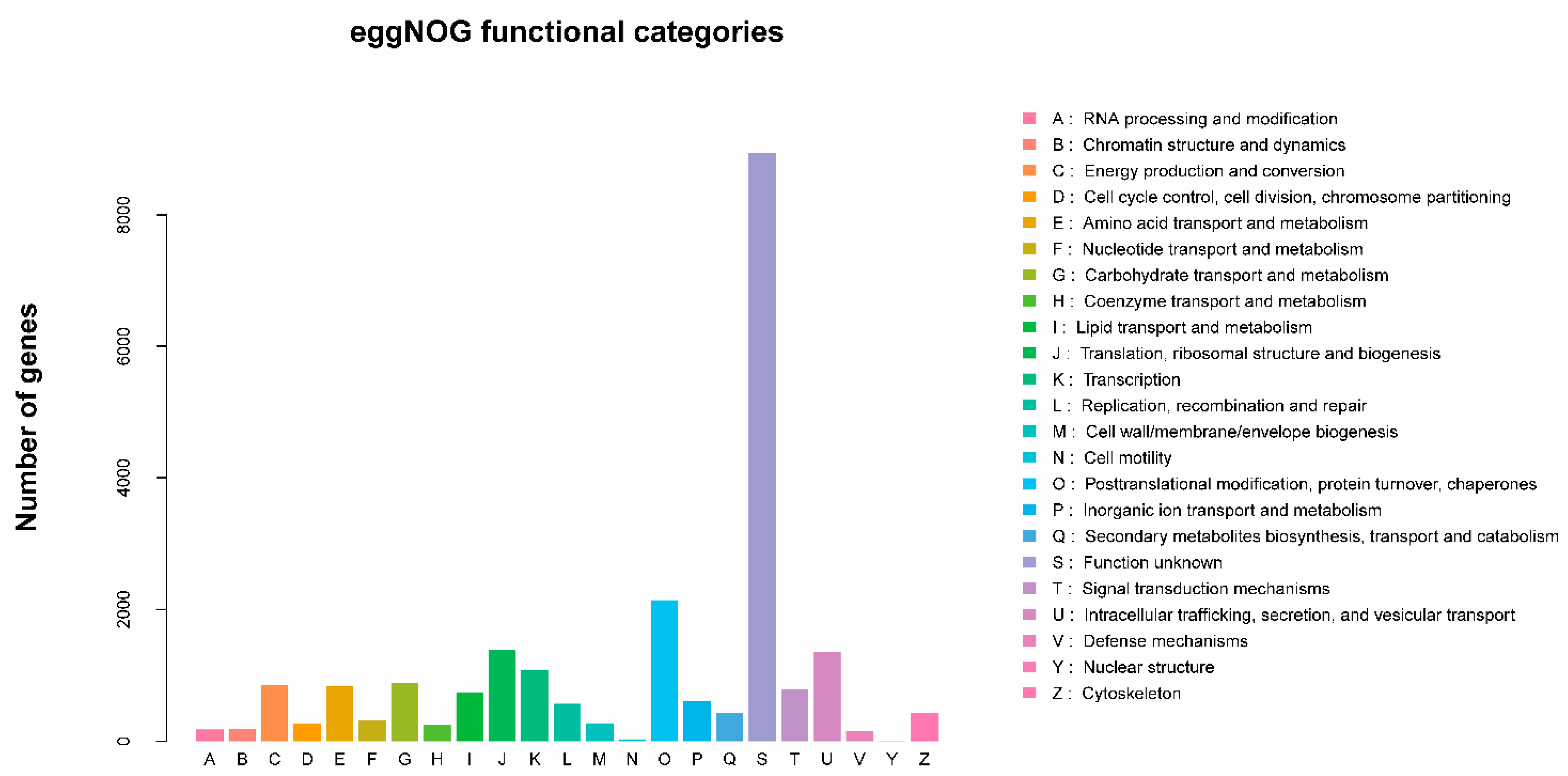
2.2. Annotation Information for Unigenes
2.3. Identification and Analysis of Putative OBPs
2.4. Identification and Analysis of Putative CSPs
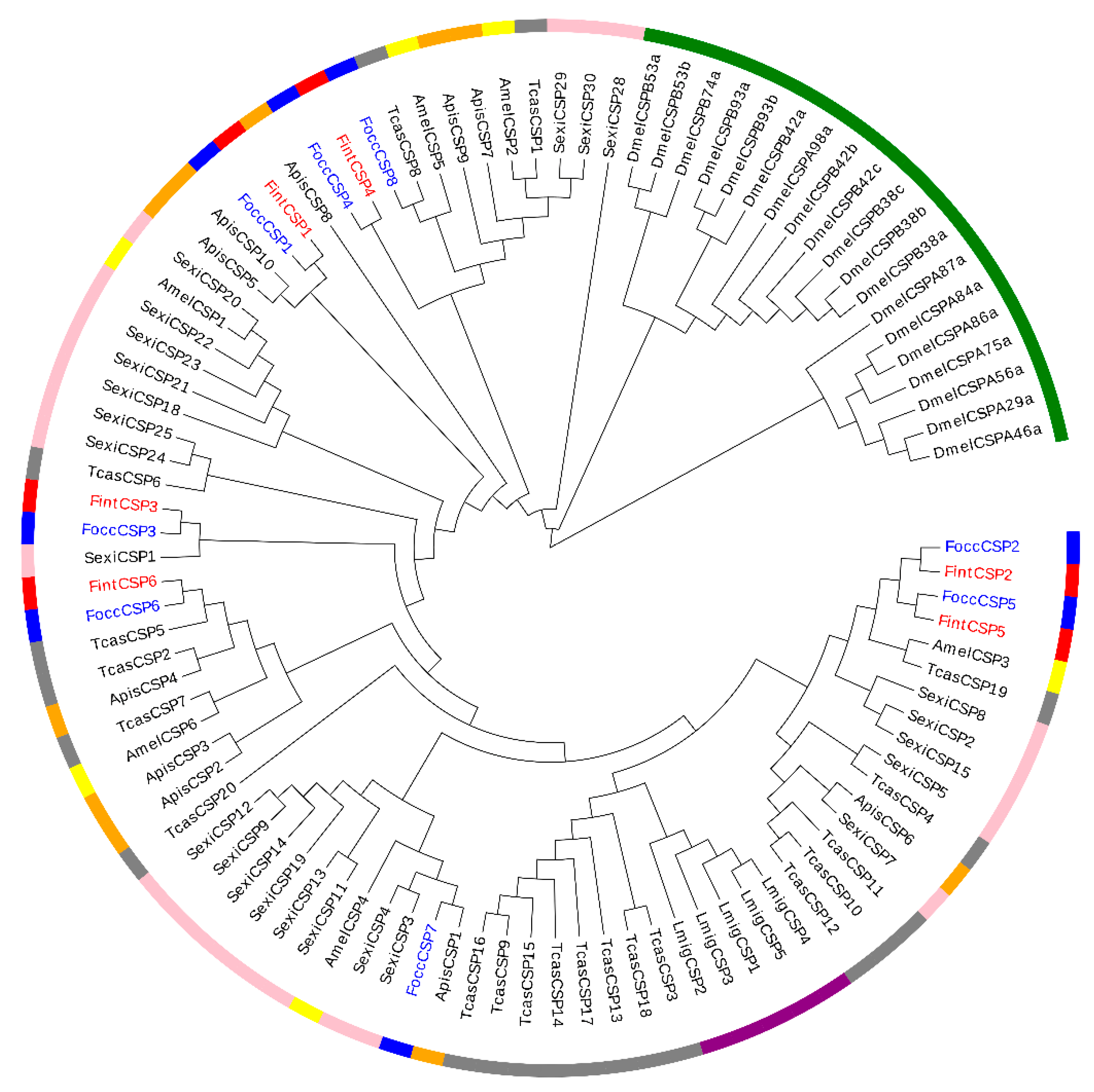
2.5. Phylogenetic Analysis of OBPs and CSPs in F. occidentalis and F. intonsa
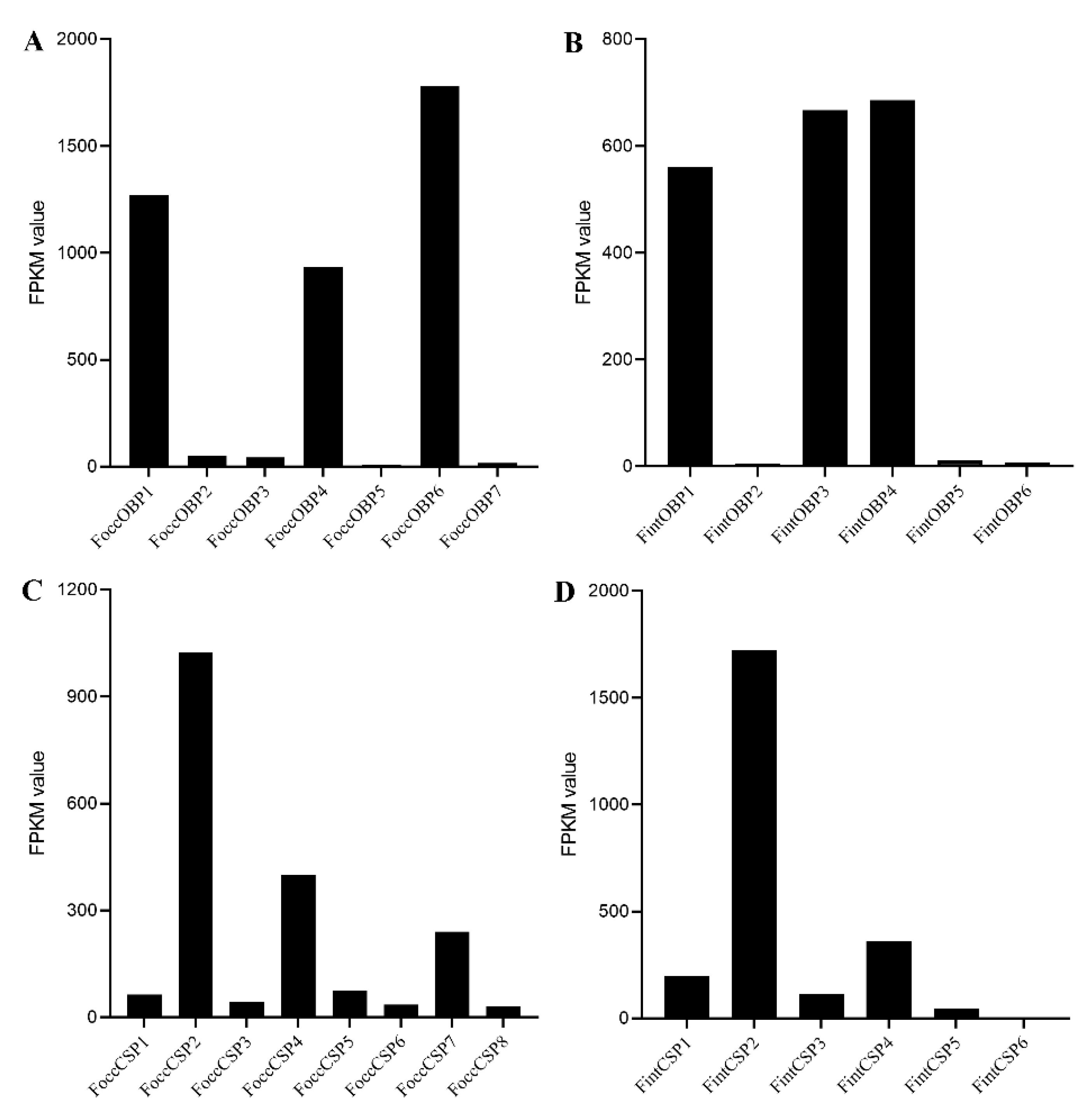
2.6. Expression Analyses of OBP and CSP Genes in Different Developmental Stages Based on Antennal Transcriptome and RT-qPCR
2.7. Modeling of Three-Dimensional (3D) Structure and Molecular Docking of Ligands
3. Discussion
4. Materials and Methods
4.1. Insect Rearing
4.2. Antennae Collection and RNA Extraction and Transcriptome Sequencing
4.3. De Novo Assembly, Unigene Annotation and Functional Classification
4.4. Identification of Putative OBP and CSP Genes
4.5. Phylogenetic Analysis of OBPs and CSPs
4.6. Expression Analyses of OBP and CSP Genes in Different Developmental Stages Based on Antennal Transcriptome and RT-qPCR
4.7. Modeling of Three-Dimensional (3D) Structure and Molecular Docking of Ligands
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bruce, T.J.A.; Wadhams, L.J.; Woodcock, C.M. Insect host location: A volatile situation. Trends Plant Sci. 2005, 10, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Reddy, G.V.P.; Guerrero, A. Interactions of insect pheromones and plant semiochemicals. Trends Plant Sci. 2004, 9, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Sandhi, R.K.; Reddy, G.V.P. A review of interactions between insect biological control agents and semiochemicals. Insects 2019, 10, 439. [Google Scholar] [CrossRef] [PubMed]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Sanchez-Gracia, A.; Vieira, F.G.; Rozas, J. Molecular evolution of the major chemosensory gene families in insects. Heredity 2009, 103, 208–216. [Google Scholar] [CrossRef]
- Olsson, S.B.; Jr, L.C.; Roelofs, W.L. The chemosensory basis for behavioral divergence involved in sympatric host shifts. I. Characterizing olfactory receptor neuron classes responding to key host volatiles. J. Comp. Physiol. 2006, 192, 279–288. [Google Scholar] [CrossRef][Green Version]
- Wang, B.; Dong, W.; Li, H.; D’Onofrio, C.; Bai, P.; Chen, R.; Yang, L.; Wu, J.; Wang, X.; Wang, B.; et al. Molecular basis of (E)-beta-farnesene-mediated aphid location in the predator Eupeodes corollae. Curr. Biol. 2022, 32, 951–962. [Google Scholar] [CrossRef]
- Campanini, E.B.; de Brito, R.A. Molecular evolution of odorant-binding proteins gene family in two closely related Anastrepha fruit flies. BMC Evol. Biol. 2016, 16, 198. [Google Scholar] [CrossRef]
- Pelosi, P.; Zhu, J.; Knoll, W. Odorant-binding proteins as sensing elements for odour monitoring. Sensors 2018, 18, 3248. [Google Scholar] [CrossRef]
- Liu, G.X.; Xuan, N.; Rajashekar, B.; Arnaud, P.; Offmann, B.; Picimbon, J.F. Comprehensive history of CSP genes: Evolution, phylogenetic distribution and functions. Genes 2020, 11, 413. [Google Scholar] [CrossRef]
- Robertson, H.M. Molecular evolution of the major arthropod chemoreceptor gene families. Annu. Rev. Entomol. 2019, 64, 227–242. [Google Scholar] [CrossRef] [PubMed]
- Cassau, S.; Krieger, J. The role of SNMPs in insect olfaction. Cell Tissue Res. 2021, 383, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Iovinella, I.; Felicioli, A.; Dani, F.R. Soluble proteins of chemical communication: An overview across arthropods. Front. Physiol. 2014, 5, 320. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Zhou, J.J.; Ban, L.P.; Calvello, M. Soluble proteins in insect chemical communication. Cell Mol. Life Sci. 2006, 63, 1658–1676. [Google Scholar] [CrossRef] [PubMed]
- Scaloni, A.; Monti, M.; Angeli, S.; Pelosi, P. Structural analysis and disulfide-bridge pairing of two odorant-binding proteins from Bombyx mori. Biochem. Biophys. Res. Commun. 1999, 266, 386–391. [Google Scholar] [CrossRef]
- Tegoni, M.; Campanacci, V.; Cambillau, C. Structural aspects of sexual attraction and chemical communication in insects. Trends Biochem. Sci. 2004, 29, 257–264. [Google Scholar] [CrossRef]
- Zhou, J.-J.; Huang, W.; Zhang, G.-A.; Pickett, J.A.; Field, L.M. “Plus-C” odorant-binding protein genes in two Drosophila species and the malaria mosquito Anopheles gambiae. Gene 2004, 327, 117–129. [Google Scholar] [CrossRef]
- Xu, P.X.; Zwiebel, L.J.; Smith, D.P. Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Mol. Biol. 2003, 12, 549–560. [Google Scholar] [CrossRef]
- Lagarde, A.; Spinelli, S.; Qiao, H.; Tegoni, M.; Pelosi, P.; Cambillau, C. Crystal structure of a novel type of odorant-binding protein from Anopheles gambiae, belonging to the C-plus class. Biochem. J. 2011, 437, 423–430. [Google Scholar] [CrossRef]
- Spinelli, S.; Lagarde, A.; Iovinella, I.; Legrand, P.; Tegoni, M.; Pelosi, P.; Cambillau, C. Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules. Insect Biochem. Mol. Biol. 2012, 42, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Angeli, S.; Ceron, F.; Scaloni, A.; Monti, M.; Monteforti, G.; Minnocci, A.; Petacchi, R.; Pelosi, P. Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur. J. Biochem. 1999, 262, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Lartigue, A.; Campanacci, V.; Roussel, A.; Larsson, A.M.; Jones, T.A.; Tegoni, M.; Cambillau, C. X-ray structure and ligand binding study of a moth chemosensory protein. J. Biol. Chem. 2002, 277, 32094–32098. [Google Scholar] [CrossRef]
- Tomaselli, S.; Crescenzi, O.; Sanfelice, D.; Eiso, A.B.; Wechselberger, R.; Angeli, S.; Scaloni, A.; Boelens, R.; Tancredi, T.; Pelosi, P.; et al. Solution structure of a chemosensory protein from the desert locust Schistocerca gregaria. Biochemistry 2006, 45, 10606–10613. [Google Scholar] [CrossRef] [PubMed]
- Jansen, S.; Chmelik, J.; Zidek, L.; Padrta, P.; Novak, P.; Zdrahal, Z.; Picimbon, J.F.; Lofstedt, C.; Sklenar, V. Structure of Bombyx mori chemosensory protein 1 in solution. Arch. Insect Biochem. Physiol. 2007, 66, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Brito, N.F.; Moreira, M.F.; Melo, A.C.A. A look inside odorant-binding proteins in insect chemoreception. J. Insect Physiol. 2016, 95, 51–65. [Google Scholar] [CrossRef]
- Zheng, J.; Li, J.; Han, L.; Wang, Y.; Wu, W.; Qi, X.; Tao, Y.; Zhang, L.; Zhang, Z.; Chen, Z. Crystal structure of the Locusta migratoria odorant binding protein. Biochem. Biophys. Res. Commun. 2015, 456, 737–742. [Google Scholar] [CrossRef]
- Jia, Q.; Zeng, H.; Zhang, J.; Gao, S.; Xiao, N.; Tang, J.; Dong, X.; Xie, W. The crystal structure of the Spodoptera litura chemosensory protein CSP8. Insects 2021, 12, 602. [Google Scholar] [CrossRef]
- Venthur, H.; Mutis, A.; Zhou, J.-J.; Quiroz, A. Ligand binding and homology modelling of insect odorant-binding proteins. Physiol. Entomol. 2014, 39, 183–198. [Google Scholar] [CrossRef]
- Wang, Q.; Wen, X.; Lu, Y.; Wen, J. Comparison and functional analysis of chemosensory protein genes from Eucryptorrhynchus scrobiculatus Motschulsky and Eucryptorrhynchus brandti Harold. Front. Physiol. 2021, 12, 661310. [Google Scholar] [CrossRef]
- PD, K.J.; Kempraj, V.; Aurade, R.M.; Kumar Roy, T.; KS, S.; Verghese, A. Computational reverse chemical ecology: Virtual screening and predicting behaviorally active semiochemicals for Bactrocera dorsalis. BMC Genom. 2014, 15, 209. [Google Scholar] [CrossRef]
- González-González, A.; Palma-Millanao, R.; Yáñez, O.; Rojas, M.; Mutis, A.; Venthur, H.; Quiroz, A.; Ramírez, C.C. Virtual screening of plant volatile compounds reveals a high affinity of Hylamorpha elegans (Coleoptera: Scarabaeidae) odorant-binding proteins for sesquiterpenes from its native host. J. Insect Sci. 2016, 16, 30. [Google Scholar] [CrossRef] [PubMed]
- Hickner, P.V.; Rivaldi, C.L.; Johnson, C.M.; Siddappaji, M.; Raster, G.J.; Syed, Z. The making of a pest: Insights from the evolution of chemosensory receptor families in a pestiferous and invasive fly, Drosophila suzukii. BMC Genom. 2016, 17, 648. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Huang, L.Q.; Ning, C.; Wang, C.Z. Two single-point mutations shift the ligand selectivity of a pheromone receptor between two closely related moth species. eLife Sci. 2017, 6, e29100. [Google Scholar] [CrossRef]
- Reitz, S.R.; Gao, Y.L.; Kirk, W.D.J.; Hoddle, M.S.; Leiss, K.A.; Funderburk, J.E. Invasion biology, ecology, and management of western flower thrips. Annu. Rev. Entomol. 2020, 65, 17–37. [Google Scholar] [CrossRef]
- Nakahara, S.; Foottit, R.G. Frankliniella intonsa (Trybom) (Thysanoptera:Thripidae), an invasive insect in North America. Proc. Entomol. Soc. Wash. 2007, 109, 733–734. [Google Scholar] [CrossRef]
- Whitfield, A.E.; Ullman, D.E.; German, T.L. Tospovirus-thrips interactions. Annu. Rev. Phytopathol. 2005, 43, 459–489. [Google Scholar] [CrossRef]
- Okuda, S.; Okuda, M.; Matsuura, S.; Okazaki, S.; Iwai, H. Competence of Frankliniella occidentalis and Frankliniella intonsa strains as vectors for Chrysanthemum stem necrosis virus. Eur. J. Plant Pathol. 2013, 136, 355–362. [Google Scholar] [CrossRef]
- Wang, C.L.; Lin, F.C.; Chiu, Y.C.; Shih, H.T. Species of Frankliniella Trybom (Thysanoptera: Thripidae) from the Asian-Pacific area. Zool. Stud. 2010, 49, 824–838. [Google Scholar]
- Lewis, T. Thrips as Crop Pests; CAB International: New York, NY, USA, 1997. [Google Scholar]
- Gerin, C.; Hance, T.; Van Impe, G. Impact of flowers on the demography of western flower thrips Frankliniella occidentalis (Thysan., Thripidae). J. Appl. Entomol. 1999, 123, 569–574. [Google Scholar] [CrossRef]
- Gao, Y.F.; Gong, Y.J.; Cao, L.J.; Chen, J.C.; Gao, Y.L.; Mirab-balou, M.; Chen, M.; Hoffmann, A.A.; Wei, S.J. Geographical and interspecific variation in susceptibility of three common thrips species to the insecticide, spinetoram. J. Pest Sci. 2021, 94, 93–99. [Google Scholar] [CrossRef]
- Zhang, P.J.; Zhu, X.Y.; Lu, Y.B. Behavioural and chemical evidence of a male-produced aggregation pheromone in the flower thrips Frankliniella intonsa. Physiol. Entomol. 2011, 36, 317–320. [Google Scholar] [CrossRef]
- Geng, S.S.; Li, X.W.; Zhang, J.M.; Zhang, Z.J.; Lu, Y.B. Research on field application of aggregation pheromones of Frankliniella occidentalis and Frankliniella intonsa and their roles in interspecific interaction. Acta Entomol. Sin. 2017, 60, 1447–1456. [Google Scholar] [CrossRef]
- Wu, S.Y.; Xing, Z.L.; Ma, T.T.; Xu, D.W.; Li, Y.Y.; Lei, Z.R.; Gao, Y.L. Competitive interaction between Frankliniella occidentalis and locally present thrips species: A global review. J. Pest Sci. 2021, 94, 5–16. [Google Scholar] [CrossRef]
- Fan, Z.; Qian, L.; Chen, Y.; Fan, R.; He, S.; Gao, Y.; Gui, F. Effects of elevated CO2 on activities of protective and detoxifying enzymes in Frankliniella occidentalis and F. intonsa under spinetoram stress. Pest Manag. Sci. 2021, 78, 274–286. [Google Scholar] [CrossRef]
- Bhuyain, M.M.H.; Lim, U.T. Relative susceptibility to pesticides and environmental conditions of Frankliniella intonsa and F.occidentalis (Thysanoptera: Thripidae), an underlying reason for their asymmetrical occurrence. PLoS ONE 2020, 15, e0237876. [Google Scholar] [CrossRef]
- Bhuyain, M.M.H.; Lim, U.T. Interference and exploitation competition between Frankliniella occidentalis and F. intonsa (Thysanoptera: Thripidae) in laboratory assays. Fla. Entomol. 2019, 102, 322–328. [Google Scholar] [CrossRef]
- Qian, L.; Chen, F.; Liu, J.; He, S.; Liu, J.; Li, Z.; Gui, F. Effects of elevated CO2 on life-history traits of three successive generations of Frankliniella occidentalis and F.intonsa on kidney bean, Phaseolus vulgaris. Entomol. Exp. Appl. 2017, 165, 50–61. [Google Scholar] [CrossRef]
- Ullah, M.S.; Lim, U.T. Life history characteristics of Frankliniella occidentalis and Frankliniella intonsa (Thysanoptera: Thripidae) in constant and fluctuating temperatures. J. Econ. Entomol. 2015, 108, 1000–1009. [Google Scholar] [CrossRef]
- Li, X.; Geng, S.; Zhang, Z.; Zhang, J.; Li, W.; Huang, J.; Lin, W.; Bei, Y.; Lu, Y. Species-specific aggregation pheromones contribute to coexistence in two closely related thrips species. Bull. Entomol. Res. 2019, 109, 119–126. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Lei, Z.R. Identification, expression profiling and fluorescence-based binding assays of a chemosensory protein gene from the western flower thrips, Frankliniella occidentalis. PLoS ONE 2015, 10, e0117726. [Google Scholar] [CrossRef]
- Zeng, F.-F.; Zhou, S.-S.; Ding, Y.-H.; Wang, M.-Q. Analysis of an antennal cDNA library and the expression patterns of two olfactory genes in Frankliniella occidentalis (Thysanoptera: Thripidae). Appl. Entomol. Zool. 2015, 50, 109–116. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Wu, S.Y.; Lei, Z.R. Cloning, sequence snalysis and expression profile of an odorant binding protein gene in western flower thrips (Frankliniella occidentalis). Sci. Agric. Sin. 2016, 49, 1106–1116. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Sheng-Yong, W.U.; Lei, Z.R. cDNA cloning, expression profiling and immunolocalization of a chemosensory protein in the western flower thrips, Frankliniella occidentalis (Thysanoptera:Thtipidae). Acta Entomol. Sin. 2015, 58, 1–14. [Google Scholar]
- Zhang, Z.K.; Hu, H.; Ma, R. Cloning, sequence analysis and expression characteristics of FoccOBP3 in western flower thrips Frankliniella occidentalis. Chin. J. Biol. Control 2021, 37, 495–507. [Google Scholar] [CrossRef]
- Rihani, K.; Ferveur, J.-F.; Briand, L. The 40-year mystery of insect odorant-binding proteins. Biomolecules 2021, 11, 509. [Google Scholar] [CrossRef]
- Zeng, Y.; Yang, Y.T.; Wu, Q.J.; Wang, S.L.; Xie, W.; Zhang, Y.J. Genome-wide analysis of odorant-binding proteins and chemosensory proteins in the sweet potato whitefly, Bemisia tabaci. Insect Sci. 2019, 26, 620–634. [Google Scholar] [CrossRef]
- Gu, S.-H.; Wu, K.-M.; Guo, Y.-Y.; Field, L.M.; Pickett, J.A.; Zhang, Y.-J.; Zhou, J.-J. Identification and expression profiling of odorant binding proteins and chemosensory proteins between two wingless morphs and a winged morph of the cotton aphid Aphis gossypii Glover. PLoS ONE 2013, 8, e73524. [Google Scholar] [CrossRef]
- Wang, Q.; Zhou, J.J.; Liu, J.T.; Huang, G.Z.; Xu, W.Y.; Zhang, Q.; Chen, J.L.; Zhang, Y.J.; Li, X.C.; Gu, S.H. Integrative transcriptomic and genomic analysis of odorant binding proteins and chemosensory proteins in aphids. Insect Mol. Biol. 2019, 28, 1–22. [Google Scholar] [CrossRef]
- Zhou, S.-S.; Sun, Z.; Ma, W.; Chen, W.; Wang, M.-Q. De novo analysis of the Nilaparvata lugens (Stal) antenna transcriptome and expression patterns of olfactory genes. Comp. Biochem. Physiol. Part D Genom. Proteom. 2014, 9, 31–39. [Google Scholar] [CrossRef]
- He, M.; He, P. Molecular characterization, expression profiling, and binding properties of odorant binding protein genes in the whitebacked planthopper, Sogatella furcifera. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2014, 174, 1–8. [Google Scholar] [CrossRef]
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 2018, 93, 184–200. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yuan, X.; Qian, P.; Cheng, J.; Zhang, C.; Gurr, G.; Zhu, Z. Identification and expression profiling of putative chemosensory protein genes in two rice planthoppers, Laodelphax striatellus (Fallen) and Sogatella furcifera (Horvath). J. Asia Pac. Entomol. 2015, 18, 771–778. [Google Scholar] [CrossRef]
- Picimbon, J.-F. Biochemistry and evolution of OBP and CSP proteins. In Insect Pheromone Biochemistry and Molecular Biology; Blomquist, G., Vogt, R., Eds.; Academic Press: San Diego, CA, USA, 2003; pp. 539–566. [Google Scholar] [CrossRef]
- Qiao, H.; He, X.; Schymura, D.; Ban, L.; Field, L.; Dani, F.R.; Michelucci, E.; Caputo, B.; della Torre, A.; Iatrou, K.; et al. Cooperative interactions between odorant-binding proteins of Anopheles gambiae. Cell Mol. Life Sci. 2011, 68, 1799–1813. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.Y.; Zhang, P.J.; Lu, Y.b. Isolation and identification of the aggregation pheromone released by male adults of Frankliniella intonsa(Thysanoptera:Thripidae). Acta Entomol. Sin. 2012, 55, 376–385. [Google Scholar]
- Guo, W.; Ren, D.; Zhao, L.; Jiang, F.; Song, J.; Wang, X.; Kang, L. Identification of odorant-binding proteins (OBPs) and functional analysis of phase-related OBPs in the migratory locust. Front. Physiol. 2018, 9, 984. [Google Scholar] [CrossRef]
- Terry, I.; Schneider, M. Copulatory behaviour and mating frequency of the western flower thrips, Frankliniella occidentalis (Insecta:Thysanoptera). Zool. (J. Pure Appl. Zool.) 1993, 4, 339–354. [Google Scholar]
- Steenbergen, M.; Abd-El-Haliem, A.; Bleeker, P.; Dicke, M.; Escobar-Bravo, R.; Cheng, G.; Haring, M.A.; Kant, M.R.; Kappers, I.; Klinkhamer, P.G.L.; et al. Thrips advisor: Exploiting thrips-induced defences to combat pests on crops. J. Exp. Bot. 2018, 69, 1837–1848. [Google Scholar] [CrossRef]
- Hamilton, J.G.C.; Hall, D.R.; Kirk, W.D.J. Identification of a male-produced aggregation pheromone in the western flower thrips Frankliniella occidentalis. J. Chem. Ecol. 2005, 31, 1369–1379. [Google Scholar] [CrossRef]
- Teerling, C.R.; Pierce, H.D.; Borden, J.H.; Gillespie, D.R. Identification and bioactivity of alarm pheromone in the western flower thrips, Frankliniella occidentalis. J. Chem. Ecol. 1993, 19, 681–697. [Google Scholar] [CrossRef]
- Okimoto, N.; Futatsugi, N.; Fuji, H.; Suenaga, A.; Morimoto, G.; Yanai, R.; Ohno, Y.; Narumi, T.; Taiji, M. High-performance drug discovery: Computational screening by combining docking and molecular dynamics smulations. PLoS Comput. Biol. 2009, 5, e1000528. [Google Scholar] [CrossRef]
- Zhang, J.; Mao, K.; Ren, Z.; Jin, R.; Zhang, Y.; Cai, T.; He, S.; Li, J.; Wan, H. Odorant binding protein 3 is associated with nitenpyram and sulfoxaflor resistance in Nilaparvata lugens. Int. J. Biol. Macromol. 2022, 209, 1352–1358. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.-Y.; Yang, K.; Liu, Y.; Xu, W.; Anderson, A.; Dong, S.-L. Two general-odorant binding proteins in Spodoptera litura are differentially tuned to sex pheromones and plant odorants. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2015, 180, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Ullah, R.M.K.; Quershi, S.R.; Adeel, M.M.; Abdelnabby, H.; Waris, M.I.; Duan, S.G.; Wang, M.Q. An odorant binding protein (SaveOBP9) involved in chemoreception of the wheat aphid Sitobion avenae. Int. J. Mol. Sci. 2020, 21, 8331. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Daniel, H.H. Fast and sensitive protein alignment using DIAMOND. Nat. Meth. 2015, 12, 59. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]




| Species | Gene Name | Signal Peptide (aa) | Amino Acids (aa) | ORF (bp) | Full ORF | Accession No. |
|---|---|---|---|---|---|---|
| Frankliniella occidentalis | FoccOBP1 | 22 | 203 | 610 | No | OP380934 |
| FoccOBP2 | 19 | 231 | 696 | Yes | OP380936 | |
| FoccOBP3 | 21 | 156 | 471 | Yes | OP380938 | |
| FoccOBP4 | 20 | 138 | 417 | Yes | OP380940 | |
| FoccOBP5 | 20 | 155 | 465 | Yes | OP380942 | |
| FoccOBP6 | 20 | 156 | 471 | Yes | OP380944 | |
| FoccOBP7 | 19 | 189 | 570 | Yes | OP380946 | |
| FoccCSP1 | 20 | 129 | 390 | Yes | OP380946 | |
| FoccCSP2 | 19 | 134 | 405 | Yes | OP380949 | |
| FoccCSP3 | 22 | 125 | 375 | Yes | OP380951 | |
| FoccCSP4 | 20 | 142 | 426 | Yes | OP380953 | |
| FoccCSP5 | 20 | 129 | 390 | Yes | OP380955 | |
| FoccCSP6 | 22 | 156 | 471 | Yes | OP380957 | |
| FoccCSP7 | 19 | 140 | 423 | Yes | OP380959 | |
| FoccCSP8 | 27 | 121 | 366 | Yes | OP380960 | |
| Frankliniella intonsa | FintOBP1 | 23 | 174 | 529 | No | OP380935 |
| FintOBP2 | 20 | 255 | 768 | Yes | OP380937 | |
| FintOBP3 | 21 | 156 | 471 | Yes | OP380939 | |
| FintOBP4 | 20 | 138 | 417 | Yes | OP380941 | |
| FintOBP5 | 19 | 148 | 444 | Yes | OP380943 | |
| FintOBP6 | 21 | 157 | 474 | Yes | OP380945 | |
| FintCSP1 | 20 | 129 | 390 | Yes | OP380948 | |
| FintCSP2 | 19 | 135 | 405 | Yes | OP380950 | |
| FintCSP3 | 22 | 124 | 375 | Yes | OP380952 | |
| FintCSP4 | 20 | 142 | 426 | Yes | OP380954 | |
| FintCSP5 | 20 | 129 | 390 | Yes | OP380956 | |
| FintCSP6 | 19 | 157 | 474 | Yes | OP380958 |
| Gene Name | Total Score | Query Cover | E-Value | Per. Identify% | |
|---|---|---|---|---|---|
| FoccOBP1 | FintOBP1 | 249 | 75% | 2 × 10−90 | 89.47 |
| FoccOBP2 | FintOBP2 | 384 | 99% | 2 × 10−141 | 76.72 |
| FoccOBP3 | FintOBP3 | 296 | 100% | 8 × 10−110 | 91.72 |
| FoccOBP4 | FintOBP4 | 231 | 100% | 1 × 10−84 | 84.17 |
| FoccOBP5 | FintOBP5 | 218 | 75% | 5 × 10−79 | 91.38 |
| FoccOBP6 | FintOBP6 | 265 | 87% | 1 × 10−97 | 93.43 |
| FoccCSP1 | FintCSP1 | 265 | 100% | 3 × 10−98 | 98.46 |
| FoccCSP2 | FintCSP2 | 267 | 100% | 5 × 10−99 | 98.52 |
| FoccCSP3 | FintCSP3 | 189 | 99% | 1 × 10−68 | 83.87 |
| FoccCSP4 | FintCSP4 | 283 | 100% | 3 × 10−105 | 98.59 |
| FoccCSP5 | FintCSP5 | 212 | 84% | 2 × 10−77 | 91.82 |
| Gene Name | N(S)2MB | (R)-LA | ||
|---|---|---|---|---|
| Mean Binding Energy (kJ/mol) | Residues Forming H-Bond with Ligand | Mean Binding Energy (kJ/mol) | Residues Forming H-Bond with Ligand | |
| FoccOBP1 | −21.80 | TYR74 | −21.13 | TYR74 |
| FoccOBP2 | −22.89 | TYR143 | −21.63 | VAL110 |
| FoccOBP3 | −25.06 | LYS25, MET28 | −23.56 | LYS25 |
| FoccOBP4 | −24.94 | MET136 | −23.05 | MET136 |
| FoccOBP5 | −23.35 | LYS29 | −20.17 | PHE76 |
| FoccOBP6 | −25.61 | TYR87 | −23.43 | TYR87 |
| FoccOBP7 | −24.73 | LEU169 | −24.02 | LEU169 |
| FintOBP1 | −25.48 | VAL150 | −22.22 | TYR38 |
| FintOBP2 | −23.30 | TRP124 | −25.40 | TRP124 |
| FintOBP3 | −22.89 | GLY135 | −19.54 | TYR31 |
| FintOBP4 | −26.99 | GLU23, LEU24 | −22.84 | GLU23 |
| FintOBP5 | −22.38 | LYS31 | −18.03 | ASN70 |
| FintOBP6 | −25.82 | ARG79 | −22.38 | ARG79 |
| FoccCSP1 | −23.39 | −18.79 | THR106 | |
| FoccCSP2 | −24.85 | SER71 | −21.21 | TYR31, GLN87 |
| FoccCSP3 | −25.73 | ARG86 | −23.30 | ARG86 |
| FoccCSP4 | −20.42 | SER57 | −20.67 | PHE26, SER57 |
| FoccCSP5 | −26.90 | −19.62 | SER66 | |
| FoccCSP6 | −23.72 | SER66 | −20.67 | ARG74 |
| FoccCSP7 | −26.94 | TYR129 | −22.22 | TYR129 |
| FoccCSP8 | −19.96 | ASN91 | −19.54 | TYR107 |
| FintCSP1 | −23.43 | THR106 | −20.71 | TYR31 |
| FintCSP2 | −25.82 | GLN87, TYR31 | −21.59 | TYR31 |
| FintCSP3 | −25.27 | −21.59 | ARG86 | |
| FintCSP4 | −19.50 | VAL31 | −19.37 | SER57 |
| FintCSP5 | −22.59 | THR86 | −21.55 | ARG74 |
| FintCSP6 | −21.63 | GLN111 | −21.21 | GLN111 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Cheng, J.; Chen, L.; Huang, J.; Zhang, Z.; Zhang, J.; Ren, X.; Hafeez, M.; Zhou, S.; Dong, W.; et al. Comparison and Functional Analysis of Odorant-Binding Proteins and Chemosensory Proteins in Two Closely Related Thrips Species, Frankliniella occidentalis and Frankliniella intonsa (Thysanoptera: Thripidae) Based on Antennal Transcriptome Analysis. Int. J. Mol. Sci. 2022, 23, 13900. https://doi.org/10.3390/ijms232213900
Li X, Cheng J, Chen L, Huang J, Zhang Z, Zhang J, Ren X, Hafeez M, Zhou S, Dong W, et al. Comparison and Functional Analysis of Odorant-Binding Proteins and Chemosensory Proteins in Two Closely Related Thrips Species, Frankliniella occidentalis and Frankliniella intonsa (Thysanoptera: Thripidae) Based on Antennal Transcriptome Analysis. International Journal of Molecular Sciences. 2022; 23(22):13900. https://doi.org/10.3390/ijms232213900
Chicago/Turabian StyleLi, Xiaowei, Jianghui Cheng, Limin Chen, Jun Huang, Zhijun Zhang, Jinming Zhang, Xiaoyun Ren, Muhammad Hafeez, Shuxing Zhou, Wanying Dong, and et al. 2022. "Comparison and Functional Analysis of Odorant-Binding Proteins and Chemosensory Proteins in Two Closely Related Thrips Species, Frankliniella occidentalis and Frankliniella intonsa (Thysanoptera: Thripidae) Based on Antennal Transcriptome Analysis" International Journal of Molecular Sciences 23, no. 22: 13900. https://doi.org/10.3390/ijms232213900
APA StyleLi, X., Cheng, J., Chen, L., Huang, J., Zhang, Z., Zhang, J., Ren, X., Hafeez, M., Zhou, S., Dong, W., & Lu, Y. (2022). Comparison and Functional Analysis of Odorant-Binding Proteins and Chemosensory Proteins in Two Closely Related Thrips Species, Frankliniella occidentalis and Frankliniella intonsa (Thysanoptera: Thripidae) Based on Antennal Transcriptome Analysis. International Journal of Molecular Sciences, 23(22), 13900. https://doi.org/10.3390/ijms232213900












