The Discovery of Novel Ferulic Acid Derivatives Incorporating Substituted Isopropanolamine Moieties as Potential Tobacco Mosaic Virus Helicase Inhibitors
Abstract
1. Introduction
2. Results and Discussion
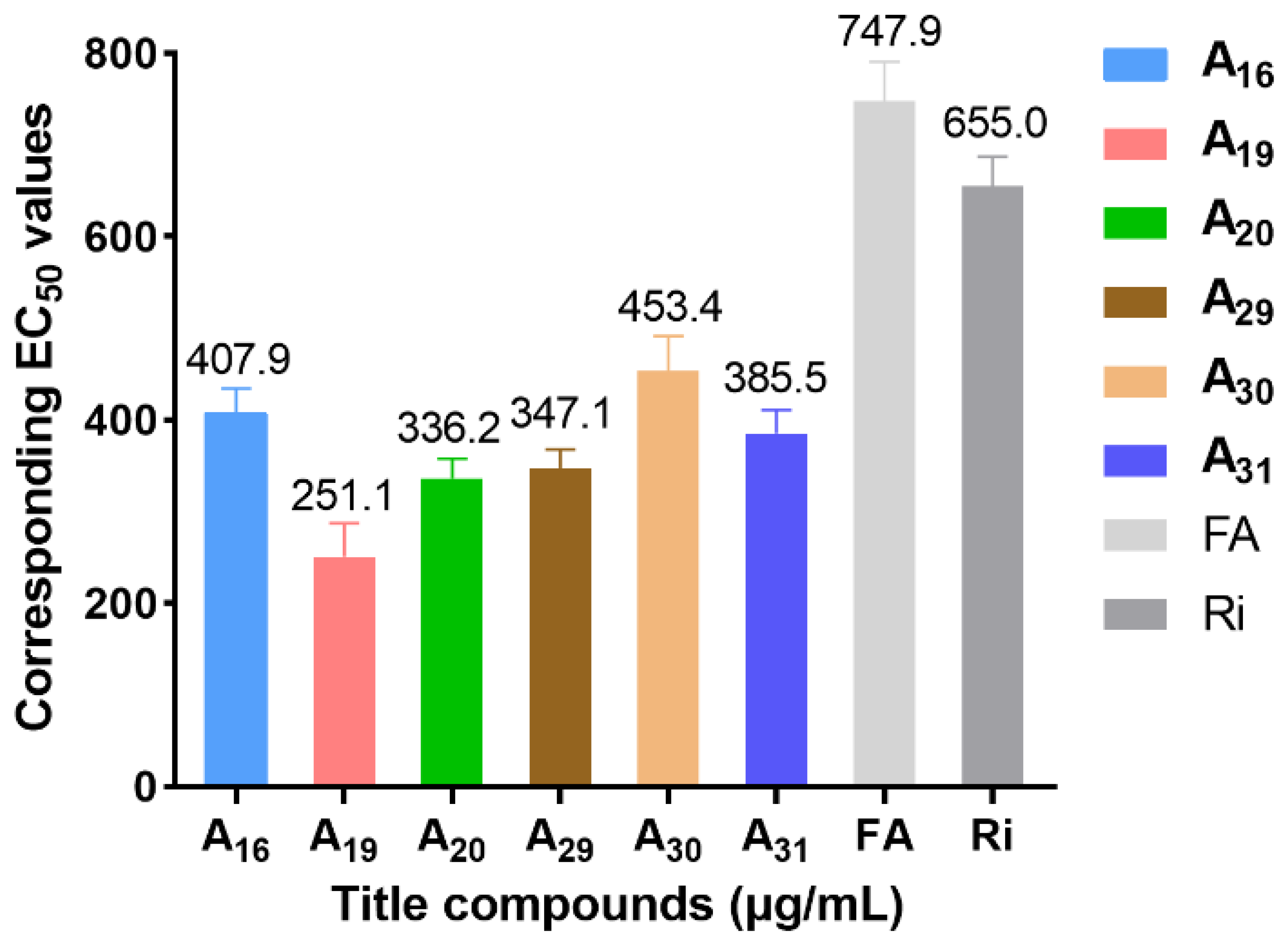
2.1. Chemistry and Antiviral Activity In Vivo
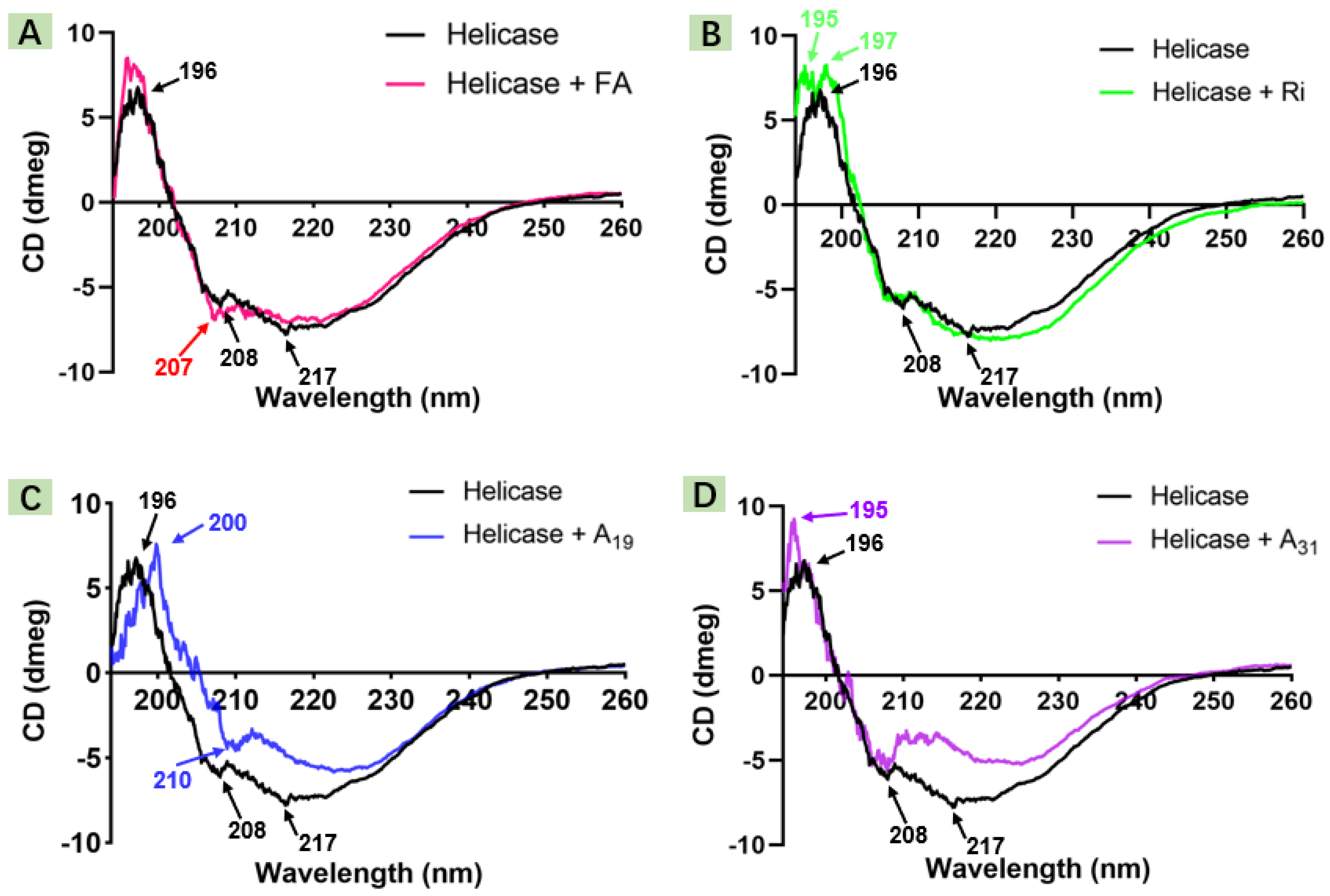
2.2. Circular Dichroism Spectroscopic Study
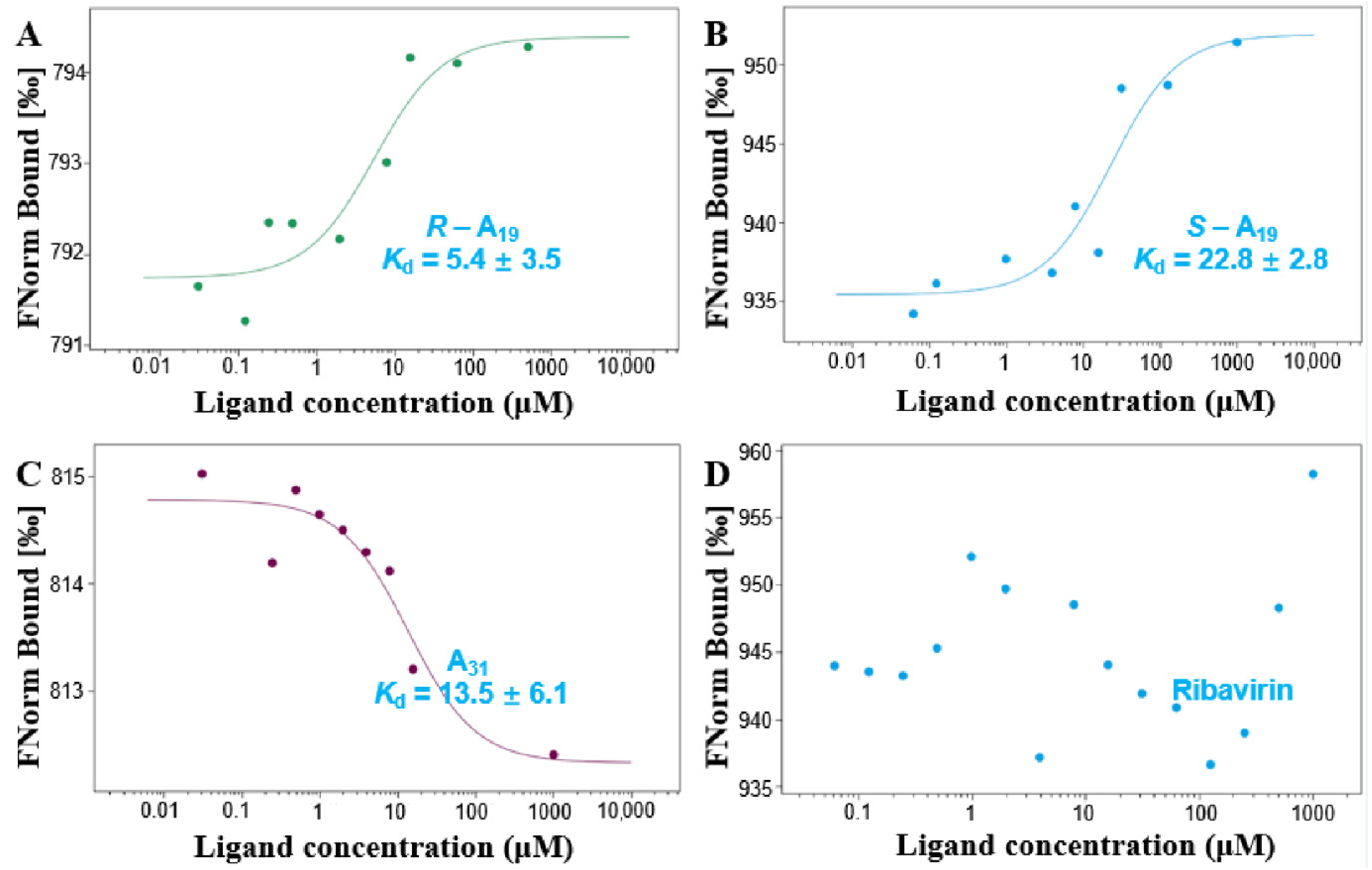
2.3. Interaction Analysis
2.4. Inhibitory Activity of Helicase ATPase
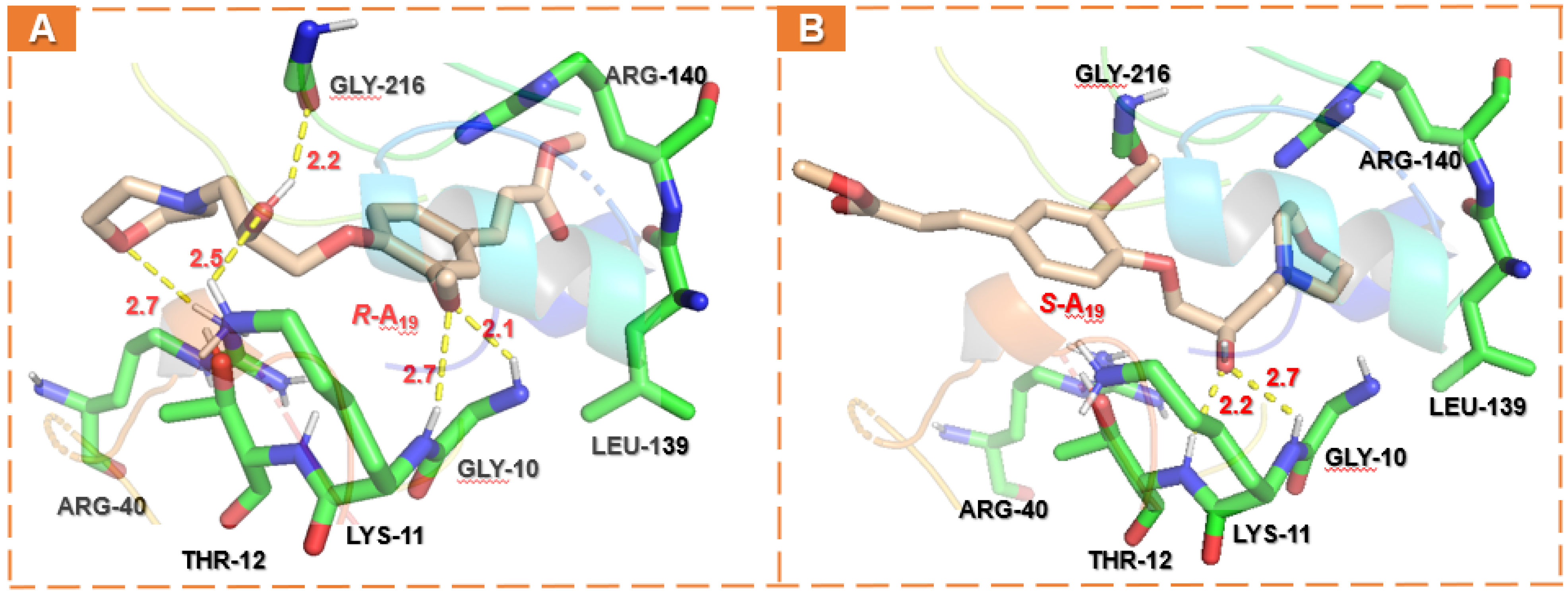
2.5. Docking Analysis
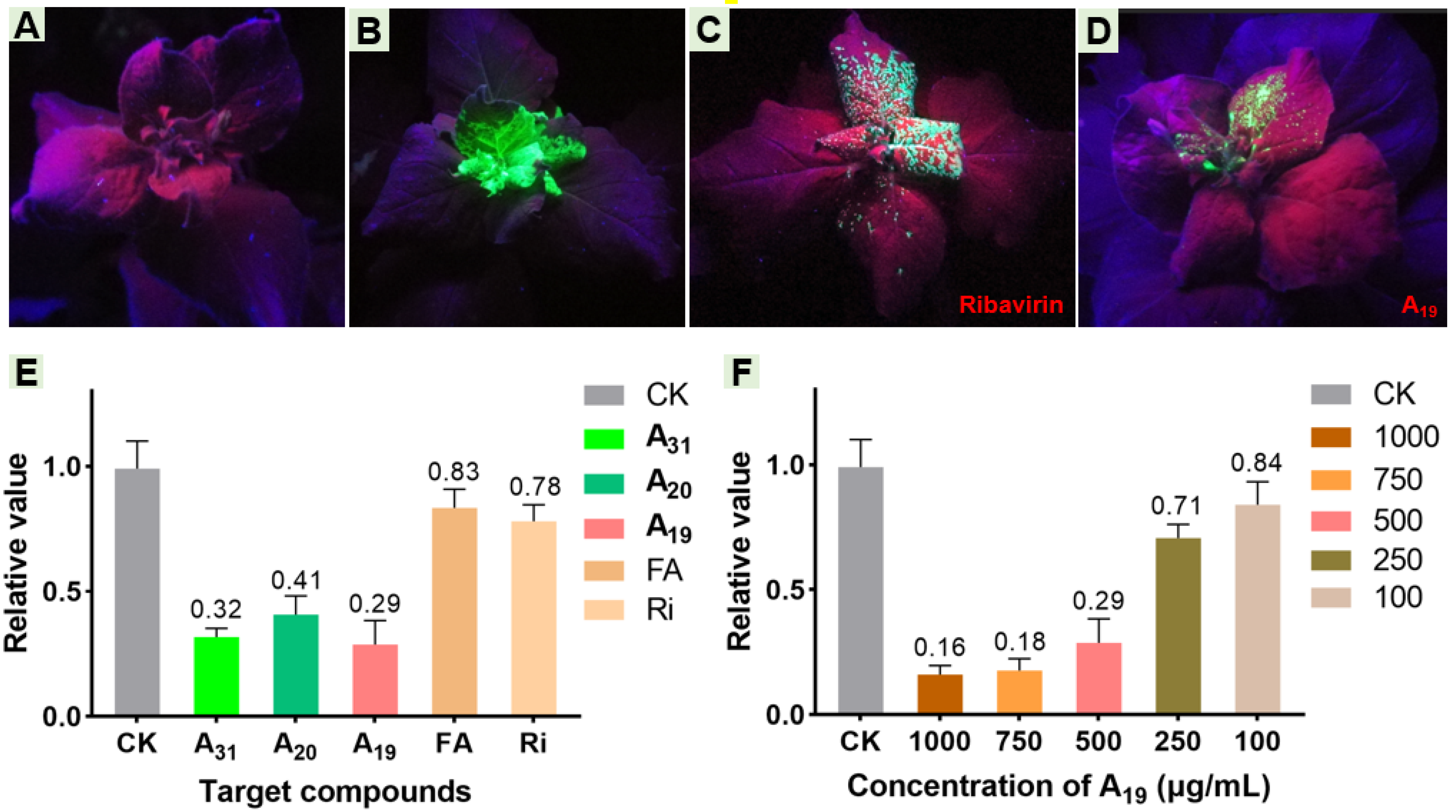
2.6. Effect of Target Compounds on GFP-Labeled TMV and Relative Expression Levels of Helicase In Vivo
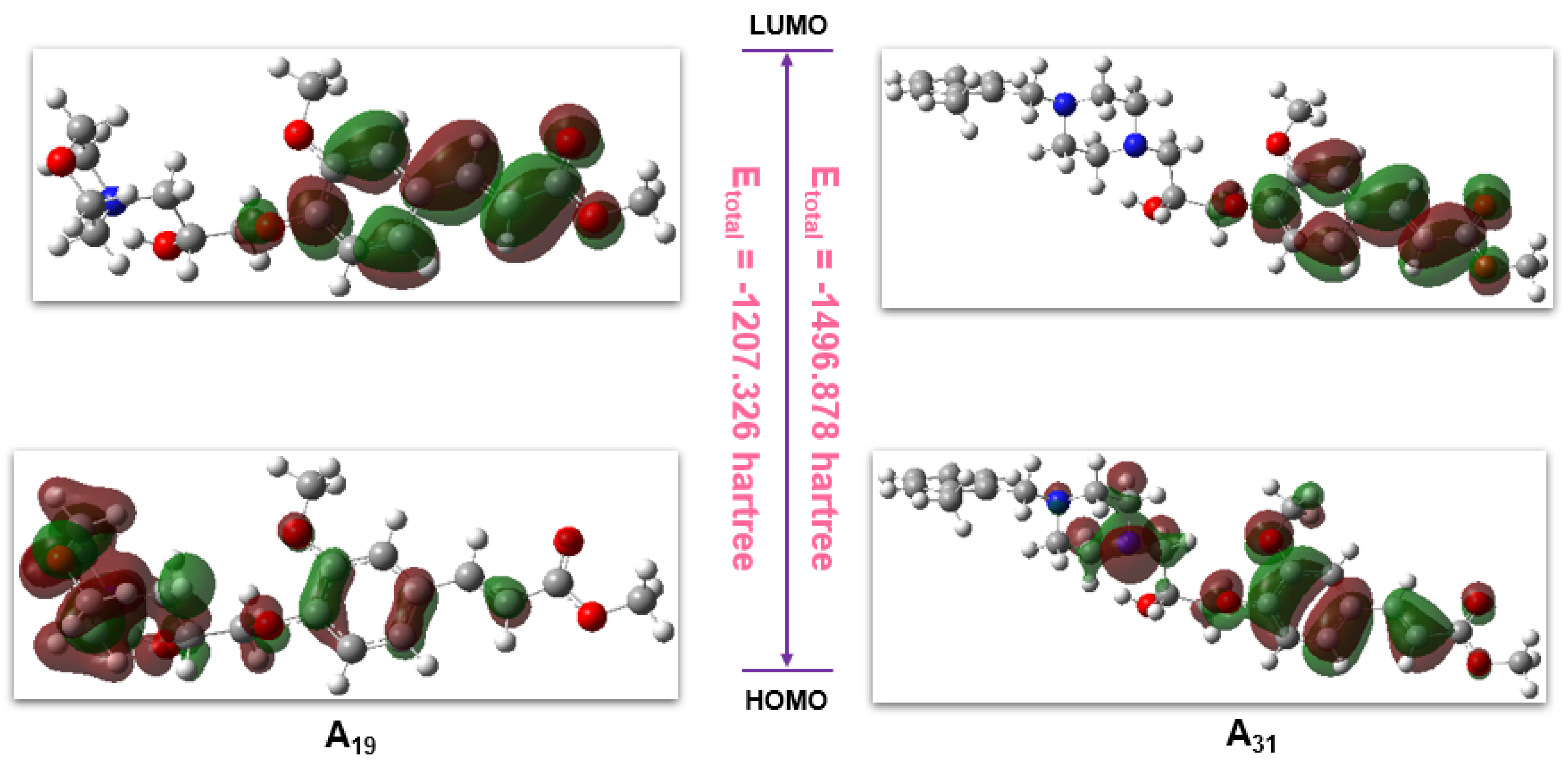
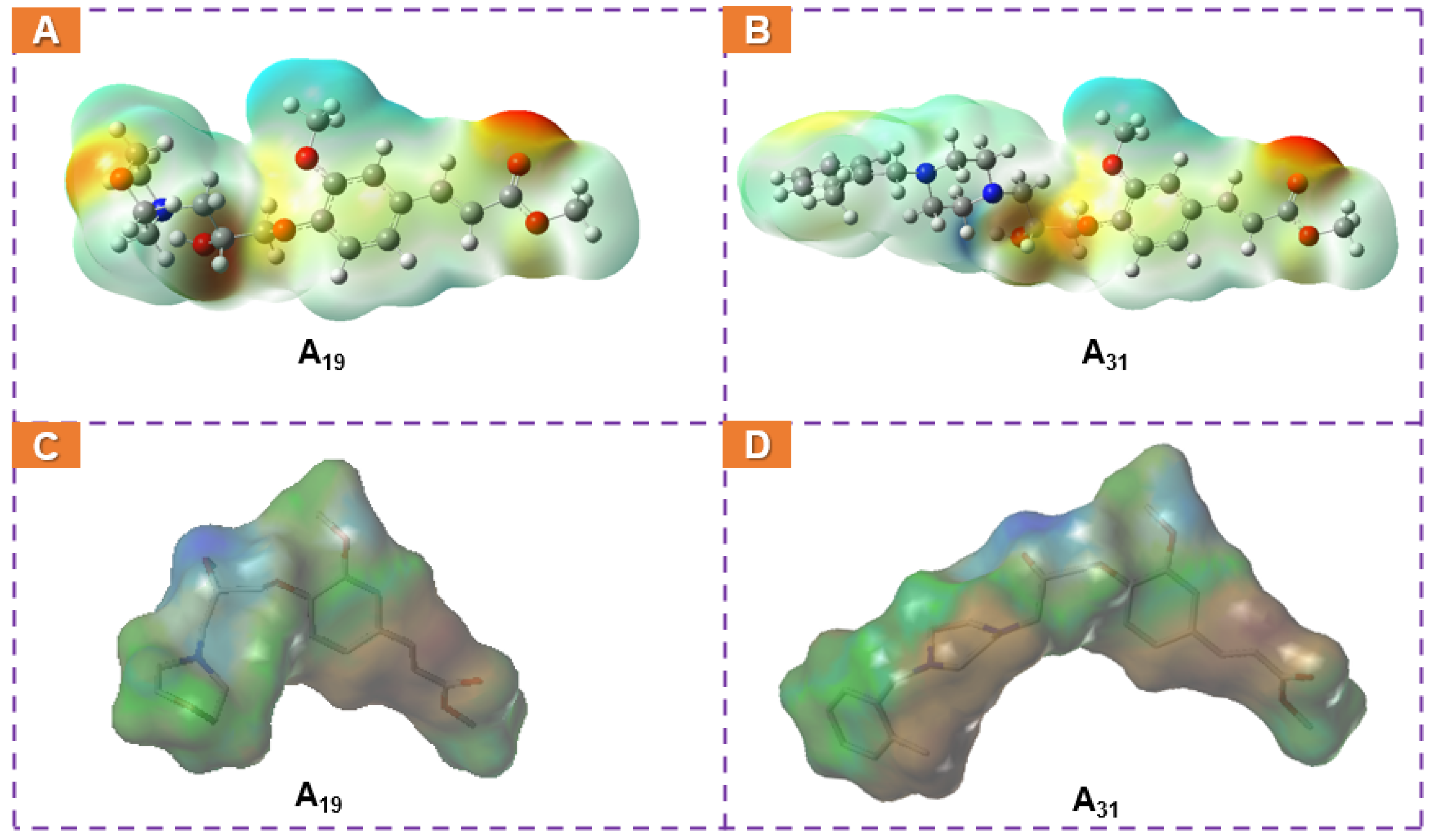
2.7. Computational Analysis
2.8. Antibacterial Activity
3. Materials and Methods
3.1. Instruments and Chemicals
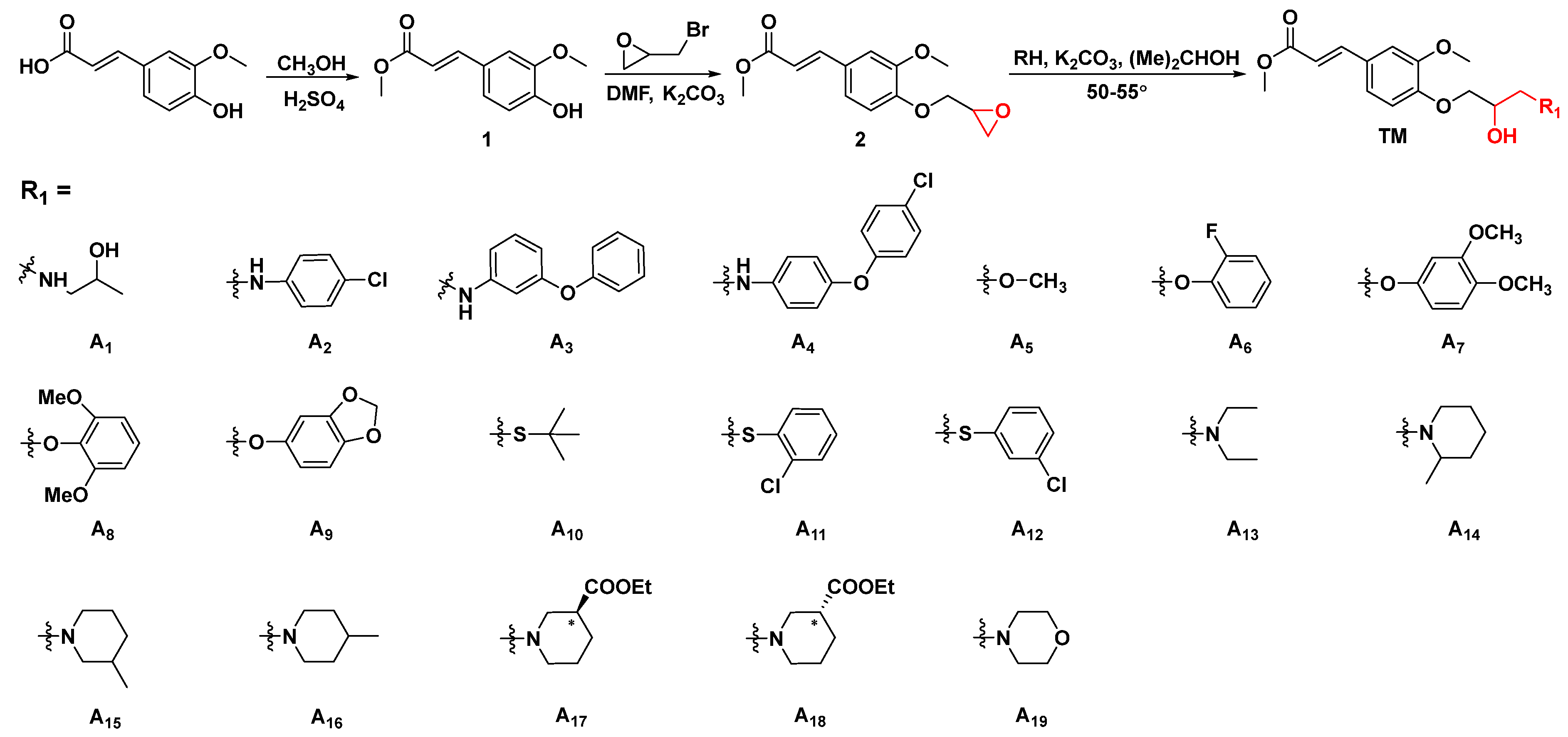
3.2. General Procedures for Preparing Ferulic Acid Derivatives [41,42,43]
3.2.1. General Procedures for Preparing Intermediate
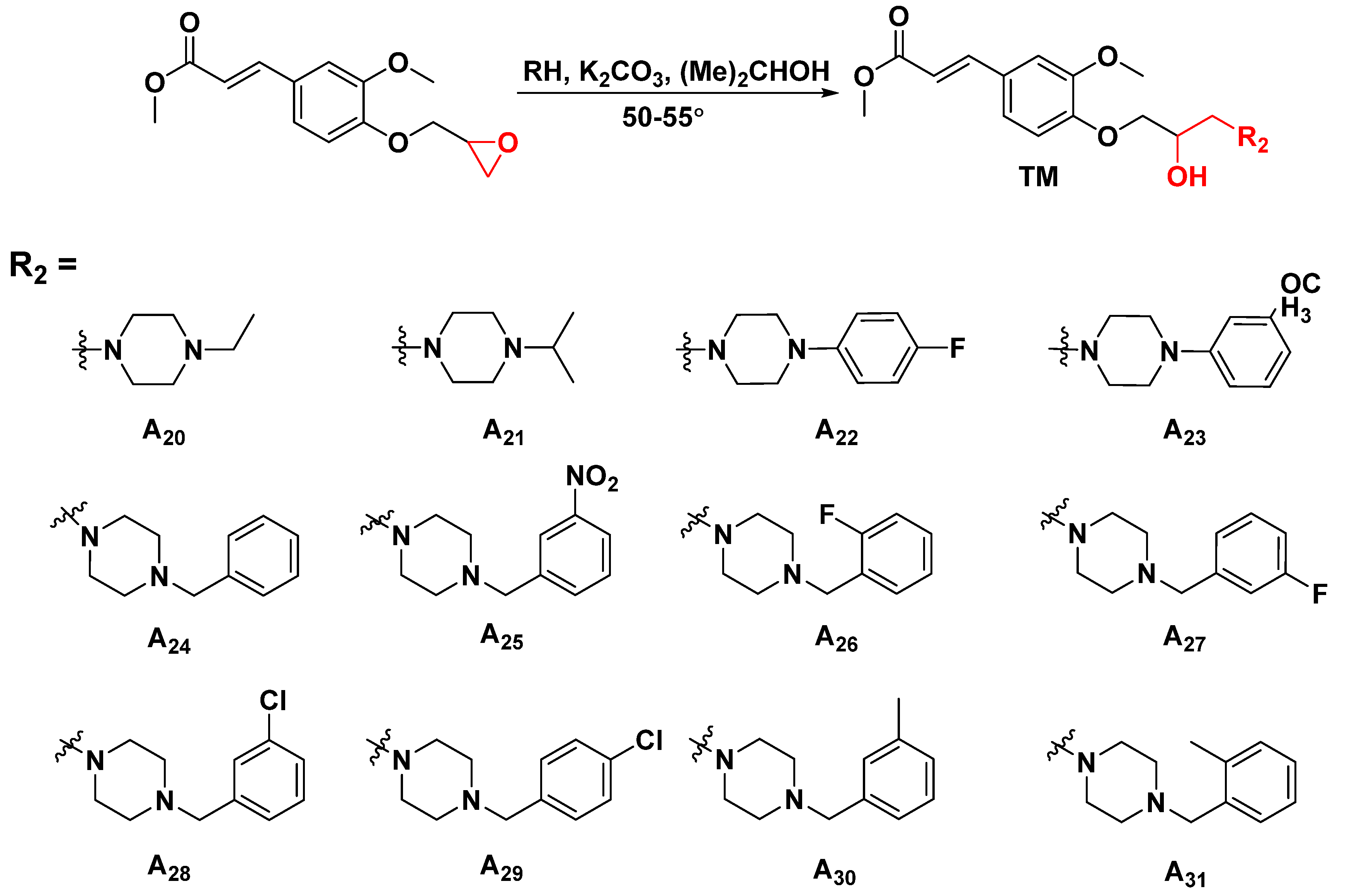
3.2.2. General Synthetic Procedures for Title Compounds A1–A31
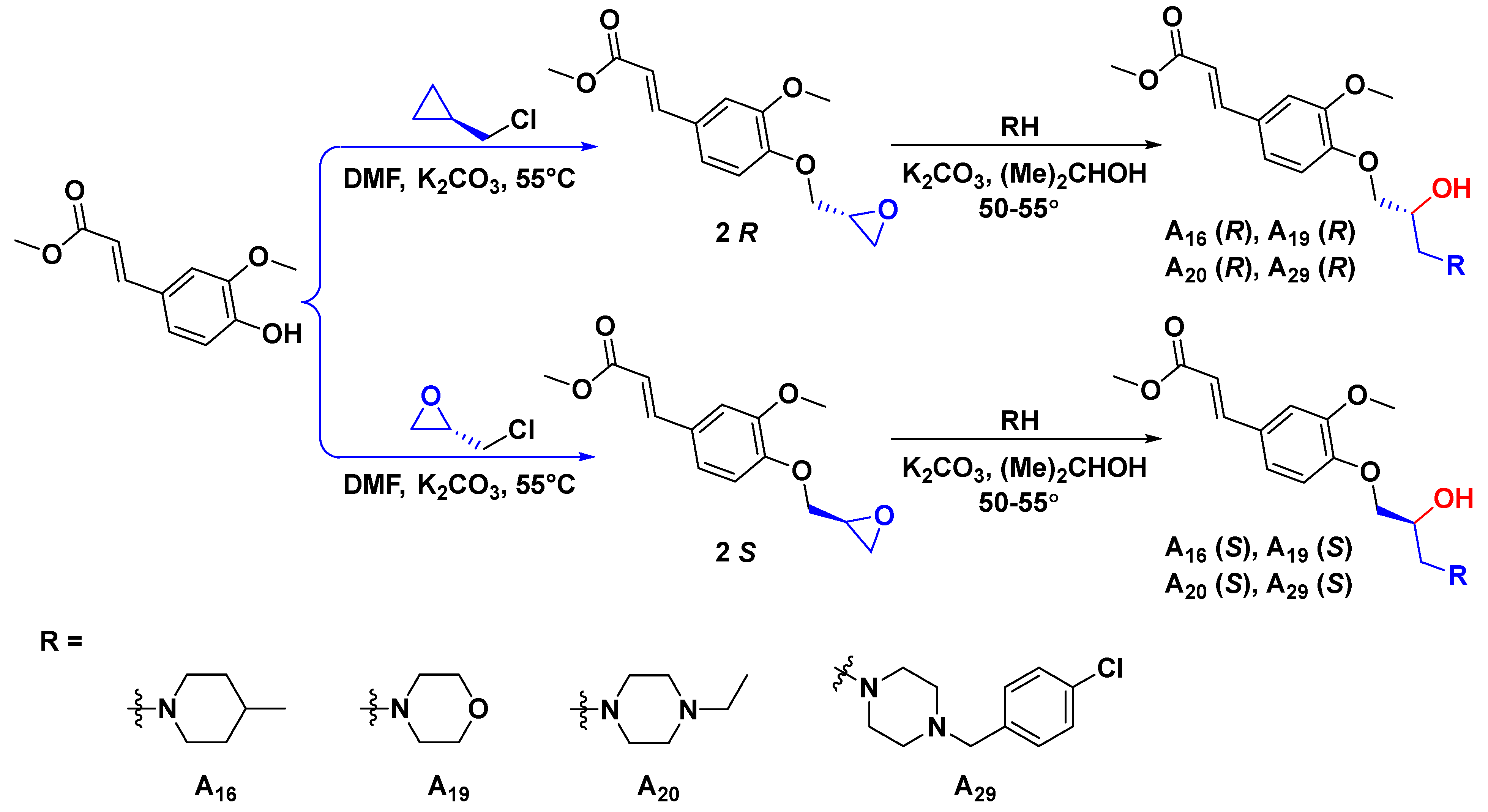
3.2.3. General Synthetic Procedures for Title Compounds R-A16, S-A16, R-A19, S-A19, R-A20, S-A20, R-A29, and S-A29
3.3. Biological Assay
3.4. pPIC9K-HIS-TMV-Helicase Expression and Purification
3.5. Secondary Structural Analyzation
3.6. Binding Analysis between Antiviral Compounds and TMV Helicase
3.7. TMV Helicase ATPase Activity
3.8. Homologous Modeling of TMV Helicase 3D Structure and Docking
3.9. Determination of Helicase Relative Gene Expression by RT-qPCR
3.10. Observation the Movement of GFP-Labeled TMV
3.11. DFT Calculation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Ferulic acid | FA |
| Ribavirin | Ri |
| Tobacco mosaic virus | TMV |
| Circular dichroism | CD |
| Quantitative real-time polymerase chain reaction | RT-qPCR |
| Microscale thermophoresis | MST |
| Density functional theory | DFT |
| Xanthomonas oryzae pv. oryzae | Xoo |
| Xanthomonas. axonopodis pv citri | Xac |
References
- Chakraborty, S.; Newton, A.C. Climate change, plant diseases and food security: An overview. Plant Pathol. 2011, 60, 2–14. [Google Scholar] [CrossRef]
- Harrison, B.D.; Mayo, M.A.; Baulcombe, D.C. Virus resistance in transgenic plants that express cucumber mosaic virus satellite RNA. Nature 1987, 328, 799–802. [Google Scholar] [CrossRef]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, P. Progress of modern agricultural chemistry and future prospects. Pest Manag. Sci. 2016, 72, 433–455. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zou, J.Y.; Xu, C.J.; You, S.Y.; Deng, Z.Y.; Chen, G.H.; Liu, Y.X.; Wang, Q.M. Preparation and Anti-Tobacco Mosaic Virus Activities of Crocetin Diesters. J. Agric. Food Chem. 2021, 69, 13637–13643. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Shi, J.; Yu, L.; Liu, D.Y.; Gan, X.H.; Song, B.A.; Hu, D.Y. Design, Synthesis, Antiviral Bioactivity and Defense Mechanisms of Novel Dithioacetal Derivatives Bearing a Strobilurin Moiety. J. Agric. Food Chem. 2018, 66, 5335–5345. [Google Scholar] [CrossRef]
- Abdelhaleem, M. Helicases: An overview. Methods Mol. Biol. 2010, 587, 1–12. [Google Scholar] [CrossRef]
- Crute, J.J.; Grygon, C.A.; Hargrave, K.D.; Simoneau, B.; Faucher, A.M.; Bolger, G.; Kibler, P.; Liuzzi, M.; Cordingley, M.G. Herpes Simplex virus helicase-primase inhibitors are active in animal models of human disease. Nat. Med. 2002, 8, 386–391. [Google Scholar] [CrossRef]
- Walstrom, K.M.; Dozono, J.M.; von Hippel, P.H. Kinetics of the RNA-DNA helicase activity of Escherichia coli transcription termination factor rho. 2. Processivity, ATP consumption, and RNA binding±. Biochemistry 1997, 36, 7993–8004. [Google Scholar] [CrossRef] [PubMed]
- Missel, A.; Souza, A.E.; Norskau, G.; Goringer, H.U. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell Biol. 1997, 17, 4895–4903. [Google Scholar] [CrossRef]
- Bilgin, D.D.; Liu, Y.; Schiff, M.; Dinesh-Kumar, S.P. P58(IPK), a plant ortholog of double-stranded RNA-dependent protein kinase PKR inhibitor, functions in viral pathogenesis. Dev. Cell 2003, 4, 651–661. [Google Scholar] [CrossRef]
- Caruthers, J.M.; McKay, D.B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002, 12, 123–133. [Google Scholar] [CrossRef]
- Wang, X.; Kelman, Z.; Culver, J.N. Helicase ATPase activity of the Tobacco mosaic virus 126-kDa protein modulates replicase complex assembly. Virology 2010, 402, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.S.; Liu, J.; Cheng, N.H.; Folimonov, A.; Hou, Y.M.; Bao, Y.; Katagi, C.; Carter, S.A.; Nelson, R.S. The Tobacco mosaic virus 126-kDa protein associated with virus replication and movement suppresses RNA silencing. Mol. Plant Microbe Interact. 2004, 17, 583–592. [Google Scholar] [CrossRef]
- Yerukhimovich, M.M.; Marohnic, C.C.; Frick, D.N. Role of the conserved DECH-Box cysteine in coupling hepatitis C virus helicase-catalyzed ATP hydrolysis to RNA unwinding. Biochemistry 2018, 57, 6247–6255. [Google Scholar] [CrossRef]
- Corby, M.J.; Stoneman, M.R.; Biener, G.; Paprocki, J.D.; Kolli, R.; Raicu, V.; Frick, D.N. Quantitative microspectroscopic imaging reveals viral and cellular RNA helicase interactions in live cells. J. Biol. Chem. 2017, 292, 11165–11177. [Google Scholar] [CrossRef]
- Frick, D.N.; Virdi, R.S.; Vuksanovic, N.; Dahal, N.; Silvaggi, N.R. Molecular basis for ADP-Ribose binding to the Mac1 domain of SARS-CoV-2 nsp3. Biochemistry 2020, 59, 2608–2615. [Google Scholar] [CrossRef]
- Bassetto, M.; Leyssen, P.; Neyts, J.; Yerukhimovich, M.M.; Frick, D.N.; Courtney-Smith, M.; Brancale, A. In silico identification, design and synthesis of novel piperazine-based antiviral agents targeting the hepatitis C virus helicase. Eur. J. Med. Chem. 2017, 125, 1115–1131. [Google Scholar] [CrossRef]
- Shah, M.A.; Rasul, A.; Yousaf, R.; Haris, M.; Faheem, H.I.; Hamid, A.; Khan, H.; Khan, A.H.; Aschner, M.; Batiha, G.E. Combination of natural antivirals and potent immune invigorators: A natural remedy to combat COVID-19. Phytother. Res. 2021, 35, 6530–6551. [Google Scholar] [CrossRef]
- Greger, H. Comparative phytochemistry of flavaglines (= rocaglamides), a group of highly bioactive flavolignans from Aglaia species (Meliaceae). Phytochem. Rev. 2022, 21, 725–764. [Google Scholar] [CrossRef]
- Guo, P.B.; Wang, Z.W.; Li, G.; Liu, Y.X.; Xie, Y.F.; Wang, Q.M. First discovery of polycarpine, polycarpaurines A and C, and their derivatives as novel antiviral and antiphytopathogenic fungus agents. J. Agric. Food Chem. 2016, 64, 4264–4272. [Google Scholar] [CrossRef]
- Scalbert, A.; Manach, C.; Morand, C.; Remesy, C.; Jimenez, L. Dietary polyphenols and the prevention of diseases. Crit. Rev. Food Sci. Nutr. 2005, 45, 287–306. [Google Scholar] [CrossRef]
- Vishnoi, S.; Agrawal, V.; Kasana, V.K. Synthesis and Structure-Activity Relationships of Substituted Cinnamic Acids and Amide Analogues: A New Class of Herbicides. J. Agric. Food Chem. 2009, 57, 3261–3265. [Google Scholar] [CrossRef]
- Piazzon, A.; Vrhovsek, U.; Masuero, D.; Mattivi, F.; Mandoj, F.; Nardini, M. Antioxidant Activity of Phenolic Acids and Their Metabolites: Synthesis and Antioxidant Properties of the Sulfate Derivatives of Ferulic and Caffeic Acids and of the Acyl Glucuronide of Ferulic Acid. J. Agric. Food Chem. 2012, 60, 12312–12323. [Google Scholar] [CrossRef]
- Bisogno, F.; Mascoti, L.; Sanchez, C.; Garibotto, F.; Kurina-Sanz, M.; Giannini, F.; Enriz, R. Structure-Antifungal Activity Relationship of Cinnamic Acid Derivatives. J. Agric. Food Chem. 2007, 55, 10635–10640. [Google Scholar] [CrossRef]
- Kim, H.Y.; Park, J.; Lee, K.H.; Lee, D.U.; Kwak, J.H.; Kim, Y.S. Ferulic acid protects against carbon tetrachloride-induced liver injury in mice. Toxicology 2011, 282, 104–111. [Google Scholar] [CrossRef]
- Folkman, J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat. Med. 1995, 1, 27–31. [Google Scholar] [CrossRef]
- Corona, A.; Wycisk, K.; Talarico, C.; Manelfi, C.; Milia, J.; Cannalire, R.; Esposito, F.; Gribbon, P.; Zaliani, A.; Iaconis, D.; et al. Natural Compounds Inhibit SARS-CoV-2 nsp13 Unwinding and ATPase Enzyme Activities. ACS Pharmacol. Transl. Sci. 2022, 5, 226–239. [Google Scholar] [CrossRef]
- Ma, X.; Guo, Z.H.; Zhang, Z.Q.; Li, X.H.; Liu, Y.L.; Zhao, L.; Wang, X.F. Ferulic Acid Protects against Porcine Parvovirus Infection-Induced Apoptosis by Suppressing the Nuclear Factor-κB Inflammasome Axis and Toll-Like Receptor 4 via Nonstructural Protein 1. Evid.-Based Complement. Alt. 2020, 2020, 3943672. [Google Scholar] [CrossRef]
- Fuzimoto, A.D.; Isidoro, C. The antiviral and coronavirus-host protein pathways inhibiting properties of herbs and natural compounds—Additional weapons in the fight against the COVID-19 pandemic? J. Tradit. Complement. Med. 2020, 10, 405–419. [Google Scholar] [CrossRef]
- Mani, J.S.; Johnson, J.B.; Steel, J.C.; Broszczak, D.A.; Neilsen, P.M.; Walsh, K.B.; Naiker, M. Natural product-derived phytochemicals as potential agents against coronaviruses: A review. Virus Res. 2020, 284, 197989. [Google Scholar] [CrossRef] [PubMed]
- Gan, X.H.; Wang, Z.X.; Hu, D.Y. Synthesis of Novel Antiviral Ferulic Acid-Eugenol and Isoeugenol Hybrids Using Various Link Reactions. J. Agric. Food Chem. 2021, 69, 13724–13733. [Google Scholar] [CrossRef] [PubMed]
- Gan, X.H.; Hu, D.Y.; Wang, Y.J.; Yu, L.; Song, B.A. Novel trans-Ferulic Acid Derivatives Containing a Chalcone Moiety as Potential Activator for Plant Resistance Induction. J. Agric. Food Chem. 2017, 65, 4367–4377. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Liu, H.W.; Long, Z.Q.; Li, Z.X.; Zhu, J.J.; Wang, P.Y.; Qi, P.Y.; Liu, L.W.; Yang, S. Rational Optimization of 1,2,3-Triazole-Tailored Carbazoles As Prospective Antibacterial Alternatives with Significant In Vivo Control Efficiency and Unique Mode of Action. J. Agric. Food Chem. 2021, 69, 4615–4627. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.Y.; Zhang, T.H.; Feng, Y.M.; Wang, M.W.; Shao, W.B.; Zeng, D.; Jin, L.H.; Wang, P.Y.; Zhou, X.; Yang, S. Exploring an Innovative Strategy for Suppressing Bacterial Plant Disease: Excavated Novel Isopropanolamine-Tailored Pterostilbene Derivatives as Potential Antibiofilm Agents. J. Agric. Food Chem. 2022, 70, 4899–4911. [Google Scholar] [CrossRef]
- Yi, J.; Bai, R.; An, Y.; Liu, T.T.; Liang, J.H.; Tian, X.G.; Huo, X.K.; Feng, L.; Ning, J.; Sun, C.P.; et al. A natural inhibitor from Alisma orientale against human carboxylesterase 2: Kinetics, circular dichroism spectroscopic analysis, and docking simulation. Int. J. Biol. Macromol. 2019, 133, 184–189. [Google Scholar] [CrossRef]
- Li, X.Y.; Liu, J.; Yang, X.; Ding, Y.; Wu, J.; Hu, D.Y.; Song, B.A. Studies of binding interactions between Dufulin and southern rice black-streaked dwarf virus P9-1. Bioorg. Med. Chem. 2015, 23, 3629–3637. [Google Scholar] [CrossRef]
- Zhou, X.; Ye, H.J.; Gao, X.H.; Feng, Y.M.; Shao, W.B.; Qi, P.Y.; Wu, Z.B.; Liu, L.W.; Wang, P.Y.; Yang, S. The discovery of natural 4-demethylepipodophyllotoxin from renewable Dysosma versipellis species as a novel bacterial cell division inhibitor for controlling intractable diseases in rice. Ind. Crop. Prod. 2021, 174, 114182. [Google Scholar] [CrossRef]
- Chen, M.J.; Li, Z.; Shao, X.S.; Maienfisch, P. Bioisosteric-Replacement-Driven Lead Optimization of Tyclopyrazoflor. J. Agric. Food Chem. 2022, 70, 11123–11137. [Google Scholar] [CrossRef]
- Zhang, X.M.; Xu, H.; Su, H.F.; Yang, X.L.; Sun, T.D.; Lu, X.X.; Shi, F.S.; Duan, H.X.; Liu, X.L.; Ling, Y. Design, Synthesis, and Biological Activity of Novel Fungicides Containing a 1,2,3,4-Tetrahydroquinoline Scaffold and Acting as Laccase Inhibitors. J. Agric. Food Chem. 2022, 70, 1776–1787. [Google Scholar] [CrossRef]
- Xu, Y.M.; Liang, P.Y.; Rashid, H.U.; Wu, L.C.; Xie, P.; Wang, H.D.; Zhang, S.Y.; Wang, L.S.; Jiang, J. Design, synthesis, and biological evaluation of matrine derivatives possessing piperazine moiety as antitumor agents. Med. Chem. Res. 2019, 28, 1618–1627. [Google Scholar] [CrossRef]
- Houlding, T.K.; Tchabanenko, K.; Rahman, M.T.; Rebrov, E.V. Direct amide formation using radio frequency heating. Org. Biomol. Chem. 2013, 11, 4171–4177. [Google Scholar] [CrossRef]
- Yasueda, Y.K.; Tamura, T.; Fujisawa, A.; Kuwata, K.K.; Tsukiji, S.Y.; Kiyonaka, S.; Hamachi, I. A Set of Organelle-Localizable Reactive Molecules for Mitochondrial Chemical Proteomics in Living Cells and Brain Tissues. J. Am. Chem. Soc. 2016, 138, 7592–7602. [Google Scholar] [CrossRef]
- Li, Z.X.; Fang, H.S.; Shao, W.B.; Wu, Z.B.; Wang, P.Y.; Yang, S. Design, synthesis, and anti-TMV bioactivities of nucleobase phosphonate analogs. Phosphorus Sulfur Silicon Relat. Elem. 2017, 192, 1279–1285. [Google Scholar] [CrossRef]
- Wang, P.Y.; Wang, M.W.; Zeng, D.; Xiang, M.; Rao, J.R.; Liu, Q.Q.; Liu, L.W.; Wu, Z.B.; Li, Z.; Song, B.A.; et al. Rational Optimization and Action Mechanism of Novel Imidazole (or Imidazolium)-Labeled 1,3,4-Oxadiazole Thioethers as Promising Antibacterial Agents against Plant Bacterial Diseases. J. Agric. Food Chem. 2019, 67, 3535–3545. [Google Scholar] [CrossRef]
- Chen, L.; Zhou, X.S.; Fan, W.F.; Zhang, Y.X. Expression, purification and characterization of a recombinant Lipomyces starkey dextranase in Pichia pastoris. Protein Expres. Purif. 2008, 58, 87–93. [Google Scholar] [CrossRef]
- Kato, M.; Ishibashi, K.; Kobayashi, C.; Ishikawa, M.; Katoh, E. Expression, purification, and functional characterization of an N-terminal fragment of the tomato mosaic virus resistance protein Tm-1. Protein. Expr. Purif. 2013, 89, 1–6. [Google Scholar] [CrossRef]
- Wan, X.B.; Yang, T.P.; Cuesta, A.; Pang, X.M.; Balius, T.E.; Irwin, J.J.; Shoichet, B.K.; Taunton, J. Discovery of Lysine-Targeted eIF4E Inhibitors through Covalent Docking. J. Am. Chem. Soc. 2020, 142, 4960–4964. [Google Scholar] [CrossRef]
- Li, Y.T.; Ye, S.W.; Hu, Z.L.; Hao, N.; Bo, X.; Liang, H.G.; Tian, X.R. Identification of anti-TMV active flavonoid glycosides and their mode of action on virus particles from Clematis lasiandra Maxim. Pest Manag. Sci 2021, 77, 5268–5277. [Google Scholar] [CrossRef]
- Shao, W.B.; Wang, P.Y.; Fang, Z.M.; Wang, J.J.; Guo, D.X.; Ji, J.; Zhou, X.; Qi, P.Y.; Liu, L.W.; Yang, S. Synthesis and Biological Evaluation of 1,2,4-Triazole Thioethers as Both Potential Virulence Factor Inhibitors against Plant Bacterial Diseases and Agricultural Antiviral Agents against Tobacco Mosaic Virus Infections. J. Agric. Food Chem. 2021, 69, 15108–15122. [Google Scholar] [CrossRef]

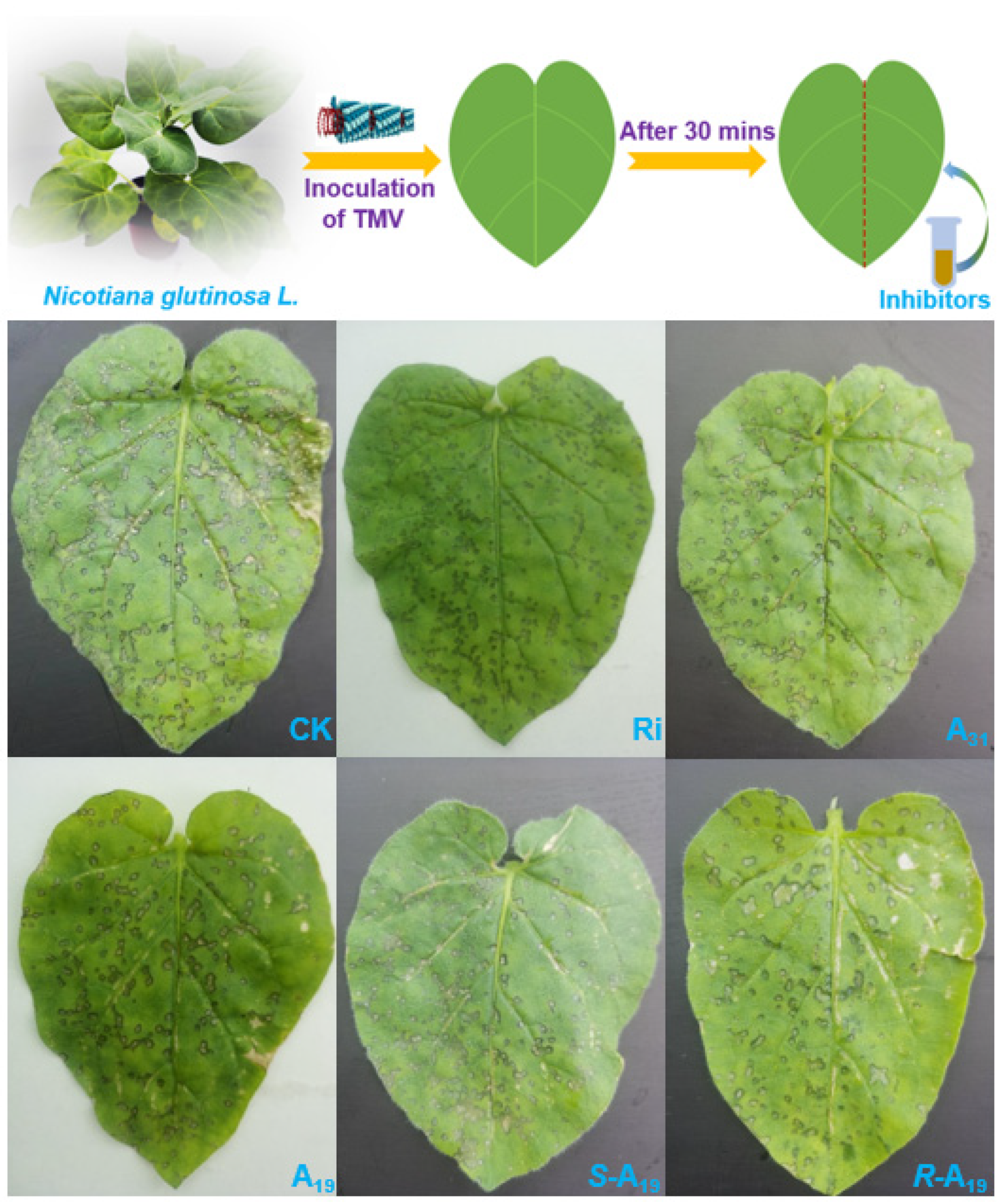

| Compound | Conc. | Inhibition Rate (%) | |
|---|---|---|---|
| (μg/mL) | Curative Effect | Protective Effect | |
| A1 | 500 | 32.7 ± 1.2 | 21.6 ± 0.2 |
| 100 | 6.4 ± 0.8 | 0 | |
| A2 | 500 | 19.2 ± 0.7 | 0 |
| 100 | 0 | 0 | |
| A3 | 500 | 38.6 ± 0.7 | 41.7 ± 2.6 |
| 100 | 19.8 ± 3.1 | 15.4 ± 3.7 | |
| A4 | 500 | 43.8 ± 1.5 | 21.4 ± 4.0 |
| 100 | 17.6 ± 1.0 | 0 | |
| A5 | 500 | 10.3 ± 3.9 | 23.1 ± 5.6 |
| 100 | 0 | 0 | |
| A6 | 500 | 6.8 ± 4.0 | 16.8 ± 0.6 |
| 100 | 0 | 0 | |
| A7 | 500 | 4.3 ± 2.3 | 47.7 ± 4.6 |
| 100 | 0 | 21.6 ± 3.9 | |
| A8 | 500 | 7.0 ± 2.2 | 42.1 ± 2.7 |
| 100 | 0 | 20.2 ± 3.4 | |
| A9 | 500 | 5.2 ± 2.9 | 11.7 ± 1.9 |
| 100 | 0 | 0 | |
| A10 | 500 | 29.0 ± 2.1 | 0 |
| 100 | 11.2 ± 2.6 | 0 | |
| A11 | 500 | 36.1 ± 4.3 | 54.2 ± 4.2 |
| 100 | 17.1 ± 2.4 | 24.5 ± 2.2 | |
| A12 | 500 | 28.4 ± 3.0 | 25.4 ± 2.4 |
| 100 | 0 | 0 | |
| A13 | 500 | 28.3 ± 0.3 | 32.7 ± 1.6 |
| 100 | 5.2 ± 0.4 | 8.9 ± 3.5 | |
| A14 | 500 | 41.5 ± 3.1 | 19.4 ± 2.6 |
| 100 | 7.1 ± 2.8 | 0 | |
| A15 | 500 | 43.1 ± 1.3 | 29.1 ± 2.7 |
| 100 | 0 | 0 | |
| A16 | 500 | 46.5 ± 1.3 | 35.3 ± 3.6 |
| 100 | 10.1 ± 2.7 | 10.2 ± 0.9 | |
| A17 | 500 | 21.7 ± 0.6 | 45.9 ± 1.7 |
| 100 | 0 | 27.5 ± 1.6 | |
| A18 | 500 | 27.1 ± 2.4 | 57.8 ± 0.7 |
| 100 | 0 | 32.1 ± 2.3 | |
| A19 | 500 | 62.7 ± 0.2 | 46.4 ± 0.4 |
| 100 | 17.6 ± 1.9 | 13.5 ± 3.6 | |
| Ferulic acid | 500 | 31.4 ± 3.3 | 43.5 ± 1.8 |
| 100 | 8.0 ± 1.7 | 16.9 ± 4.5 | |
| Ribavirin | 500 | 42.1 ± 1.4 | 39.5 ± 1.2 |
| 100 | 11.7 ± 1.8 | 8.6 ± 2.1 | |
| Compound | Conc. | Inhibition Rate (%) | |
|---|---|---|---|
| (μg/mL) | Curative Effect | Protective Effect | |
| A20 | 500 | 56.9 ± 2.2 | 55.2 ± 3.0 |
| 100 | 18.4 ± 1.7 | 20.1 ± 1.3 | |
| A21 | 500 | 45.2 ± 1.0 | 50.6 ± 1.2 |
| 100 | 0 | 16.2 ± 2.5 | |
| A22 | 500 | 20.1 ± 1.3 | 7.9 ± 4.1 |
| 100 | 0 | 0 | |
| A23 | 500 | 31.6 ± 4.9 | 15.7 ± 3.1 |
| 100 | 9.1 ± 3.4 | 0 | |
| A24 | 500 | 44.6 ± 3.7 | 45.7 ± 4.2 |
| 100 | 11.4 ± 0.6 | 19.4 ± 3.6 | |
| A25 | 500 | 47.3 ± 1.4 | 35.3 ± 1.0 |
| 100 | 23.2 ± 1.5 | 12.4 ± 4.1 | |
| A26 | 500 | 28.7 ± 3.1 | 43.1 ± 2.5 |
| 100 | 0 | 8.1 ± 2.3 | |
| A27 | 500 | 42.6 ± 1.3 | 60.8 ± 4.2 |
| 100 | 0 | 31.6 ± 3.7 | |
| A28 | 500 | 41.8 ± 3.2 | 46.1 ± 2.1 |
| 100 | 0 | 0 | |
| A29 | 500 | 59.1 ± 0.1 | 42.1 ± 2.9 |
| 100 | 25.4 ± 2.5 | 5.3 ± 2.4 | |
| A30 | 500 | 51.1 ± 2.6 | 14.5 ± 0.8 |
| 100 | 17.4 ± 0.1 | 0 | |
| A31 | 500 | 61.7 ± 3.3 | 24.9 ± 2.7 |
| 100 | 27.1 ± 1.7 | 8.2 ± 0.9 | |
| Ferulic acid | 500 | 31.4 ± 3.3 | 43.5 ± 1.8 |
| 100 | 8.0 ± 1.7 | 16.9 ± 4.5 | |
| Ribavirin | 500 | 42.1 ± 1.4 | 39.5 ± 1.2 |
| 100 | 11.7 ± 1.8 | 8.6 ± 2.1 | |
| Compound | Conc. | Inhibition Rate (%) | |
|---|---|---|---|
| (μg/mL) | Curative Effect | Protective Effect | |
| (R/S)-A16 | 500 | 46.5 ± 1.3 | 35.3 ± 3.6 |
| 100 | 10.1 ± 2.7 | 10.2 ± 0.9 | |
| R-A16 | 500 | 55.1 ± 0.4 | 33.6 ± 1.5 |
| 100 | 20.6 ± 3.1 | 8.5 ± 3.3 | |
| S-A16 | 500 | 45.7 ± 0.9 | 15.7 ± 0.4 |
| 100 | 16.8 ± 2.7 | 0 | |
| (R/S)-A19 | 500 | 62.7 ± 0.2 | 46.4 ± 0.4 |
| 100 | 17.6 ± 1.9 | 13.5 ± 3.6 | |
| R-A19 | 500 | 67.5 ± 2.3 | 44.9 ± 2.2 |
| 100 | 26.3 ± 1.0 | 15.8 ± 1.4 | |
| S-A19 | 500 | 55.4 ± 3.5 | 32.6 ± 1.7 |
| 100 | 14.4 ± 1.2 | 8.4 ± 3.0 | |
| (R/S)-A20 | 500 | 56.9 ± 2.2 | 55.2 ± 3.0 |
| 100 | 18.4 ± 1.7 | 20.1 ± 1.3 | |
| R-A20 | 500 | 58.0 ± 3.9 | 46.7 ± 2.4 |
| 100 | 24.8 ± 2.5 | 18.8 ± 2.8 | |
| S-A20 | 500 | 49.9 ± 1.3 | 41.5 ± 1.7 |
| 100 | 8.7 ± 2.4 | 17.9 ± 3.7 | |
| (R/S)-A29 | 500 | 59.1 ± 0.1 | 42.1 ± 2.9 |
| 100 | 25.4 ± 2.5 | 5.3 ± 2.4 | |
| R-A29 | 500 | 56.7 ± 1.9 | 45.7 ± 4.1 |
| 100 | 22.5 ± 1.4 | 0 | |
| S-A29 | 500 | 45.1 ± 2.6 | 36.4 ± 2.6 |
| 100 | 13.5 ± 4.2 | 0 | |
| Ferulic acid | 500 | 31.4 ± 3.3 | 43.5 ± 1.8 |
| 100 | 8.0 ± 1.7 | 16.9 ± 4.5 | |
| Ribavirin | 500 | 42.1 ± 1.4 | 39.5 ± 1.2 |
| 100 | 11.7 ± 1.8 | 8.6 ± 2.1 | |
| Parameters | A19 | A31 |
|---|---|---|
| Etotal/hartree | −1207.326 | −1496.878 |
| EHOMO/hartree | −0.219 | −0.219 |
| ELUMO/hartree | −0.069 | −0.067 |
| △E/hartree | 0.15 | 0.152 |
| Clog P | 1.386 | 4.118 |
| TPSA | 77.47 | 71.48 |
| Compound | Inhibition Rate (%) | Compound | Inhibition Rate (%) | ||
|---|---|---|---|---|---|
| 100 μg/mL | 50 μg/mL | 100 μg/mL | 50 μg/mL | ||
| A1 | 70.5 ± 4.0 | 46.6 ± 4.5 | A19 | 0 | 0 |
| A2 | 23.9 ± 0.7 | 0 | R-A19 | 7.4 ± 1.8 | 0 |
| A3 | 0 | 0 | S-A19 | 0 | 0 |
| A4 | 0 | 0 | A20 | 0 | 0 |
| A5 | 0 | 0 | R-A20 | 0 | 0 |
| A6 | 0 | 0 | S-A20 | 0 | 0 |
| A7 | 42.8 ± 2.7 | 22.1 ± 2.4 | A21 | 58.4 ± 2.7 | 36.3 ± 3.6 |
| A8 | 0 | 0 | A22 | 41.4 ± 0.8 | 35.1 ± 5.3 |
| A9 | 0 | 0 | A23 | 0 | 0 |
| A10 | 56.1 ± 2.3 | 23.3 ± 4.7 | A24 | 12.2 ± 4.8 | 0 |
| A11 | 30.2 ± 4.6 | 18.6 ± 0.4 | A25 | 31.1 ± 2.6 | 0 |
| A12 | 39.7 ± 4.2 | 24.3 ± 3.2 | A26 | 37.1 ± 5.8 | 0 |
| A13 | 0 | 0 | A27 | 32.2 ± 3.9 | 0 |
| A14 | 42.8 ± 5.0 | 29.8 ± 4.2 | A28 | 29.8 ± 3.2 | 0 |
| A15 | 0 | 0 | A29 | 0 | 0 |
| A16 | 46.4 ± 2.7 | 34.7 ± 3.9 | R-A29 | 9.1 ± 3.0 | 0 |
| R-A16 | 40.1 ± 2.6 | 28.8 ± 1.5 | S-A29 | 6.1 ± 1.0 | 0 |
| S-A16 | 26.7 ± 4.1 | 21.0 ± 4.3 | A30 | 22.3 ± 4.8 | 0 |
| A17 | 25.8 ± 1.1 | 17.0 ± 1.2 | A31 | 38.5 ± 3.9 | 0 |
| A18 | 7.3 ± 4.1 | 0 | TC | 40.6 ± 5.4 | 19.2 ± 0.1 |
| Compound | Inhibition Rate (%) | Toxic Regression Equation | EC50 (μg/mL) | r2 | |
|---|---|---|---|---|---|
| 100 μg/mL | 50 μg/mL | ||||
| A1 | 45.5 ± 4.0 | 20.1 ± 1.7 | |||
| A2 | 29.0 ± 1.4 | 0 | |||
| A3 | 0 | 0 | |||
| A4 | 0 | 0 | |||
| A5 | 0 | 0 | |||
| A6 | 79.6 ± 1.2 | 0 | |||
| A7 | 27.3 ± 0.7 | 0 | |||
| A8 | 0 | 0 | |||
| A9 | 0 | 0 | |||
| A10 | 31.1 ± 5.6 | 11.6 ± 0.7 | |||
| A11 | 0 | 0 | |||
| A12 | 0 | 0 | |||
| A13 | 20.0 ± 0.7 | 0 | |||
| A14 | 0 | 0 | |||
| A15 | 33.4 ± 0.8 | 12.4 ± 4.8 | |||
| A16 | 0 | 0 | |||
| R-A16 | 24.2 ± 2.8 | 0 | |||
| S-A16 | 29.8 ± 3.1 | 0 | |||
| A17 | 68.2 ± 1.3 | 38.6 ± 5.8 | |||
| A18 | 69.0 ± 4.9 | 66.7 ± 1.7 | y = 2.591x + 1.012 | 34.61 ± 0.64 | 0.995 |
| A19 | 23.6 ± 0.6 | 9.2 ± 0.6 | |||
| R-A19 | 0 | 0 | |||
| S-A19 | 26.1 ± 3.8 | 17.3 ± 5.0 | |||
| A20 | 23.5 ± 1.2 | 13.1 ± 4.0 | |||
| R-A20 | 0 | 0 | |||
| S-A20 | 21.9 ± 0.7 | 0 | |||
| A21 | 0 | 0 | |||
| A22 | 60.6 ± 0.6 | 25.3 ± 2.2 | |||
| A23 | 100 | 100 | y = 7.801x − 5.505 | 22.21 ± 0.43 | 0.941 |
| A24 | 97.3 ± 0.3 | 84.9 ± 5.7 | y = 3.543x + 0.014 | 25.54 ± 0.70 | 0.984 |
| A25 | 91.8 ± 0.9 | 40.9 ± 4.6 | |||
| A26 | 97.9 ± 0.1 | 72.9 ± 1.9 | y = 1.987x + 2.276 | 23.49 ± 1.30 | 0.999 |
| A27 | 97.4 ± 0.1 | 44.7 ± 1.1 | |||
| A28 | 100 | 100 | y = 5.312x − 0.855 | 12.65 ± 0.10 | 0.992 |
| A29 | 100 | 100 | y = 3.528x + 1.145 | 12.38 ± 0.72 | 0.915 |
| R-A29 | 100 | 100 | y = 5.975x − 2.562 | 18.43 ± 0.08 | 0.812 |
| S-A29 | 100 | 100 | y = 5.713x − 0.863 | 10.62 ± 0.37 | 0.994 |
| A30 | 100 | 100 | y = 2.629x + 2.275 | 10.87 ± 0.23 | 0.955 |
| A31 | 100 | 41.1 ± 2.7 | |||
| TC | 91.3 ± 2.5 | 60.9 ± 1.5 | y = 1.098x + 0.835 | 36.47 ± 1.27 | 0.992 |
| Compound | Inhibition Rate (%) | Toxic Regression Equation | EC50 (μg/mL) | r2 | |
|---|---|---|---|---|---|
| 100 μg/mL | 50 μg/mL | ||||
| A1 | 14.3 ± 3.3 | 0 | |||
| A2 | 71.3 ± 2.7 | 44.0 ± 0.2 | |||
| A3 | 0 | 0 | |||
| A4 | 32.1 ± 1.3 | 0 | |||
| A5 | 15.3 ± 3.1 | 0 | |||
| A6 | 23.7 ± 4.4 | 21.8 ± 1.3 | |||
| A7 | 27.2 ± 4.3 | 0 | |||
| A8 | 17.4 ± 3.1 | 0 | |||
| A9 | 14.3 ± 1.7 | 0 | |||
| A10 | 100 | 94.7 ± 1.0 | y = 3.267x − 0.289 | 41.58 ± 0.65 | 0.926 |
| A11 | 0 | 0 | |||
| A12 | 86.7 ± 0.9 | 40.6 ± 3.0 | |||
| A13 | 20.1 ± 2.6 | 12.2 ± 2.2 | |||
| A14 | 46.8 ± 5.5 | 22.3 ± 4.3 | |||
| A15 | 47.8 ± 2.9 | 29.2 ± 2.0 | |||
| A16 | 68.5 ± 5.0 | 30.2 ± 3.2 | |||
| R-A16 | 61.2 ± 2.6 | 35.5 ± 4.0 | |||
| S-A16 | 64.7 ± 3.6 | 30.6 ± 3.1 | |||
| A17 | 0 | 0 | |||
| A18 | 31.0 ± 0.6 | 33.8 ± 2.9 | |||
| A19 | 23.9 ± 5.2 | 0 | |||
| R-A19 | 25.4 ± 3.9 | 0 | |||
| S-A19 | 20.1 ± 4.6 | 0 | |||
| A20 | 24.7 ± 2.0 | 11.4 ± 3.9 | |||
| R-A20 | 28.1 ± 4.1 | 0 | |||
| S-A20 | 22.5 ± 3.4 | 15.1 ± 2.8 | |||
| A21 | 28.4 ± 4.3 | 16.3 ± 3.6 | |||
| A22 | 59.1 ± 5.7 | 40.2 ± 5.7 | |||
| A23 | 100 | 95.1 ± 1.1 | y = 1.354x + 3.687 | 9.33 ± 0.16 | 0.867 |
| A24 | 100 | 100 | y = 1.332x + 3.561 | 12.03 ± 0.19 | 0.954 |
| A25 | 29.5 ± 1.3 | 19.2 ± 0.7 | |||
| A26 | 90.8 ± 0.8 | 78.2 ± 1.7 | y = 2.041x + 2.630 | 14.49 ± 0.54 | 0.867 |
| A27 | 86.7 ± 0.8 | 37.1 ± 2.6 | |||
| A28 | 100 | 83.2 ± 4.0 | y = 1.622x + 3.570 | 7.61 ± 0.22 | 0.994 |
| A29 | 100 | 100 | y = 4.209x + 0.647 | 10.66 ± 0.16 | 0.982 |
| R-A29 | 100 | 100 | y = 0.824x + 4.154 | 10.63 ± 0.51 | 0.983 |
| S-A29 | 100 | 96.4 ± 1.4 | y = 1.939x + 3.076 | 9.82 ± 0.32 | 0.992 |
| A30 | 100 | 79.1 ± 3.5 | y = 1.564x + 3.202 | 14.11 ± 0.60 | 0.992 |
| A31 | 92.8 ± 1.7 | 30.1 ± 3.0 | |||
| TC | 56.1 ± 2.3 | 32.4 ± 2.3 | y = 2.153x + 0.941 | 72.59 ± 2.73 | 0.962 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Yang, B.; Liu, H.; Ding, Y.; Fang, Z.; Shao, W.; Qi, P.; Zhou, X.; Liu, L.; Yang, S. The Discovery of Novel Ferulic Acid Derivatives Incorporating Substituted Isopropanolamine Moieties as Potential Tobacco Mosaic Virus Helicase Inhibitors. Int. J. Mol. Sci. 2022, 23, 13991. https://doi.org/10.3390/ijms232213991
Li Z, Yang B, Liu H, Ding Y, Fang Z, Shao W, Qi P, Zhou X, Liu L, Yang S. The Discovery of Novel Ferulic Acid Derivatives Incorporating Substituted Isopropanolamine Moieties as Potential Tobacco Mosaic Virus Helicase Inhibitors. International Journal of Molecular Sciences. 2022; 23(22):13991. https://doi.org/10.3390/ijms232213991
Chicago/Turabian StyleLi, Zhenxing, Binxin Yang, Hongwu Liu, Yue Ding, Zimian Fang, Wubin Shao, Puying Qi, Xiang Zhou, Liwei Liu, and Song Yang. 2022. "The Discovery of Novel Ferulic Acid Derivatives Incorporating Substituted Isopropanolamine Moieties as Potential Tobacco Mosaic Virus Helicase Inhibitors" International Journal of Molecular Sciences 23, no. 22: 13991. https://doi.org/10.3390/ijms232213991
APA StyleLi, Z., Yang, B., Liu, H., Ding, Y., Fang, Z., Shao, W., Qi, P., Zhou, X., Liu, L., & Yang, S. (2022). The Discovery of Novel Ferulic Acid Derivatives Incorporating Substituted Isopropanolamine Moieties as Potential Tobacco Mosaic Virus Helicase Inhibitors. International Journal of Molecular Sciences, 23(22), 13991. https://doi.org/10.3390/ijms232213991






