Integrative and Comprehensive Pan-Cancer Analysis of Lymphocyte-Specific Protein Tyrosine Kinase in Human Tumors
Abstract
1. Introduction
2. Results
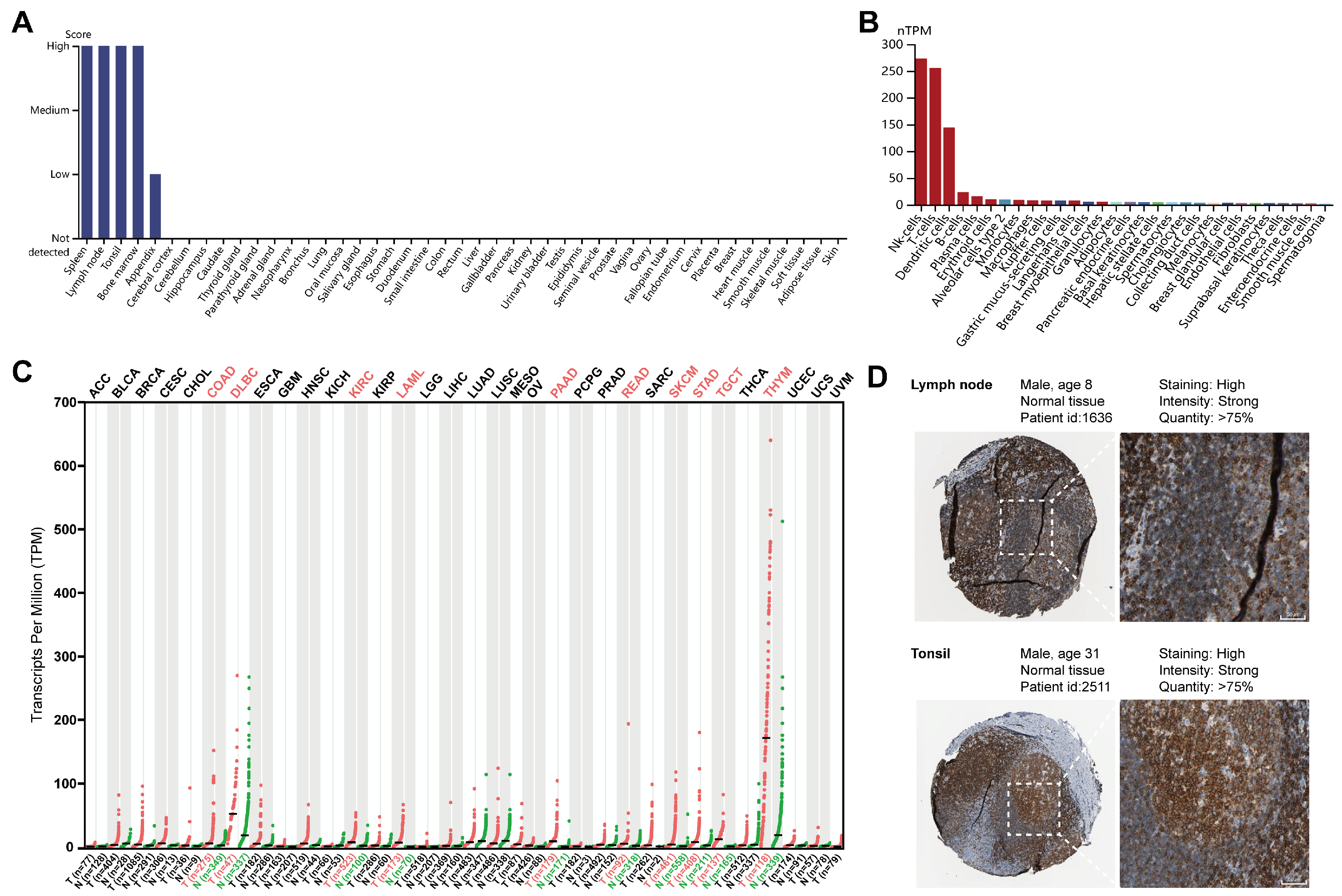
2.1. Expression Profiles of LCK in Different Human Tissues and Cells
2.2. LCK Expression and Cancer Patient’s Prognosis
2.3. LCK Promoter Methylation and Prognostic Value of CpG Islands in the Survival of Tumor Patients
2.4. Genetic Alterations and Mutations of LCK in Pan-Cancer Analysis
2.5. Genome-Wide Association of LCK in Cancers
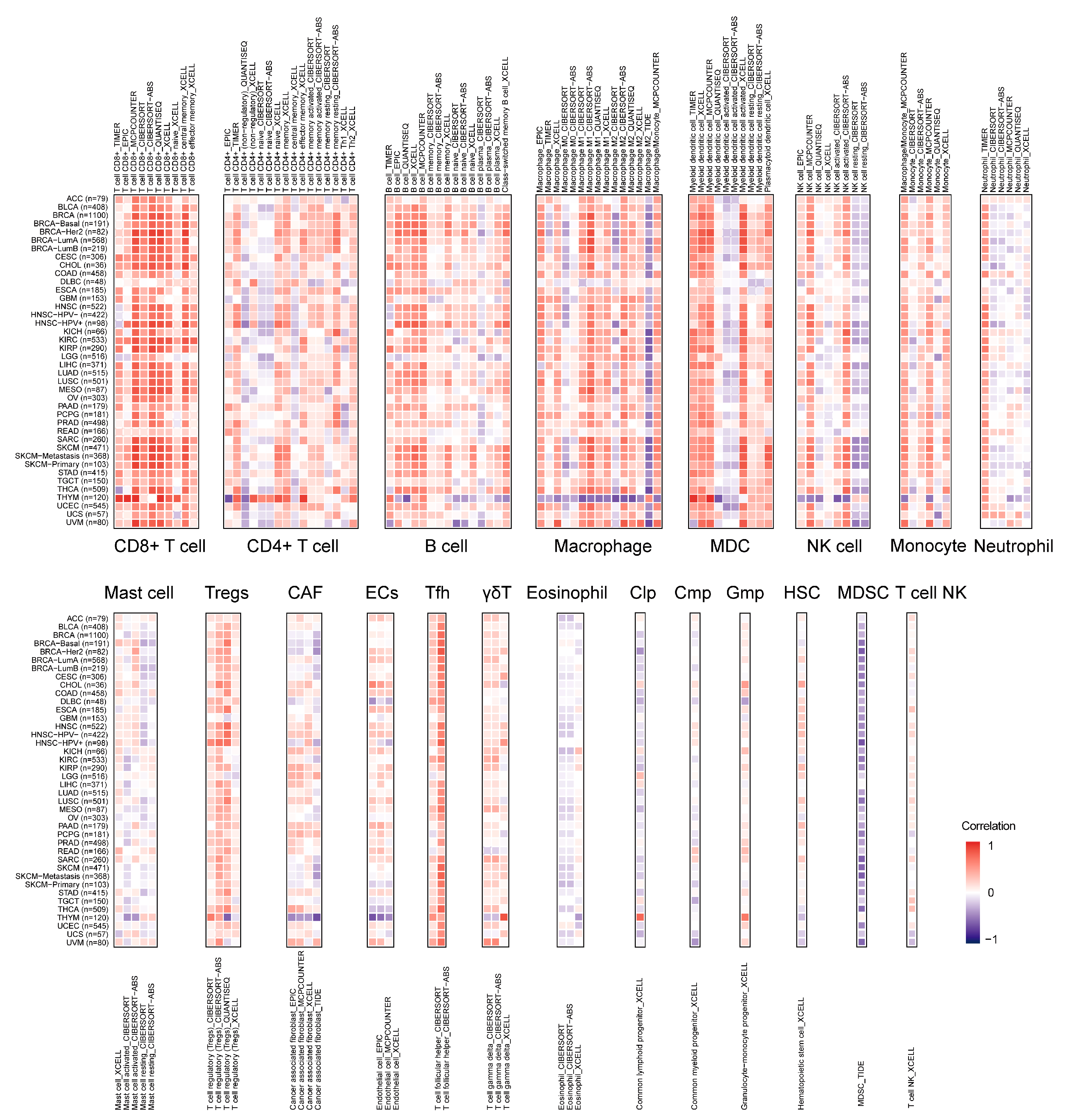
2.6. Landscape of LCK Correlating with Immune Infiltration
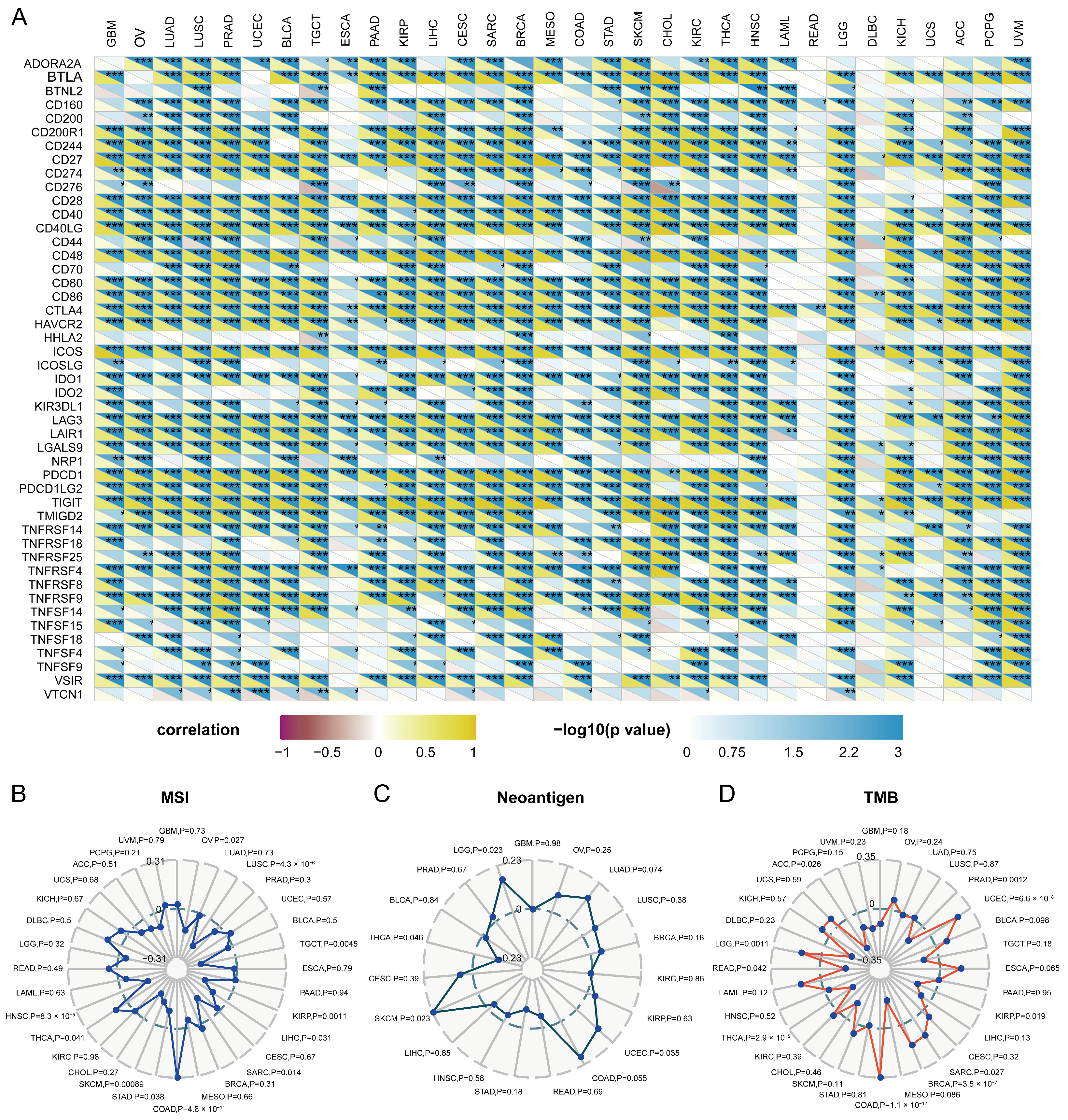
2.7. Relationship between Immune Checkpoints and LCK
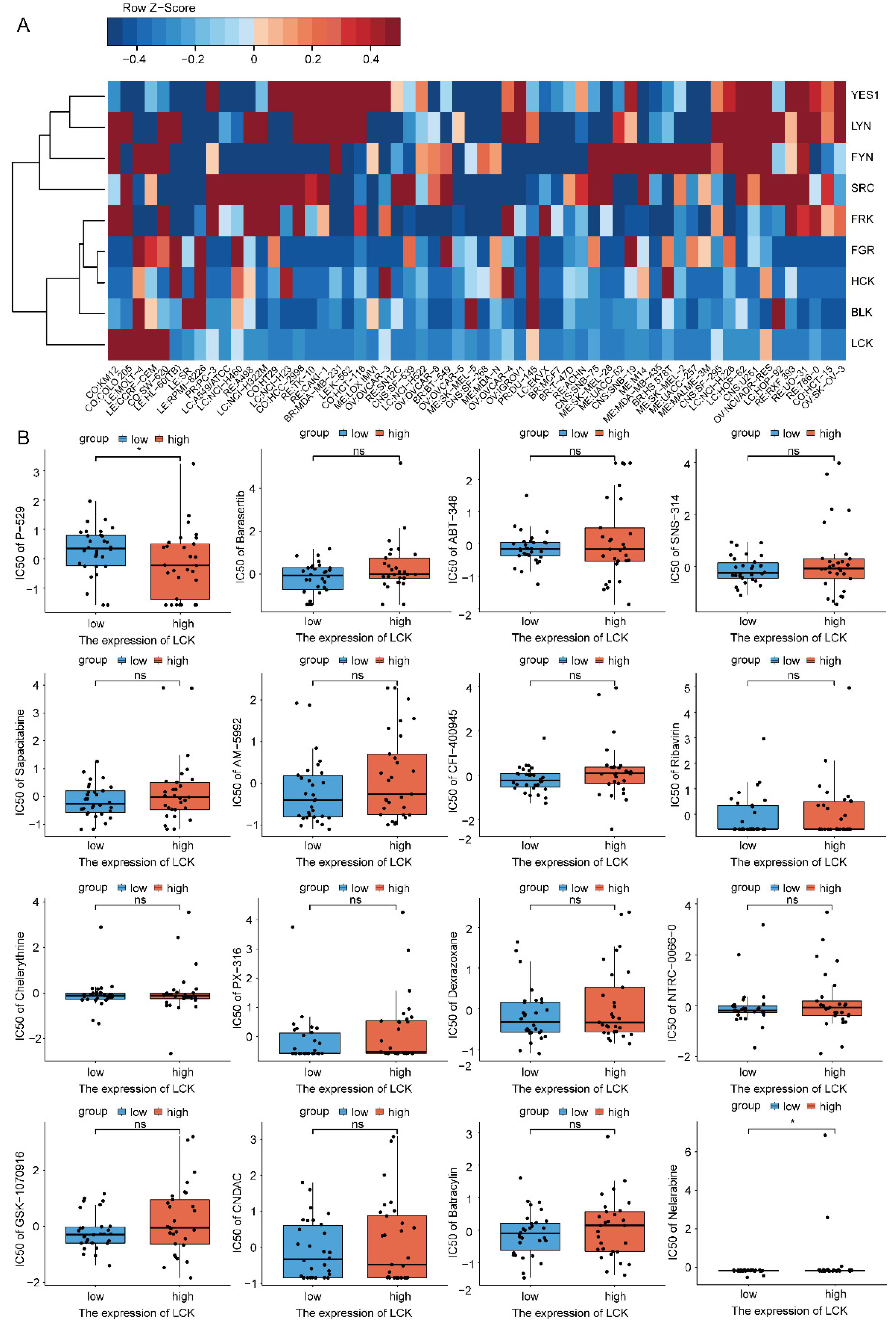
2.8. Drug Sensitivity Analysis of LCK
2.9. Protein–Protein Interaction Network and Pathway Enrichment of LCK
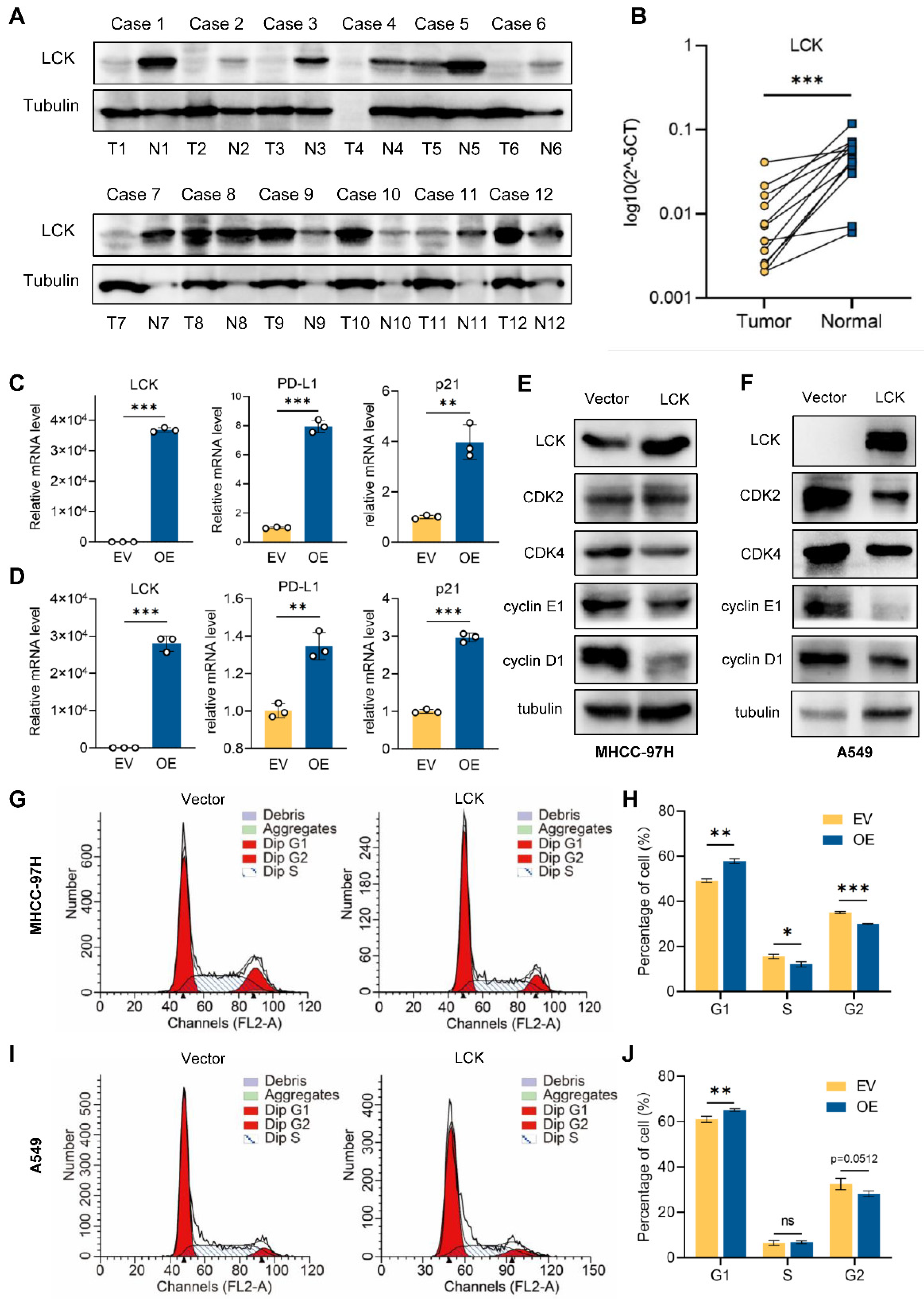
2.10. Tumor Suppressor Role of LCK in Liver and Lung Cancer
3. Discussion
4. Materials and Methods
4.1. Expression Profiles of LCK in Human Normal and Tumor Tissues
4.2. Prognostic Value of LCK in Patients with Tumors
4.3. LCK Promoter Methylation Level and CpG Sites on the Survival of Tumors
4.4. Genetic Alteration Analysis of LCK across Tumors
4.5. Genomic Correlation of LCK Expression
4.6. Immune Infiltration Analysis
4.7. Discovering Drug Sensitivity of LCK in Tumor Cells
4.8. Protein–Protein Interaction and Pathway Enrichment of LCK
4.9. Cell Culture
4.10. Transfection
4.11. Tissue Samples and Western Blotting
4.12. RT-qPCR
4.13. Cell Cycle Analysis
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Laethem, F.; Tikhonova, A.N.; Pobezinsky, L.A.; Tai, X.; Kimura, M.Y.; Le Saout, C.; Guinter, T.I.; Adams, A.; Sharrow, S.O.; Bernhardt, G.; et al. Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell 2013, 154, 1326–1341. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, G.; Li, L.; Li, D.; Dong, Z.; Jiang, P. Asparagine enhances LCK signalling to potentiate CD8(+) T-cell activation and anti-tumour responses. Nat. Cell Biol. 2021, 23, 75–86. [Google Scholar] [CrossRef]
- Serafin, V.; Capuzzo, G.; Milani, G.; Minuzzo, S.A.; Pinazza, M.; Bortolozzi, R.; Bresolin, S.; Porcu, E.; Frasson, C.; Indraccolo, S.; et al. Glucocorticoid resistance is reverted by LCK inhibition in pediatric T-cell acute lymphoblastic leukemia. Blood 2017, 130, 2750–2761. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Jing, L.; Jiang, X.; Ma, Y.; Wang, D.; Xiang, J.; Chen, X.; Wu, Z.; Yan, H.; Jia, J.; et al. CD146 bound to LCK promotes T cell receptor signaling and antitumor immune responses in mice. J. Clin. Investig. 2021, 131, e148568. [Google Scholar] [CrossRef]
- Liu, J.; Guo, Z.; Zhang, Y.; Wu, T.; Ma, Y.; Lai, W.; Guo, Z. LCK inhibitor attenuates atherosclerosis in ApoE(-/-) mice via regulating T cell differentiation and reverse cholesterol transport. J. Mol. Cell Cardiol. 2020, 139, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Liao, X.; Nie, L.; Lin, T.; Xu, H.; Yang, L.; Shen, B.; Qiu, S.; Ai, J.; Wei, Q. LCK and CD3E Orchestrate the Tumor Microenvironment and Promote Immunotherapy Response and Survival of Muscle-Invasive Bladder Cancer Patients. Front. Cell Dev. Biol. 2021, 9, 748280. [Google Scholar] [CrossRef]
- Mestermann, K.; Giavridis, T.; Weber, J.; Rydzek, J.; Frenz, S.; Nerreter, T.; Mades, A.; Sadelain, M.; Einsele, H.; Hudecek, M. The tyrosine kinase inhibitor dasatinib acts as a pharmacologic on/off switch for CAR T cells. Sci. Transl. Med. 2019, 11, eaau5907. [Google Scholar] [CrossRef]
- Gocho, Y.; Liu, J.; Hu, J.; Yang, W.; Dharia, N.V.; Zhang, J.; Shi, H.; Du, G.; John, A.; Lin, T.N.; et al. Network-based systems pharmacology reveals heterogeneity in LCK and BCL2 signaling and therapeutic sensitivity of T-cell acute lymphoblastic leukemia. Nat. Cancer 2021, 2, 284–299. [Google Scholar] [CrossRef]
- Bommhardt, U.; Schraven, B.; Simeoni, L. Beyond TCR Signaling: Emerging Functions of Lck in Cancer and Immunotherapy. Int. J. Mol. Sci. 2019, 20, 3500. [Google Scholar] [CrossRef]
- Fabarius, A.; Giehl, M.; Rebacz, B.; Kramer, A.; Frank, O.; Haferlach, C.; Duesberg, P.; Hehlmann, R.; Seifarth, W.; Hochhaus, A. Centrosome aberrations and G1 phase arrest after in vitro and in vivo treatment with the SRC/ABL inhibitor dasatinib. Haematologica 2008, 93, 1145–1154. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Y.; He, Q. Distinct characteristics of dasatinib-induced pyroptosis in gasdermin E-expressing human lung cancer A549 cells and neuroblastoma SH-SY5Y cells. Oncol. Lett. 2020, 20, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Kumar Singh, P.; Kashyap, A.; Silakari, O. Exploration of the therapeutic aspects of Lck: A kinase target in inflammatory mediated pathological conditions. Biomed. Pharmacother. 2018, 108, 1565–1571. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Huang, T.; Chen, X.; Lu, Y. A comprehensive analysis of the expression and regulation network of lymphocyte-specific protein tyrosine kinase in breast cancer. Transl. Cancer Res. 2021, 10, 1519–1536. [Google Scholar] [CrossRef] [PubMed]
- Weisse, J.; Rosemann, J.; Muller, L.; Kappler, M.; Eckert, A.W.; Glass, M.; Misiak, D.; Huttelmaier, S.; Ballhausen, W.G.; Hatzfeld, M.; et al. Identification of lymphocyte cell-specific protein-tyrosine kinase (LCK) as a driver for invasion and migration of oral cancer by tumor heterogeneity exploitation. Mol. Cancer 2021, 20, 88. [Google Scholar] [CrossRef]
- Huang, Y.; Li, S.; Liu, Q.; Wang, Z.; Li, S.; Liu, L.; Zhao, W.; Wang, K.; Zhang, R.; Wang, L.; et al. The LCK-14-3-3zeta-TRPM8 axis regulates TRPM8 function/assembly and promotes pancreatic cancer malignancy. Cell Death Dis. 2022, 13, 524. [Google Scholar] [CrossRef] [PubMed]
- Liew, K.; Yu, G.Q.S.; Wei Pua, L.J.; Wong, L.Z.; Tham, S.Y.; Hii, L.W.; Lim, W.M.; OuYong, B.M.; Looi, C.K.; Mai, C.W.; et al. Parallel genome-wide RNAi screens identify lymphocyte-specific protein tyrosine kinase (LCK) as a targetable vulnerability of cell proliferation and chemoresistance in nasopharyngeal carcinoma. Cancer Lett. 2021, 504, 81–90. [Google Scholar] [CrossRef]
- Nakahira, K.; Morita, A.; Kim, N.S.; Yanagihara, I. Phosphorylation of FOXP3 by LCK downregulates MMP9 expression and represses cell invasion. PLoS ONE 2013, 8, e77099. [Google Scholar] [CrossRef] [PubMed]
- Morgan, A.E.; Davies, T.J.; Mc Auley, M.T. The role of DNA methylation in ageing and cancer. Proc. Nutr. Soc. 2018, 77, 412–422. [Google Scholar] [CrossRef]
- Iengar, P. An analysis of substitution, deletion and insertion mutations in cancer genes. Nucleic Acids Res. 2012, 40, 6401–6413. [Google Scholar] [CrossRef]
- Sotomayor-Vivas, C.; Hernandez-Lemus, E.; Dorantes-Gilardi, R. Linking protein structural and functional change to mutation using amino acid networks. PLoS ONE 2022, 17, e0261829. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Chen, H.; Xie, P.; Ma, R.; He, J.; Zhang, H. Bioinformatics analysis for the role of CALR in human cancers. PLoS ONE 2021, 16, e0261254. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Severson, E.; Pignon, J.C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef]
- Elkamhawy, A.; Ali, E.M.H.; Lee, K. New horizons in drug discovery of lymphocyte-specific protein tyrosine kinase (Lck) inhibitors: A decade review (2011-2021) focussing on structure-activity relationship (SAR) and docking insights. J. Enzym. Inhib. Med. Chem. 2021, 36, 1574–1602. [Google Scholar] [CrossRef] [PubMed]
- Boman, C.; Zerdes, I.; Martensson, K.; Bergh, J.; Foukakis, T.; Valachis, A.; Matikas, A. Discordance of PD-L1 status between primary and metastatic breast cancer: A systematic review and meta-analysis. Cancer Treat. Rev. 2021, 99, 102257. [Google Scholar] [CrossRef] [PubMed]
- Zamoyska, R.; Basson, A.; Filby, A.; Legname, G.; Lovatt, M.; Seddon, B. The influence of the src-family kinases, Lck and Fyn, on T cell differentiation, survival and activation. Immunol. Rev. 2003, 191, 107–118. [Google Scholar] [CrossRef]
- Wan, R.; Wu, J.; Ouyang, M.; Lei, L.; Wei, J.; Peng, Q.; Harrison, R.; Wu, Y.; Cheng, B.; Li, K.; et al. Biophysical basis underlying dynamic Lck activation visualized by ZapLck FRET biosensor. Sci. Adv. 2019, 5, eaau2001. [Google Scholar] [CrossRef]
- Betapudi, V.; Shukla, M.; Alluri, R.; Merkulov, S.; McCrae, K.R. Novel role for p56/Lck in regulation of endothelial cell survival and angiogenesis. FASEB J. 2016, 30, 3515–3526. [Google Scholar] [CrossRef]
- Goldman, F.D.; Ballas, Z.K.; Schutte, B.C.; Kemp, J.; Hollenback, C.; Noraz, N.; Taylor, N. Defective expression of p56lck in an infant with severe combined immunodeficiency. J. Clin. Investig. 1998, 102, 421–429. [Google Scholar] [CrossRef]
- Gorska, M.M.; Alam, R. A mutation in the human Uncoordinated 119 gene impairs TCR signaling and is associated with CD4 lymphopenia. Blood 2012, 119, 1399–1406. [Google Scholar] [CrossRef]
- Salmond, R.J.; Filby, A.; Pirinen, N.; Magee, A.I.; Zamoyska, R. Mislocalization of Lck impairs thymocyte differentiation and can promote development of thymomas. Blood 2011, 117, 108–117. [Google Scholar] [CrossRef]
- Harr, M.W.; Caimi, P.F.; McColl, K.S.; Zhong, F.; Patel, S.N.; Barr, P.M.; Distelhorst, C.W. Inhibition of Lck enhances glucocorticoid sensitivity and apoptosis in lymphoid cell lines and in chronic lymphocytic leukemia. Cell Death Differ. 2010, 17, 1381–1391. [Google Scholar] [CrossRef] [PubMed]
- Till, K.J.; Allen, J.C.; Talab, F.; Lin, K.; Allsup, D.; Cawkwell, L.; Bentley, A.; Ringshausen, I.; Duckworth, A.D.; Pettitt, A.R.; et al. Lck is a relevant target in chronic lymphocytic leukaemia cells whose expression variance is unrelated to disease outcome. Sci. Rep. 2017, 7, 16784. [Google Scholar] [CrossRef] [PubMed]
- Dolina, J.S.; Van Braeckel-Budimir, N.; Thomas, G.D.; Salek-Ardakani, S. CD8(+) T Cell Exhaustion in Cancer. Front. Immunol. 2021, 12, 715234. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hu, Y.; Shao, M.; Teng, X.; Jiang, P.; Wang, X.; Wang, H.; Cui, J.; Yu, J.; Liang, Z.; et al. Dasatinib enhances anti-leukemia efficacy of chimeric antigen receptor T-cells by inhibiting cell differentiation and exhaustion. J. Hematol. Oncol. 2021, 14, 113. [Google Scholar] [CrossRef]
- Li, S.; Gao, J.; Xu, Q.; Zhang, X.; Huang, M.; Dai, X.; Huang, K.; Liu, L. A Signature-Based Classification of Gastric Cancer That Stratifies Tumor Immunity and Predicts Responses to PD-1 Inhibitors. Front. Immunol. 2021, 12, 693314. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, X.; Sun, N.; Zhao, Z.; He, J. Comprehensive Analysis of the Tumor Microenvironment in Cutaneous Melanoma associated with Immune Infiltration. J. Cancer 2020, 11, 3858–3870. [Google Scholar] [CrossRef]
- Raynor, J.L.; Chi, H. LCK senses asparagine for T cell activation. Nat. Cell Biol. 2021, 23, 7–8. [Google Scholar] [CrossRef]
- Liu, X.; Zhan, Y.; Xu, W.; Liu, X.; Geng, Y.; Liu, L.; Da, J.; Wang, J.; Zhang, X.; Jin, H.; et al. Prognostic and immunological role of Fam20C in pan-cancer. Biosci. Rep. 2021, 41, BSR20201920. [Google Scholar] [CrossRef]
- Chakraborty, G.; Rangaswami, H.; Jain, S.; Kundu, G.C. Hypoxia regulates cross-talk between Syk and Lck leading to breast cancer progression and angiogenesis. J. Biol. Chem. 2006, 281, 11322–11331. [Google Scholar] [CrossRef]
- Wu, T.; Wang, X.; Li, J.; Song, X.; Wang, Y.; Wang, Y.; Zhang, L.; Li, Z.; Tian, J. Identification of Personalized Chemoresistance Genes in Subtypes of Basal-Like Breast Cancer Based on Functional Differences Using Pathway Analysis. PLoS ONE 2015, 10, e0131183. [Google Scholar] [CrossRef]
- Koster, A.; Landgraf, S.; Leipold, A.; Sachse, R.; Gebhart, E.; Tulusan, A.H.; Ronay, G.; Schmidt, C.; Dingermann, T. Expression of oncogenes in human breast cancer specimens. Anticancer Res. 1991, 11, 193–201. [Google Scholar]
- Clarke, C.N.; Lee, M.S.; Wei, W.; Manyam, G.; Jiang, Z.Q.; Lu, Y.; Morris, J.; Broom, B.; Menter, D.; Vilar-Sanchez, E.; et al. Proteomic features of colorectal cancer identify tumor subtypes independent of oncogenic mutations and independently predict relapse-free survival. Ann. Surg. Oncol. 2017, 24, 4051–4058. [Google Scholar] [CrossRef] [PubMed]
- Janikowska, G.; Janikowski, T.; Pyka-Pajak, A.; Mazurek, U.; Janikowski, M.; Gonciarz, M.; Lorenc, Z. Potential biomarkers for the early diagnosis of colorectal adenocarcinoma—Transcriptomic analysis of four clinical stages. Cancer Biomark. 2018, 22, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Veillette, A.; Foss, F.M.; Sausville, E.A.; Bolen, J.B.; Rosen, N. Expression of the lck tyrosine kinase gene in human colon carcinoma and other non-lymphoid human tumor cell lines. Oncogene Res. 1987, 1, 357–374. [Google Scholar] [PubMed]
- Mahabeleshwar, G.H.; Kundu, G.C. Tyrosine kinase p56lck regulates cell motility and nuclear factor kappaB-mediated secretion of urokinase type plasminogen activator through tyrosine phosphorylation of IkappaBalpha following hypoxia/reoxygenation. J. Biol. Chem. 2003, 278, 52598–52612. [Google Scholar] [CrossRef]
- Lin, J.; Yu, M.; Xu, X.; Wang, Y.; Xing, H.; An, J.; Yang, J.; Tang, C.; Sun, D.; Zhu, Y. Identification of biomarkers related to CD8(+) T cell infiltration with gene co-expression network in clear cell renal cell carcinoma. Aging 2020, 12, 3694–3712. [Google Scholar] [CrossRef]
- Sha, D.; Jin, Z.; Budczies, J.; Kluck, K.; Stenzinger, A.; Sinicrope, F.A. Tumor Mutational Burden as a Predictive Biomarker in Solid Tumors. Cancer Discov. 2020, 10, 1808–1825. [Google Scholar] [CrossRef]
- Hu, J.; Qiu, D.; Yu, A.; Hu, J.; Deng, H.; Li, H.; Yi, Z.; Chen, J.; Zu, X. YTHDF1 Is a Potential Pan-Cancer Biomarker for Prognosis and Immunotherapy. Front. Oncol. 2021, 11, 607224. [Google Scholar] [CrossRef]
- Arcaroli, J.J.; Touban, B.M.; Tan, A.C.; Varella-Garcia, M.; Powell, R.W.; Eckhardt, S.G.; Elvin, P.; Gao, D.; Messersmith, W.A. Gene array and fluorescence in situ hybridization biomarkers of activity of saracatinib (AZD0530), a Src inhibitor, in a preclinical model of colorectal cancer. Clin. Cancer Res. 2010, 16, 4165–4177. [Google Scholar] [CrossRef]
- Chan, C.M.; Jing, X.; Pike, L.A.; Zhou, Q.; Lim, D.J.; Sams, S.B.; Lund, G.S.; Sharma, V.; Haugen, B.R.; Schweppe, R.E. Targeted inhibition of Src kinase with dasatinib blocks thyroid cancer growth and metastasis. Clin. Cancer Res. 2012, 18, 3580–3591. [Google Scholar] [CrossRef]
- Heilmann, T.; Rumpf, A.L.; Roscher, M.; Tietgen, M.; Will, O.; Gerle, M.; Damm, T.; Borzikowsky, C.; Maass, N.; Gluer, C.C.; et al. Dasatinib prevents skeletal metastasis of osteotropic MDA-MB-231 cells in a xenograft mouse model. Arch. Gynecol. Obstet. 2020, 301, 1493–1502. [Google Scholar] [CrossRef]
- Levitt, J.M.; Yamashita, H.; Jian, W.; Lerner, S.P.; Sonpavde, G. Dasatinib is preclinically active against Src-overexpressing human transitional cell carcinoma of the urothelium with activated Src signaling. Mol. Cancer Ther. 2010, 9, 1128–1135. [Google Scholar] [CrossRef] [PubMed]
- Morton, J.P.; Karim, S.A.; Graham, K.; Timpson, P.; Jamieson, N.; Athineos, D.; Doyle, B.; McKay, C.; Heung, M.Y.; Oien, K.A.; et al. Dasatinib inhibits the development of metastases in a mouse model of pancreatic ductal adenocarcinoma. Gastroenterology 2010, 139, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Huang, W.C.; Zhang, L.; Zhang, C.; Lowery, F.J.; Ding, Z.; Guo, H.; Wang, H.; Huang, S.; Sahin, A.A.; et al. SRC family kinases as novel therapeutic targets to treat breast cancer brain metastases. Cancer Res. 2013, 73, 5764–5774. [Google Scholar] [CrossRef]
- Araujo, J.C.; Trudel, G.C.; Saad, F.; Armstrong, A.J.; Yu, E.Y.; Bellmunt, J.; Wilding, G.; McCaffrey, J.; Serrano, S.V.; Matveev, V.B.; et al. Docetaxel and dasatinib or placebo in men with metastatic castration-resistant prostate cancer (READY): A randomised, double-blind phase 3 trial. Lancet Oncol. 2013, 14, 1307–1316. [Google Scholar] [CrossRef]
- Chee, C.E.; Krishnamurthi, S.; Nock, C.J.; Meropol, N.J.; Gibbons, J.; Fu, P.; Bokar, J.; Teston, L.; O’Brien, T.; Gudena, V.; et al. Phase II study of dasatinib (BMS-354825) in patients with metastatic adenocarcinoma of the pancreas. Oncologist 2013, 18, 1091–1092. [Google Scholar] [CrossRef] [PubMed]
- Danson, S.; Mulvey, M.R.; Turner, L.; Horsman, J.; Escott, K.; Coleman, R.E.; Ahmedzai, S.H.; Bennett, M.I.; Andrew, D. An exploratory randomized-controlled trial of the efficacy of the Src-kinase inhibitor saracatinib as a novel analgesic for cancer-induced bone pain. J. Bone Oncol. 2019, 19, 100261. [Google Scholar] [CrossRef]
- Evans, T.R.J.; Van Cutsem, E.; Moore, M.J.; Bazin, I.S.; Rosemurgy, A.; Bodoky, G.; Deplanque, G.; Harrison, M.; Melichar, B.; Pezet, D.; et al. Phase 2 placebo-controlled, double-blind trial of dasatinib added to gemcitabine for patients with locally-advanced pancreatic cancer. Ann. Oncol. 2017, 28, 354–361. [Google Scholar] [CrossRef]
- Gangadhar, T.C.; Clark, J.I.; Karrison, T.; Gajewski, T.F. Phase II study of the Src kinase inhibitor saracatinib (AZD0530) in metastatic melanoma. Investig. New Drugs 2013, 31, 769–773. [Google Scholar] [CrossRef]
- Gucalp, A.; Sparano, J.A.; Caravelli, J.; Santamauro, J.; Patil, S.; Abbruzzi, A.; Pellegrino, C.; Bromberg, J.; Dang, C.; Theodoulou, M.; et al. Phase II trial of saracatinib (AZD0530), an oral SRC-inhibitor for the treatment of patients with hormone receptor-negative metastatic breast cancer. Clin. Breast Cancer 2011, 11, 306–311. [Google Scholar] [CrossRef]
- Herold, C.I.; Chadaram, V.; Peterson, B.L.; Marcom, P.K.; Hopkins, J.; Kimmick, G.G.; Favaro, J.; Hamilton, E.; Welch, R.A.; Bacus, S.; et al. Phase II trial of dasatinib in patients with metastatic breast cancer using real-time pharmacodynamic tissue biomarkers of Src inhibition to escalate dosing. Clin. Cancer Res. 2011, 17, 6061–6070. [Google Scholar] [CrossRef] [PubMed]
- Kalinsky, K.; Lee, S.; Rubin, K.M.; Lawrence, D.P.; Iafrarte, A.J.; Borger, D.R.; Margolin, K.A.; Leitao, M.M., Jr.; Tarhini, A.A.; Koon, H.B.; et al. A phase 2 trial of dasatinib in patients with locally advanced or stage IV mucosal, acral, or vulvovaginal melanoma: A trial of the ECOG-ACRIN Cancer Research Group (E2607). Cancer 2017, 123, 2688–2697. [Google Scholar] [CrossRef] [PubMed]
- Mackay, H.J.; Au, H.J.; McWhirter, E.; Alcindor, T.; Jarvi, A.; MacAlpine, K.; Wang, L.; Wright, J.J.; Oza, A.M. A phase II trial of the Src kinase inhibitor saracatinib (AZD0530) in patients with metastatic or locally advanced gastric or gastro esophageal junction (GEJ) adenocarcinoma: A trial of the PMH phase II consortium. Investig. New Drugs 2012, 30, 1158–1163. [Google Scholar] [CrossRef]
- Posadas, E.M.; Ahmed, R.S.; Karrison, T.; Szmulewitz, R.Z.; O’Donnell, P.H.; Wade, J.L., 3rd; Shen, J.; Gururajan, M.; Sievert, M.; Stadler, W.M. Saracatinib as a metastasis inhibitor in metastatic castration-resistant prostate cancer: A University of Chicago Phase 2 Consortium and DOD/PCF Prostate Cancer Clinical Trials Consortium Study. Prostate 2016, 76, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Powles, T.; Brown, J.; Larkin, J.; Jones, R.; Ralph, C.; Hawkins, R.; Chowdhury, S.; Boleti, E.; Bhal, A.; Fife, K.; et al. A randomized, double-blind phase II study evaluating cediranib versus cediranib and saracatinib in patients with relapsed metastatic clear-cell renal cancer (COSAK). Ann. Oncol. 2016, 27, 880–886. [Google Scholar] [CrossRef]
- Pusztai, L.; Moulder, S.; Altan, M.; Kwiatkowski, D.; Valero, V.; Ueno, N.T.; Esteva, F.J.; Avritscher, R.; Qi, Y.; Strauss, L.; et al. Gene signature-guided dasatinib therapy in metastatic breast cancer. Clin. Cancer Res. 2014, 20, 5265–5271. [Google Scholar] [CrossRef]
- Schott, A.F.; Barlow, W.E.; Van Poznak, C.H.; Hayes, D.F.; Moinpour, C.M.; Lew, D.L.; Dy, P.A.; Keller, E.T.; Keller, J.M.; Hortobagyi, G.N. Phase II studies of two different schedules of dasatinib in bone metastasis predominant metastatic breast cancer: SWOG S0622. Breast Cancer Res. Treat. 2016, 159, 87–95. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Mizuno, H.; Kitada, K.; Nakai, K.; Sarai, A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009, 2, 18. [Google Scholar] [CrossRef]
- Lanczky, A.; Gyorffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Modhukur, V.; Iljasenko, T.; Metsalu, T.; Lokk, K.; Laisk-Podar, T.; Vilo, J. MethSurv: A web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018, 10, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Yi, J. Comprehensive analysis of LASS6 expression and prognostic value in ovarian cancer. J. Ovarian Res. 2021, 14, 117. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of drug sensitivity in cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef]
- Reinhold, W.C.; Sunshine, M.; Liu, H.; Varma, S.; Kohn, K.W.; Morris, J.; Doroshow, J.; Pommier, Y. CellMiner: A web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012, 72, 3499–3511. [Google Scholar] [CrossRef]
- Devi, M.S.; Meiguilungpou, R.; Sharma, A.L.; Anjali, C.; Devi, K.M.; Singh, L.S.; Singh, T.R. Spindlin docking protein (SPIN.DOC) interaction with SPIN1 (a histone code reader) regulates Wnt signaling. Biochem. Biophys. Res. Commun. 2019, 511, 498–503. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, M.; Li, Y.; Guo, Y.; Zhu, W.; Jiang, J. Integrative and Comprehensive Pan-Cancer Analysis of Lymphocyte-Specific Protein Tyrosine Kinase in Human Tumors. Int. J. Mol. Sci. 2022, 23, 13998. https://doi.org/10.3390/ijms232213998
Han M, Li Y, Guo Y, Zhu W, Jiang J. Integrative and Comprehensive Pan-Cancer Analysis of Lymphocyte-Specific Protein Tyrosine Kinase in Human Tumors. International Journal of Molecular Sciences. 2022; 23(22):13998. https://doi.org/10.3390/ijms232213998
Chicago/Turabian StyleHan, Mingwei, Yiming Li, Yixiao Guo, Wanwan Zhu, and Jianli Jiang. 2022. "Integrative and Comprehensive Pan-Cancer Analysis of Lymphocyte-Specific Protein Tyrosine Kinase in Human Tumors" International Journal of Molecular Sciences 23, no. 22: 13998. https://doi.org/10.3390/ijms232213998
APA StyleHan, M., Li, Y., Guo, Y., Zhu, W., & Jiang, J. (2022). Integrative and Comprehensive Pan-Cancer Analysis of Lymphocyte-Specific Protein Tyrosine Kinase in Human Tumors. International Journal of Molecular Sciences, 23(22), 13998. https://doi.org/10.3390/ijms232213998





