Identification of Epigenetic Interactions between MicroRNA-30c-5p and DNA Methyltransferases in Neuropathic Pain
Abstract
1. Introduction
2. Results
2.1. The miR-30c-5p Inhibitor Prevents Neuropathic Pain after Sciatic Nerve Injury in Rats
2.2. Changes in Global DNA Methylation Induced by Sciatic Nerve Injury within Spinal Dorsal Horn and Dorsal Root Ganglia Neurons and Its Modification by Treatment with miR-30c-5p-Inhibitor
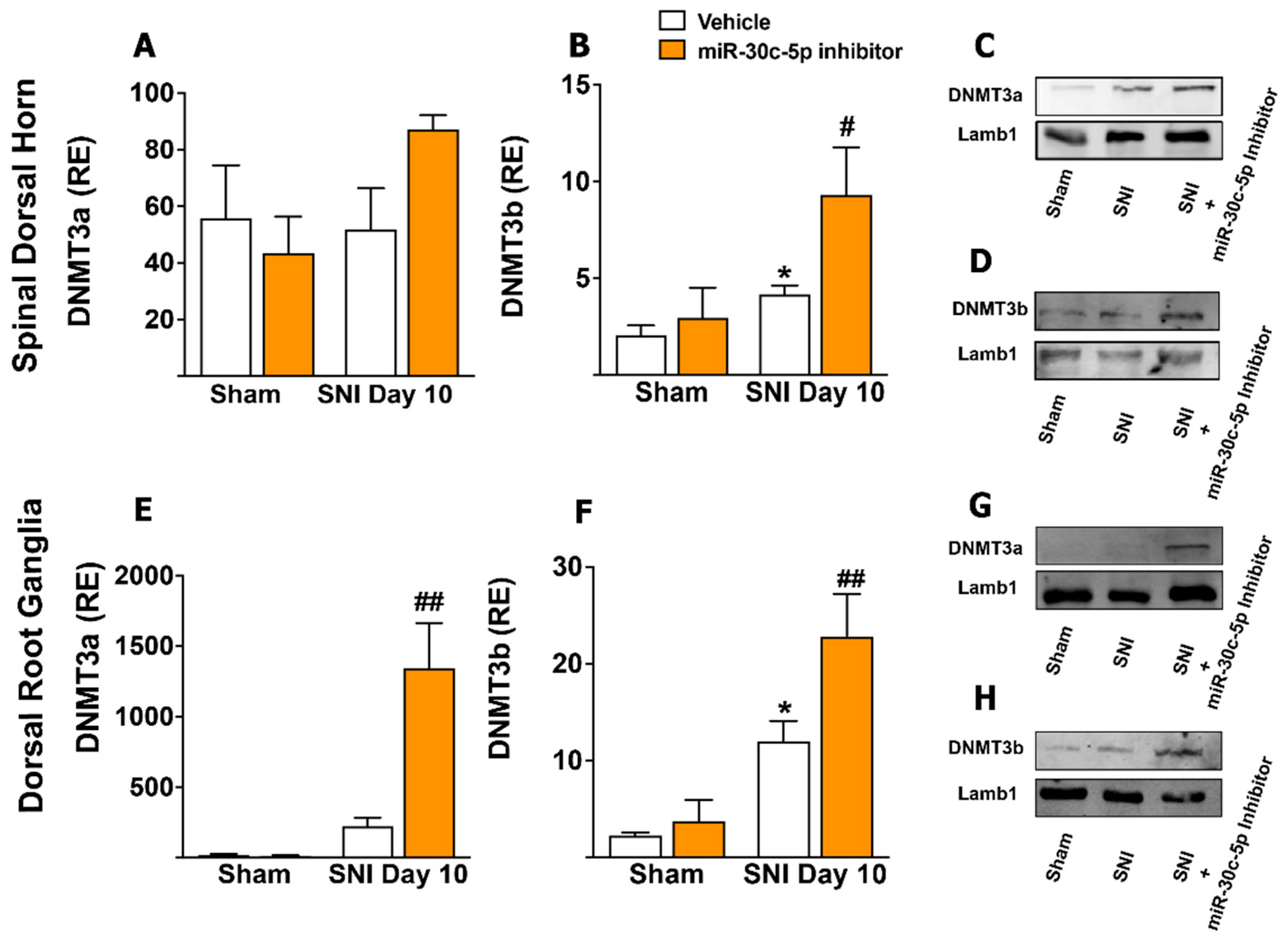
2.3. Changes in the Expression of DNA Methyltransferases Induced by Sciatic Nerve Injury within Spinal Dorsal Horn and Dorsal Root Ganglia Neurons and Its Modification by miR-30c-5p-Inhibitor Treatment
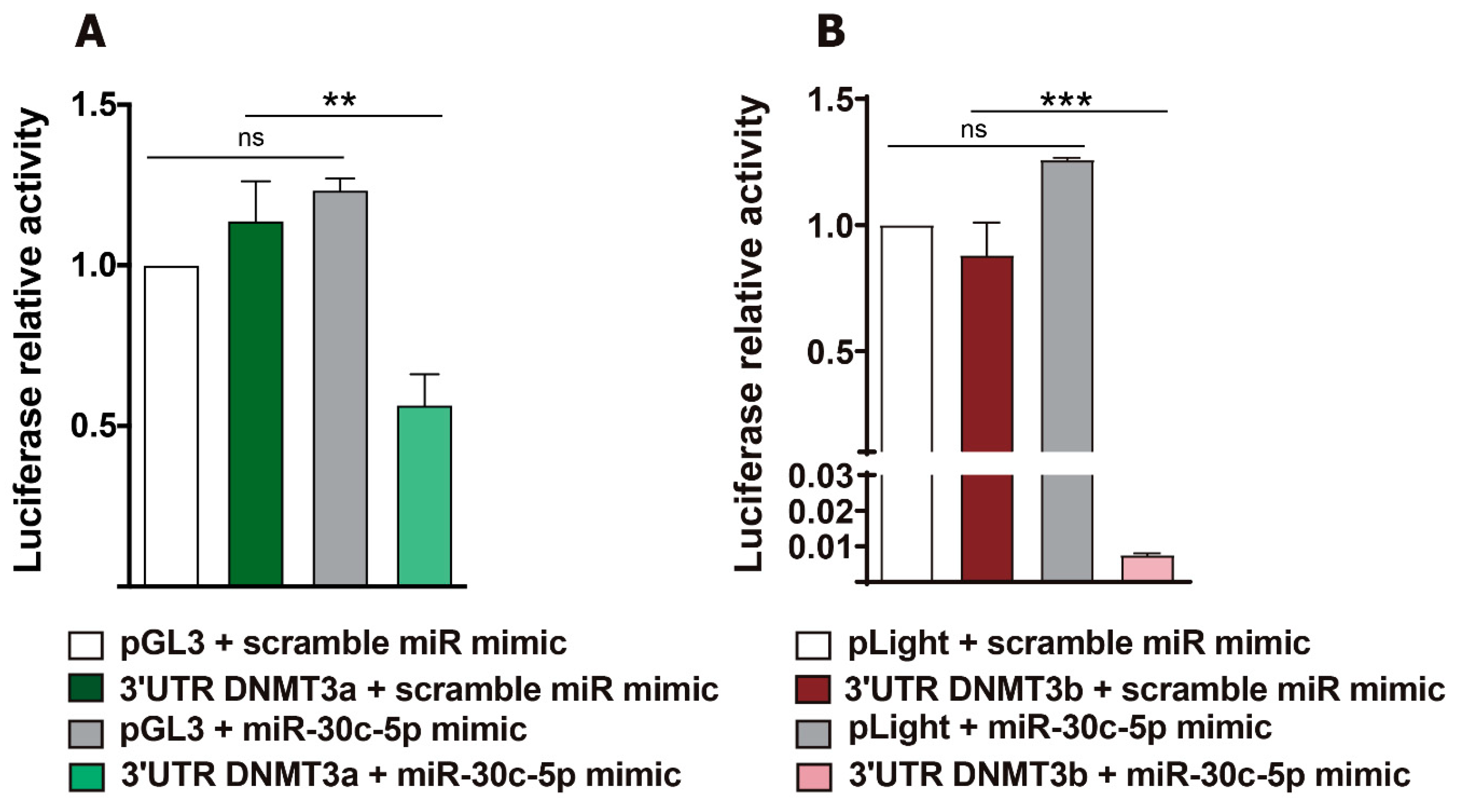
2.4. Luciferase Reporter Assays Showed Post-Transcriptional Regulation of De Novo DNMT3a and DNMT3b by miR-30c-5p in HeLa Cells
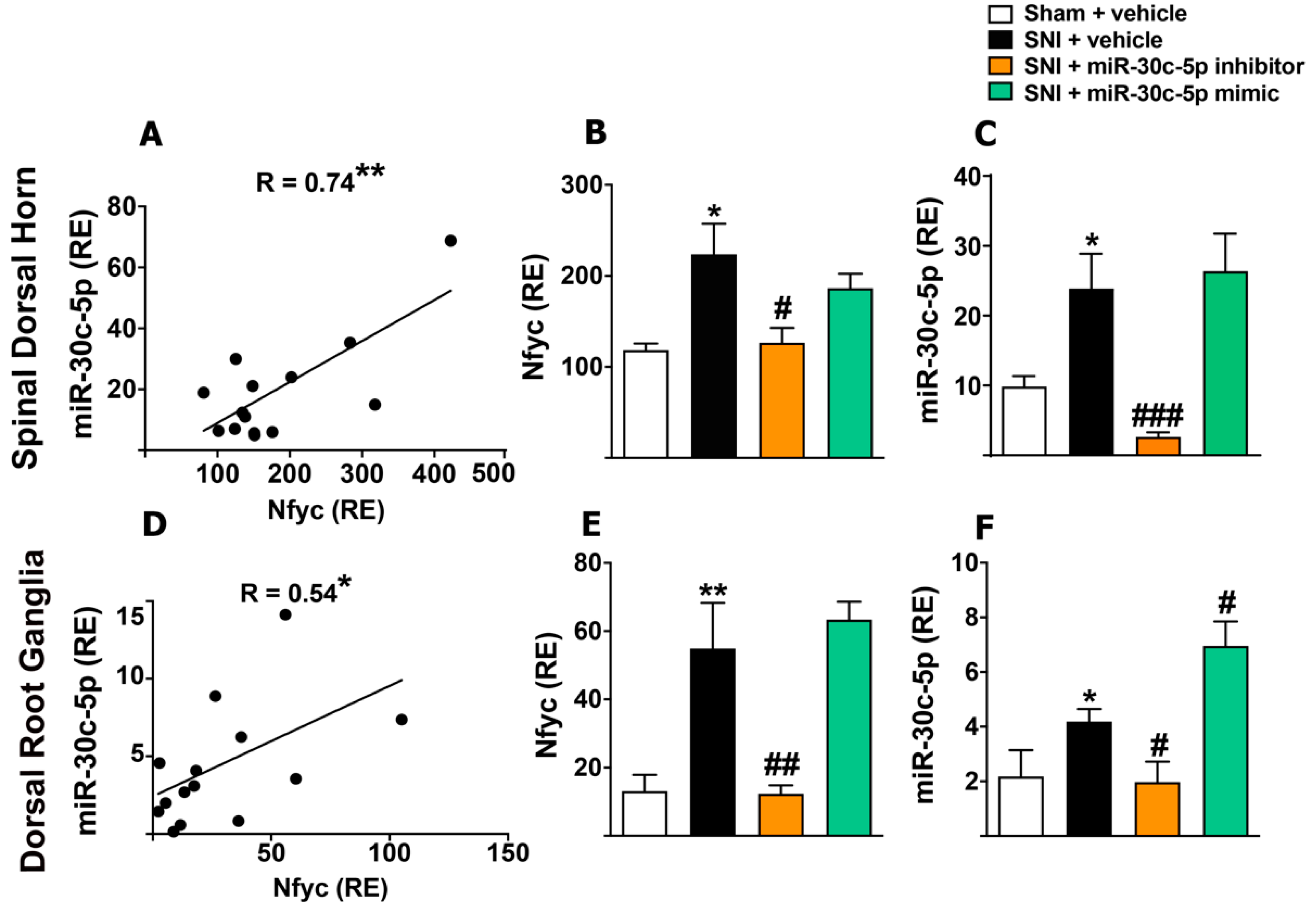
2.5. Modulation of miR-30c-5p in Rats Subjected to Sciatic Nerve Injury Results in Methylation Changes of Its Host Gene Nfyc
2.6. Modulation of miR-30c-5p in Rats Subjected to Sciatic Nerve Injury Results in Methylation Changes of Its Target TGF-β1
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Study Design
4.3. Neuropathic Pain Model
4.4. Treatment and Experimental Groups
4.5. Immunofluorescence and Confocal Microscopy: 5′-MeC Detection
4.6. RNA Extraction and Real-Time RT-PCR
4.7. Western Blot
4.8. Methylation Sensitive qPCR (ms-qPCR)
4.9. Cell Culture and Luciferase Assay
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kuner, R.; Flor, H. Structural plasticity and reorganisation in chronic pain. Nat. Rev. Neurosci. 2017, 18, 20–30. [Google Scholar] [CrossRef]
- Price, T.J.; Basbaum, A.I.; Bresnahan, J.; Chambers, J.F.; De Koninck, Y.; Edwards, R.R.; Ji, R.R.; Katz, J.; Kavelaars, A.; Levine, J.D.; et al. Transition to chronic pain: Opportunities for novel therapeutics. Nat. Rev. Neurosci. 2018, 19, 383–384. [Google Scholar] [CrossRef]
- Scholz, J.; Finnerup, N.B.; Attal, N.; Aziz, Q.; Baron, R.; Bennett, M.I.; Benoliel, R.; Cohen, M.; Cruccu, G.; Davis, K.D.; et al. Classification Committee of the Neuropathic Pain Special Interest Group (NeuPSIG). The IASP classification of chronic pain for ICD-11: Chronic neuropathic pain. Pain 2019, 160, 53–59. [Google Scholar] [CrossRef]
- Colloca, L.; Ludman, T.; Bouhassira, D.; Baron, R.; Dickenson, A.H.; Yarnitsky, D.; Freeman, R.; Truini, A.; Attal, N.; Finnerup, N.B.; et al. Neuropathic pain. Nat. Rev. Dis. Primers 2017, 16, 17002. [Google Scholar] [CrossRef]
- Attal, N.; Bouhassira, D. Translational neuropathic pain research. Pain 2019, 160, S23–S28. [Google Scholar] [CrossRef]
- Finnerup, N.B.; Kuner, R.; Jensen, T.S. Neuropathic Pain: From Mechanisms to Treatment. Physiol. Rev. 2021, 101, 259–301. [Google Scholar] [CrossRef]
- Descalzi, G.; Ikegami, D.; Ushijima, T.; Nestler, E.J.; Zachariou, V.; Narita, M. Epigenetic mechanisms of chronic pain. Trends Neurosci. 2015, 38, 237–246. [Google Scholar] [CrossRef]
- López-González, M.J.; Landry, M.; Favereaux, A. MicroRNA and chronic pain: From mechanisms to therapeutic potential. Pharmacol. Ther. 2017, 180, 1–15. [Google Scholar] [CrossRef]
- Penas, C.; Navarro, X. Epigenetic modifications associated to neuroinflammation and neuropathic pain after neural trauma. Front. Cell. Neurosci. 2018, 12, 158. [Google Scholar] [CrossRef]
- Torres-Perez, J.V.; Irfan, J.; Febrianto, M.R.; Di Giovanni, S.; Nagy, I. Histone post-translational modifications as potential therapeutic targets for pain management. Trends Pharmacol. Sci. 2021, 42, 897–911. [Google Scholar] [CrossRef]
- Topham, L.; Gregoire, S.; Kang, H.; Salmon-Divon, M.; Lax, E.; Millecamps, M.; Szyf, M.; Stone, L.S. The transition from acute to chronic pain: Dynamic epigenetic reprogramming of the mouse prefrontal cortex up to 1 year after nerve injury. Pain 2020, 161, 2394–2409. [Google Scholar] [CrossRef]
- Ye, G.; Zhang, Y.; Zhao, J.; Chen, Y.; Kong, L.; Sheng, C.; Yuan, L. miR-384-5p ameliorates neuropathic pain by targeting SCN3A in a rat model of chronic constriction injury. Neurol. Res. 2020, 42, 299–307. [Google Scholar] [CrossRef]
- Huang, A.; Ji, L.; Huang, Y.; Yu, Q.; Li, Y. miR-185-5p alleviates CCI-induced neuropathic pain by repressing NLRP3 inflammasome through dual targeting MyD88 and CXCR4. Int. Immunopharmacol. 2022, 104, 108508. [Google Scholar] [CrossRef]
- Tramullas, M.; Francés, R.; De la Fuente, R.; Velategui, S.; Carcelén, M.; García, R.; Llorca, J.; Hurlé, MA. MicroRNA-30c-5p modulates neuropathic pain in rodents. Sci. Transl. Med. 2018, 10, eaao6299. [Google Scholar] [CrossRef]
- Holla, S.; Balaji, K.N. Epigenetics and miRNA during bacteria-induced host immune responses. Epigenomics 2015, 7, 1197–1212. [Google Scholar] [CrossRef]
- Mancini, M.; Grasso, M.; Muccillo, L.; Babbio, F.; Precazzini, F.; Castiglioni, I.; Zanetti, V.; Rizzo, F.; Pistore, C.; De Marino, MG.; et al. DNMT3A epigenetically regulates key microRNAs involved in epithelial-to-mesenchymal transition in prostate cancer. Carcinogenesis 2021, 42, 1449–1460. [Google Scholar] [CrossRef]
- Han, X.; Zhen, S.; Ye, Z.; Lu, J.; Wang, L.; Li, P.; Li, J.; Zheng, X.; Li, H.; Chen, W.; et al. A Feedback Loop Between miR-30a/c-5p and DNMT1 Mediates Cisplatin Resistance in Ovarian Cancer Cells. Cell. Physiol. Biochem. 2017, 4, 973–986. [Google Scholar] [CrossRef]
- Wang, S.; Wu, W.; Claret, F.X. Mutual regulation of microRNAs and DNA methylation in human cancers. Epigenetics 2017, 4, 187–197. [Google Scholar] [CrossRef]
- Tao, J.; Xia, L.; Cai, Z.; Liang, L.; Chen, Y.; Meng, J.; Wang, Z. Interaction Between microRNA and DNA Methylation in Atherosclerosis. DNA Cell Biol. 2020, 40, 101–115. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, C.; Liu, X.; Zhang, B.; Wang, P.; Yang, Z.; Niu, N. The regulatory role of aberrant methylation of microRNA-34a promoter CpGs in osteosarcoma. Transl. Cancer Res. 2019, 8, 2328–2338. [Google Scholar] [CrossRef]
- Ma, Y.; Chai, N.; Jiang, Q.; Chang, Z.; Chai, Y.; Li, X.; Sun, H.; Hou, J.; Linghu, E. DNA methyltransferase mediates the hypermethylation of the microRNA 34a promoter and enhances the resistance of patient-derived pancreatic cancer cells to molecular targeting agents. Pharmacol. Res. 2020, 160, 105071. [Google Scholar] [CrossRef]
- Lafarga, M.; Tapia, O.; Romero, A.M.; Berciano, M.T. Cajal bodies in neurons. RNA Biol. 2017, 14, 712–725. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Zhang, Z.M.; Lu, R.; Wang, P.; Yu, Y.; Chen, D.; Gao, L.; Liu, S.; Ji, D.; Rothbart, S.B.; Wang, Y.; et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature 2018, 15, 387–391. [Google Scholar] [CrossRef]
- Gao, L.; Emperle, M.; Guo, Y.; Grimm, S.A.; Ren, W.; Adam, S.; Uryu, H.; Zhang, Z.M.; Chen, D.; Yin, J.; et al. Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms. Nat. Commun. 2020, 11, 3355. [Google Scholar] [CrossRef]
- Fuso, A.; Raia, T.; Orticello, M.; Lucarelli, M. The complex interplay between DNA methylation and miRNAs in gene expression regulation. Biochimie 2020, 173, 12–16. [Google Scholar] [CrossRef]
- Baskerville, S.; Bartel, D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005, 11, 241–247. [Google Scholar] [CrossRef]
- França, G.S.; Vibranovski, M.D.; Galante, P.A. Host gene constraints and genomic context impact the expression and evolution of human microRNAs. Nat. Commun. 2016, 7, 11438. [Google Scholar] [CrossRef]
- Oakes, C.C.; La Salle, S.; Robaire, B.; Trasler, J.M. Evaluation of a quantitative DNA methylation analysis technique using methylation-sensitive/dependent restriction enzymes and real-time PCR. Epigenetics 2006, 1, 146–152. [Google Scholar] [CrossRef]
- Yang, F.; Sun, W.; Luo, W.J.; Yang, Y.; Yang, F.; Wang, X.L.; Chen, J. SDF1-CXCR4 Signaling Contributes to the Transition from Acute to Chronic Pain State. Mol. Neurobiol. 2017, 54, 2763–2775. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, S.R.; Chen, H.; Pan, H.L. RE1-silencing transcription factor controls the acute-to-chronic neuropathic pain transition and Chrm2 receptor gene expression in primary sensory neurons. J. Biol. Chem. 2018, 293, 19078–19091. [Google Scholar] [CrossRef]
- Wang, S.; Du, J.; Xi, D.; Shao, F.; Qiu, M.; Shao, X.; Liang, Y.; Liu, B.; Jin, X.; Fang, J.; et al. Role of GABAAR in the Transition from Acute to Chronic Pain and the Analgesic Effect of Electroacupuncture on Hyperalgesic Priming Model Rats. Front. Neurosci. 2021, 15, 691455. [Google Scholar] [CrossRef]
- Bali, K.K.; Hackenberg, M.; Lubin, A.; Kuner, R.; Devor, M. Sources of individual variability: miRNAs that predispose to neuropathic pain identified using genome-wide sequencing. Mol. Pain 2014, 10, 22. [Google Scholar] [CrossRef]
- Chang, H.L.; Wang, H.C.; Chunag, Y.T.; Chou, C.W.; Lin, I.L.; Lai, C.S.; Chang, L.L.; Cheng, K.I. miRNA Expression Change in Dorsal Root Ganglia After Peripheral Nerve Injury. J. Mol. Neurosci. 2017, 61, 169–177. [Google Scholar] [CrossRef]
- Peng, C.; Li, L.; Zhang, M.D.; Bengtsson Gonzales, C.; Parisien, M.; Belfer, I.; Usoskin, D.; Abdo, H.; Furlan, A.; Häring, M.; et al. miR-183 cluster scales mechanical pain sensitivity by regulating basal and neuropathic pain genes. Science 2017, 356, 1168–1171. [Google Scholar] [CrossRef]
- Jeltsch, A. Molecular enzymology of mammalian DNA methyltransferases. Curr. Top. Microbiol. Immunol. 2006, 301, 203–225. [Google Scholar] [CrossRef]
- Siedlecki, P.; Zielenkiewicz, P. Mammalian DNA methyltransferases. Acta Biochim. Pol. 2006, 53, 245–256. [Google Scholar] [CrossRef]
- Pollema-Mays, S.L.; Centeno, M.V.; Apkarian, A.V.; Martina, M. Expression of DNA methyltransferases in adult dorsal root ganglia is cell-type specific and up regulated in a rodent model of neuropathic pain. Front. Cell. Neurosci. 2014, 8, 217. [Google Scholar] [CrossRef]
- Shao, C.; Gao, Y.; Jin, D.; Xu, X.; Tan, S.; Yu, H.; Zhao, Q.; Zhao, L.; Wansheng, W.; Wang, D. DNMT3a methylation in neuropathic pain. J. Pain Res. 2017, 10, 2253–2256. [Google Scholar] [CrossRef]
- Bianchi, M.; Renzini, A.; Adamo, S.; Moresi, V. Coordinated Actions of MicroRNAs with other Epigenetic Factors Regulate Skeletal Muscle Development and Adaptation. Int. J. Mol. Sci. 2017, 18, 840. [Google Scholar] [CrossRef]
- Saito, Y.; Jones, P.A. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle 2006, 5, 2220–2222. [Google Scholar] [CrossRef]
- Xu, B.; Cao, J.; Zhang, J.; Jia, S.; Wu, S.; Mo, K.; Wei, G.; Liang, L.; Miao, X.; Bekker, A.; et al. Role of MicroRNA-143 in Nerve Injury-Induced Upregulation of Dnmt3a Expression in Primary Sensory Neurons. Front. Mol. Neurosci. 2017, 10, 350. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xu, D.; Wang, T.; Zhang, Y.; Yang, X.; Wang, X.; Tang, Y. Epigenetic reduction of miR-214–3p upregulates astrocytic colony-stimulating factor- 1 and contributes to neuropathic pain induced by nerve injury. Pain 2020, 161, 96–108. [Google Scholar] [CrossRef]
- Mata-Garrido, J.; Casafont, I.; Tapia, O.; Berciano, M.T.; Lafarga, M. Neuronal accumulation of unrepaired DNA in a novel specific chromatin domain: Structural, molecular and transcriptional characterization. Acta Neuropathol Commun. 2016, 4, 41. [Google Scholar] [CrossRef]
- Lafontaine, D.LJ.; Riback, J.A.; Bascetin, R.; Brangwynne, C.P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 2021, 22, 165–182. [Google Scholar] [CrossRef]
- Jin, J.; Lian, T.; Gu, C.; Yu, K.; Gao, Y.Q.; Su, X.D. The effects of cytosine methylation on general transcription factors. Sci. Rep. 2016, 7, 6. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, Z.P.; Zheng, H.Z.; Zhang, S.; Zhang, Z.L.; Chen, Y.; You, Y.S.; Yang, M.H. Abnormal DNA methylation in the lumbar spinal cord following chronic constriction injury in rats. Neurosci. Lett. 2016, 610, 1–5. [Google Scholar] [CrossRef]
- Sun, L.; Zhao, J.Y.; Gu, X.; Liang, L.; Wu, S.; Mo, K.; Feng, J.; Guo, W.; Zhang, J.; Bekker, A.; et al. Nerve injury-induced epigenetic silencing of opioid receptors controlled by DNMT3a in primary afferent neurons. Pain 2017, 158, 637–648. [Google Scholar] [CrossRef]
- Mao, Q.; Yuan, J.; Xiong, M.; Wu, S.; Chen, L.; Bekker, A.; Tao, Y.-X.; Yang, T. Role of dorsal root ganglion K2p1.1 in peripheral nerve injury-induced neuropathic pain. Mol. Pain 2017, 13, 1744806917701135. [Google Scholar] [CrossRef]
- Tochiki, K.K.; Cunningham, J.; Hunt, S.P.; Géranton, S.M. The expression of spinal methyl-CpG-binding protein 2, DNA methyltransferases and histone deacetylases is modulated in persistent pain states. Mol. Pain 2012, 8, 14. [Google Scholar] [CrossRef]
- Niederberger, E.; Resch, E.; Parnham, M.J.; Geisslinger, G. Drugging the pain epigenome. Nat. Rev. Neurol. 2017, 13, 434–447. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Bracken, C.P.; Goodall, G.J. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011, 39, 6845–6853. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.Y.; Liang, L.; Gu, X.; Li, Z.; Wu, S.; Sun, L.; Atianjoh, F.E.; Feng, J.; Mo, K.; Jia, S.; et al. DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons. Nat. Commun. 2017, 8, 14712. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.L.; Yu, L.N.; Wang, Y.; Tang, L.H.; Peng, Y.N.; Cao, J.L.; Yan, M. Increased methylation of the MOR gene proximal promoter in primary sensory neurons plays a crucial role in the decreased analgesic effect of opioids in neuropathic pain. Mol. Pain 2014, 10, 51. [Google Scholar] [CrossRef]
- Sun, L.; Gu, X.; Pan, Z.; Guo, X.; Liu, J.; Atianjoh, F.E.; Wu, S.; Mo, K.; Xu, B.; Liang, L.; et al. Contribution of DNMT1 to neuropathic pain genesis partially through epigenetically repressing Kcna2 in primary afferent neurons. J. Neurosci. 2019, 39, 6595–6607. [Google Scholar] [CrossRef]
- Jiang, B.C.; Zhang, W.W.; Yang, T.; Guo, C.Y. Demethylation of G-protein-coupled receptor 151 promoter facilitates the binding of Krüppel-like factor 5 and enhances neuropathic pain after nerve injury in mice. J. Neurosci. 2018, 38, 10535–10551. [Google Scholar] [CrossRef]
- Jiang, B.C.; He, L.N.; Wu, X.B.; Shi, H.; Zhang, W.W.; Zhang, Z.J.; Cao, D.L.; Li, C.H.; Gu, J.; Gao, Y.J. Promoted Interaction of C/EBPα with Demethylated Cxcr3 Gene Promoter Contributes to Neuropathic Pain in Mice. J. Neurosci. 2017, 37, 685–700. [Google Scholar] [CrossRef]
- Steiman-Shimony, A.; Shtrikman, O.; Margalit, H. Assessing the functional association of intronic miRNAs with their host genes. RNA 2018, 24, 991–1004. [Google Scholar] [CrossRef]
- Megraw, M.; Sethupathy, P.; Gumireddy, K.; Jensen, S.T.; Huang, Q.; Hatzigeorgiou, A.G. Isoform Specific Gene Auto-Regulation via miRNAs: A Case Study on miR-128b and ARPP-21. Theor. Chem. Acc. 2010, 125, 593–598. [Google Scholar] [CrossRef][Green Version]
- Dluzen, D.F.; Sun, D.; Salzberg, A.C.; Jones, N.; Bushey, R.T.; Robertson, G.P.; Lazarus, P. Regulation of UDP-glucuronosyltransferase 1A1 expression and activity by microRNA 491-3p. J. Pharmacol. Exp. Ther. 2014, 348, 465–477. [Google Scholar] [CrossRef]
- Massagué, J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012, 13, 616–630. [Google Scholar] [CrossRef]
- Lantero, A.; Tramullas, M.; Díaz, A.; Hurlé, M.A. Transforming growth factor-β in normal nociceptive processing and pathological pain models. Mol. Neurobiol. 2012, 45, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Echeverry, S.; Shi, X.Q.; Haw, A.; Liu, H.; Zhang, Z.W.; Zhang, J. Transforming growth factor-beta1 impairs neuropathic pain through pleiotropic effects. Mol. Pain 2009, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Tramullas, M.; Lantero, A.; Díaz, A.; Morchón, N.; Merino, D.; Villar, A.; Buscher, D.; Merino, R.; Hurlé, J.M.; Izpisúa-Belmonte, J.C.; et al. BAMBI (bone morphogenetic protein and activin membrane-bound inhibitor) reveals the involvement of the transforming growth factor-beta family in pain modulation. J. Neurosci. 2010, 30, 1502–1511. [Google Scholar] [CrossRef] [PubMed]
- Lantero, A.; Tramullas, M.; Pílar-Cuellar, F.; Valdizán, E.; Santillán, R.; Roques, B.P.; Hurlé, M.A. TGF-β and opioid receptor signaling crosstalk results in improvement of endogenous and exogenous opioid analgesia under pathological pain conditions. J. Neurosci. 2014, 34, 5385–5395. [Google Scholar] [CrossRef]
- Decosterd, I.; Woolf, C.K. Spared nerve injury: An animal model of persistent peripheral neuropathic pain. Pain 2000, 87, 149–158. [Google Scholar] [CrossRef]
- Bonin, R.P.; Bories, C.; De Koninck, Y. A simplified up-down method (SUDO) for measuring mechanical nociception in rodents using von Frey filaments. Mol. Pain 2014, 10, 26. [Google Scholar] [CrossRef]
- Pena, E.; Berciano, M.T.; Fernandez, R.; Ojeda, J.L.; Lafarga, M. Neuronal body size correlates with the number of nucleoli and Cajal bodies, and with the organization of the splicing machinery in rat trigeminal ganglion neurons. J. Comp. Neurol. 2001, 430, 250–263. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Francés, R.; Mata-Garrido, J.; de la Fuente, R.; Carcelén, M.; Lafarga, M.; Berciano, M.T.; García, R.; Hurlé, M.A.; Tramullas, M. Identification of Epigenetic Interactions between MicroRNA-30c-5p and DNA Methyltransferases in Neuropathic Pain. Int. J. Mol. Sci. 2022, 23, 13994. https://doi.org/10.3390/ijms232213994
Francés R, Mata-Garrido J, de la Fuente R, Carcelén M, Lafarga M, Berciano MT, García R, Hurlé MA, Tramullas M. Identification of Epigenetic Interactions between MicroRNA-30c-5p and DNA Methyltransferases in Neuropathic Pain. International Journal of Molecular Sciences. 2022; 23(22):13994. https://doi.org/10.3390/ijms232213994
Chicago/Turabian StyleFrancés, Raquel, Jorge Mata-Garrido, Roberto de la Fuente, María Carcelén, Miguel Lafarga, María Teresa Berciano, Raquel García, María A. Hurlé, and Mónica Tramullas. 2022. "Identification of Epigenetic Interactions between MicroRNA-30c-5p and DNA Methyltransferases in Neuropathic Pain" International Journal of Molecular Sciences 23, no. 22: 13994. https://doi.org/10.3390/ijms232213994
APA StyleFrancés, R., Mata-Garrido, J., de la Fuente, R., Carcelén, M., Lafarga, M., Berciano, M. T., García, R., Hurlé, M. A., & Tramullas, M. (2022). Identification of Epigenetic Interactions between MicroRNA-30c-5p and DNA Methyltransferases in Neuropathic Pain. International Journal of Molecular Sciences, 23(22), 13994. https://doi.org/10.3390/ijms232213994






