Granulosa Cell Specific Loss of Adar in Mice Delays Ovulation, Oocyte Maturation and Leads to Infertility
Abstract
1. Introduction
2. Results
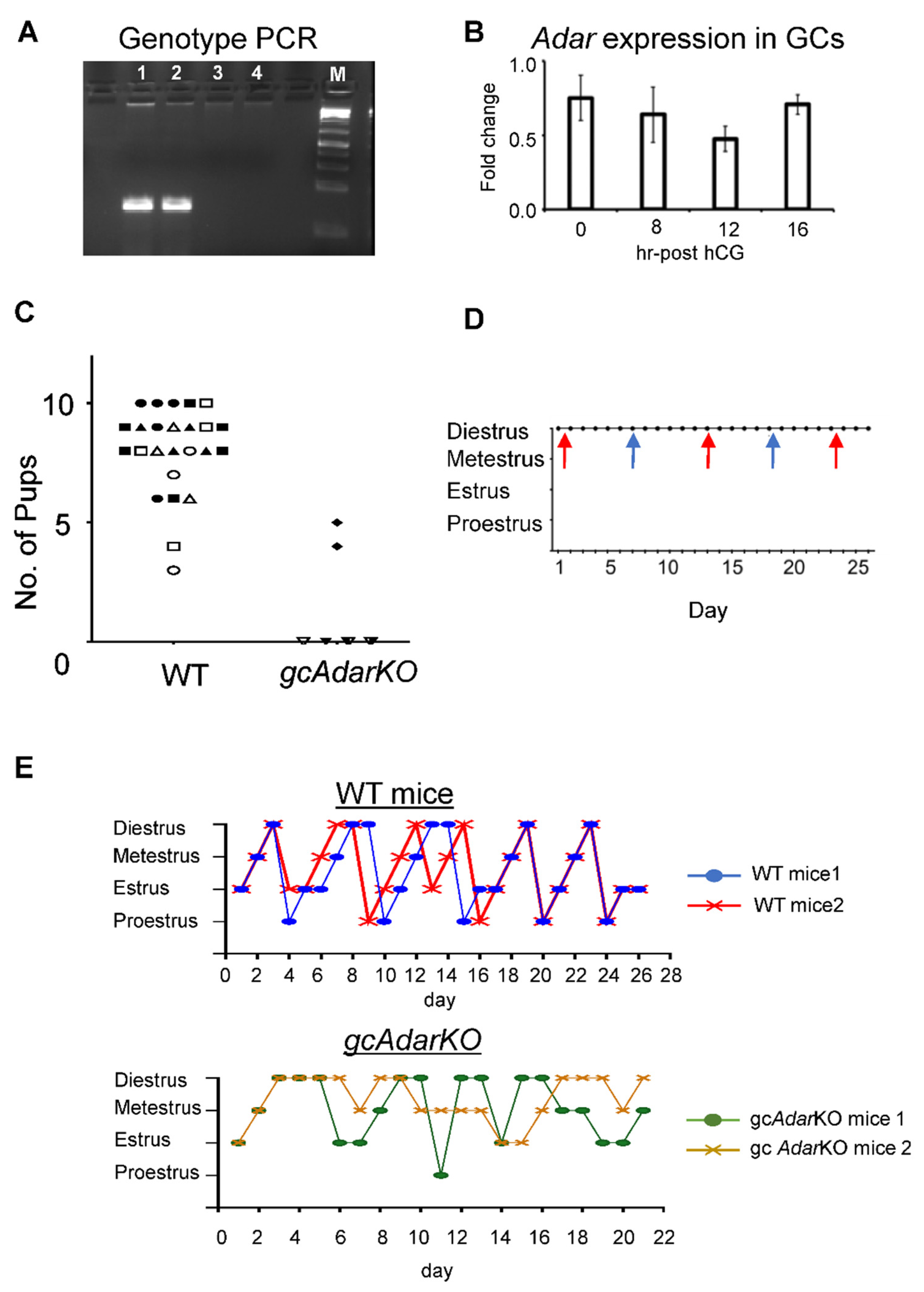
2.1. Conditional Deletion of Adar in mGC Renders Female Mice Infertile
2.2. Estrous Cyclicity Disturbances in gcAdarKO Mice
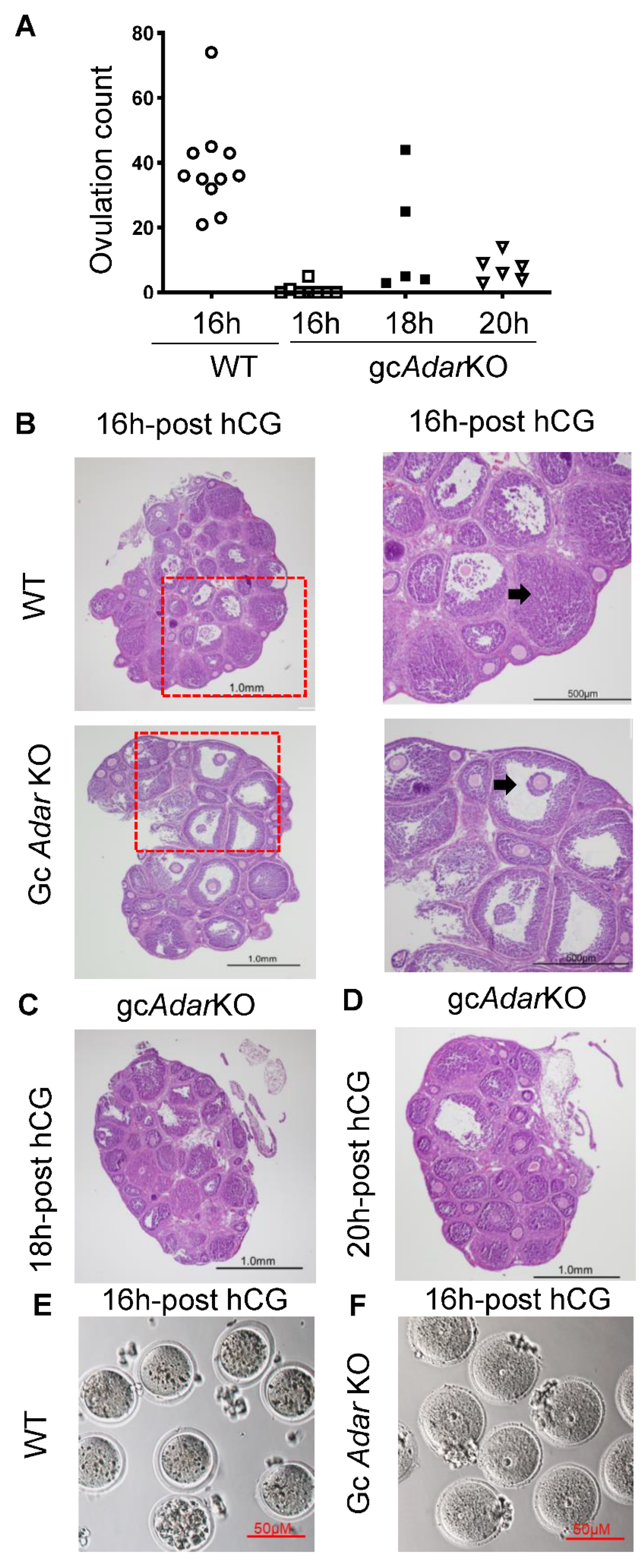
2.3. Histological Analysis of Ovaries
2.4. Delayed Ovulation in gcAdarKO Mice
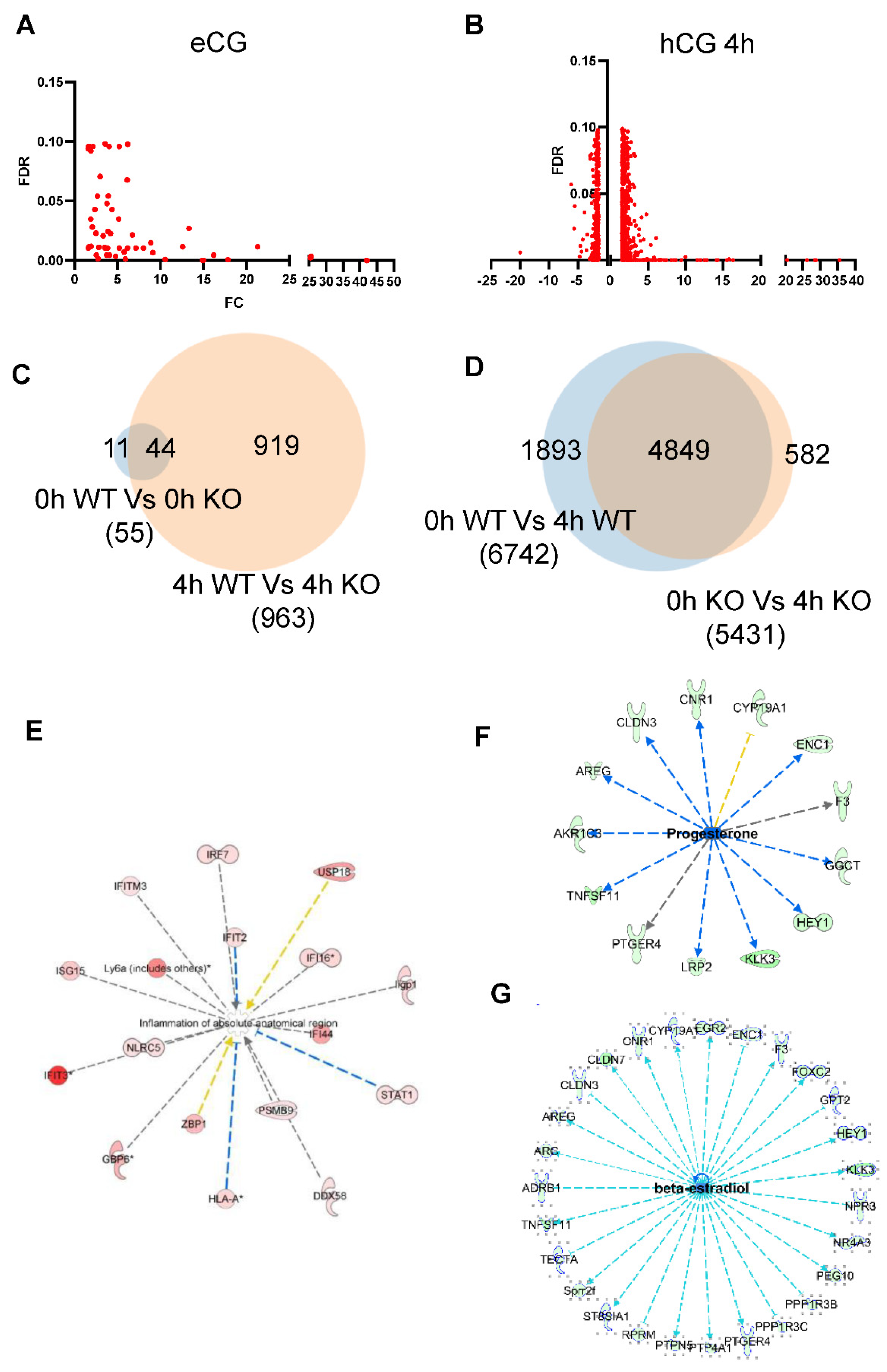
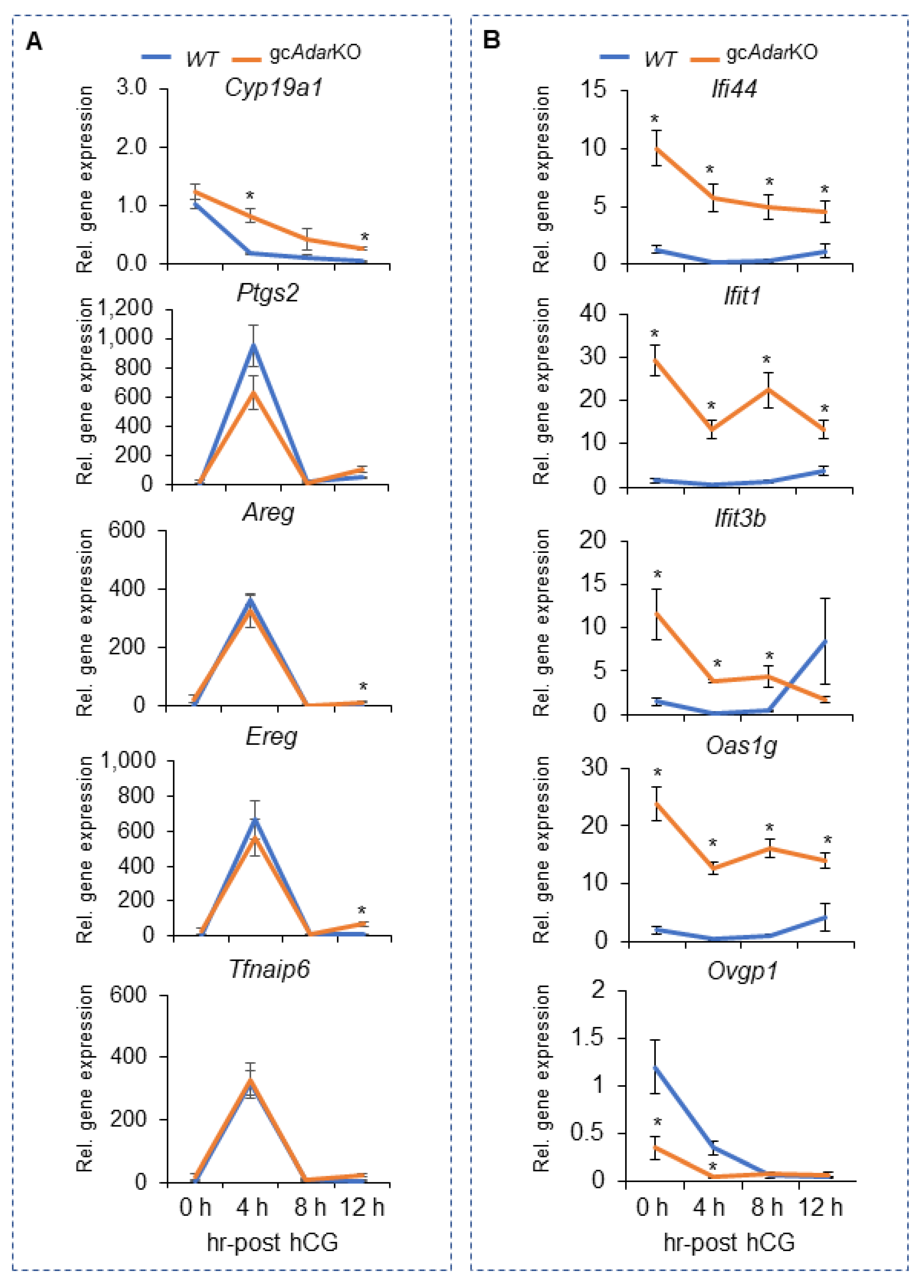
2.5. Transcriptome Analysis of mGCs from gcAdarKO Mice
2.6. Adar Regulated Pathways in mGCs
3. Discussion
4. Materials and Methods
4.1. Animals and Generation of Conditional Knockout
4.2. Assessment of Fertility
4.3. Induction of Superovulation by Gonadotropins
4.4. Histology
4.5. Isolation of Mural Granulosa Cell RNA
4.6. Quantitative RT-PCR Analysis and RNA Sequencing
4.7. Pathway Analyses
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Russell, D.L.; Robker, R.L. Molecular mechanisms of ovulation: Co-ordination through the cumulus complex. Hum. Reprod. Update 2007, 13, 289–312. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.T.; Brewer, M.S.; Chen, S.; Hong, W.; Zhu, Y. Transcriptomic signatures for ovulation in vertebrates. Gen. Comp. Endocrinol. 2017, 247, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, M.; Yang, M. Post-Translational Modifications in Oocyte Maturation and Embryo Development. Front. Cell Dev. Biol. 2021, 9, 645318. [Google Scholar] [CrossRef] [PubMed]
- Harcourt, E.M.; Kietrys, A.M.; Kool, E.T. Chemical and structural effects of base modifications in messenger RNA. Nature 2017, 541, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002, 71, 817–846. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.X.; Cho, D.S.; Wang, Q.; Lai, F.; Carter, K.C.; Nishikura, K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 2000, 6, 755–767. [Google Scholar] [CrossRef]
- Pestal, K.; Funk, C.C.; Snyder, J.M.; Price, N.D.; Treuting, P.M.; Stetson, D.B. Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development. Immunity 2015, 43, 933–944. [Google Scholar] [CrossRef]
- Tomaselli, S.; Bonamassa, B.; Alisi, A.; Nobili, V.; Locatelli, F.; Gallo, A. ADAR enzyme and miRNA story: A nucleotide that can make the difference. Int. J. Mol. Sci. 2013, 14, 22796–22816. [Google Scholar] [CrossRef]
- Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016, 17, 83–96. [Google Scholar] [CrossRef]
- Sakurai, M.; Shiromoto, Y.; Ota, H.; Song, C.; Kossenkov, A.V.; Wickramasinghe, J.; Showe, L.C.; Skordalakes, E.; Tang, H.-Y.; Speicher, D.W. ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol. 2017, 24, 534–543. [Google Scholar] [CrossRef]
- Eisenberg, E.; Levanon, E.Y. A-to-I RNA editing—Immune protector and transcriptome diversifier. Nat. Rev. Genet. 2018, 19, 473–490. [Google Scholar] [CrossRef] [PubMed]
- Ramaswami, G.; Zhang, R.; Piskol, R.; Keegan, L.P.; Deng, P.; O’connell, M.A.; Li, J.B. Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods 2013, 10, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Brachova, P.; Alvarez, N.S.; Hong, X.; Gunewardena, S.; Vincent, K.A.; Latham, K.E.; Christenson, L.K. Inosine RNA modifications are enriched at the codon wobble position in mouse oocytes and eggs. Biol. Reprod. 2019, 101, 938–949. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; Manzari, C.; Mastropasqua, F.; Aiello, I.; D’Erchia, A.M.; Pesole, G. Profiling RNA editing in human tissues: Towards the inosinome Atlas. Sci. Rep. 2015, 5, 14941. [Google Scholar] [CrossRef]
- O’Connell, M. RNA modification and the epitranscriptome; the next frontier. RNA 2015, 21, 703–704. [Google Scholar] [CrossRef][Green Version]
- Hartner, J.C.; Schmittwolf, C.; Kispert, A.; Müller, A.M.; Higuchi, M.; Seeburg, P.H. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004, 279, 4894–4902. [Google Scholar] [CrossRef]
- Ben-Shoshan, S.O.; Kagan, P.; Sultan, M.; Barabash, Z.; Dor, C.; Jacob-Hirsch, J.; Harmelin, A.; Pappo, O.; Marcu-Malina, V.; Ben-Ari, Z.; et al. ADAR1 deletion induces NFκB and interferon signaling dependent liver inflammation and fibrosis. RNA Biol. 2017, 14, 587–602. [Google Scholar] [CrossRef]
- El Azzouzi, H.; Vilaça, A.P.; Feyen, D.A.M.; Gommans, W.M.; de Weger, R.A.; Doevendans, P.A.F.; Sluijter, J.P.G. Cardiomyocyte Specific Deletion of ADAR1 Causes Severe Cardiac Dysfunction and Increased Lethality. Front. Cardiovasc. Med. 2020, 7, 30. [Google Scholar] [CrossRef]
- Wang, Q.; Miyakoda, M.; Yang, W.; Khillan, J.; Stachura, D.L.; Weiss, M.J.; Nishikura, K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 2004, 279, 4952–4961. [Google Scholar] [CrossRef]
- Richards, J.S.; Russell, D.L.; Ochsner, S.; Espey, L.L. Ovulation: New dimensions and new regulators of the inflammatory-like response. Annu. Rev. Physiol. 2002, 64, 69–92. [Google Scholar] [CrossRef]
- Duffy, D.M.; Ko, C.; Jo, M.; Brannstrom, M.; Curry, T.E. Ovulation: Parallels With Inflammatory Processes. Endocr. Rev. 2019, 40, 369–416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, L.; Yue, J.; Wei, X.; Wang, L.; Liu, X.; Gao, H.; Hou, X.; Zhao, F.; Yan, H.; et al. Genome-wide identification of RNA editing in seven porcine tissues by matched DNA and RNA high-throughput sequencing. J. Anim. Sci. Biotechnol. 2019, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; D’Erchia, A.M.; Lo Giudice, C.; Pesole, G. REDIportal: A comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 2017, 45, D750–D757. [Google Scholar] [CrossRef] [PubMed]
- Buchumenski, I.; Holler, K.; Appelbaum, L.; Eisenberg, E.; Junker, J.P.; Levanon, E.Y. Systematic identification of A-to-I RNA editing in zebrafish development and adult organs. Nucleic Acids Res. 2021, 49, 4325–4337. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Y.; Guo, J.; Cao, W.; Zhang, M.; He, J.; Li, Z. Integrated sequencing of exome and mRNA of large-sized single cells. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Li, W.; Xiong, H.; Liu, D.; Bai, Y.; Wu, K.; Zhang, X.; Yang, H.; Ma, K.; Hou, Y. Single-cell RNA sequencing reveals dynamic changes in A-to-I RNA editome during early human embryogenesis. BMC Genom. 2016, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Brachova, P.; Alvarez, N.S.; Christenson, L.K. Loss of Cnot6l Impairs Inosine RNA Modifications in Mouse Oocytes. Int. J. Mol. Sci. 2021, 22, 1191. [Google Scholar] [CrossRef]
- Edwards, R.G.; Gates, A.H. Timing of the stages of the maturation divisions, ovulation, fertilization and the first cleavage of eggs of adult mice treated with gonadotrophins. J. Endocrinol. 1959, 18, 292–304. [Google Scholar] [CrossRef]
- Sterneck, E.; Tessarollo, L.; Johnson, P.F. An essential role for C/EBPbeta in female reproduction. Genes Dev. 1997, 11, 2153–2162. [Google Scholar] [CrossRef]
- Lim, H.; Paria, B.C.; Das, S.K.; Dinchuk, J.E.; Langenbach, R.; Trzaskos, J.M.; Dey, S.K. Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell 1997, 91, 197–208. [Google Scholar] [CrossRef]
- Espey, L.L. Ovulation as an inflammatory reaction—A hypothesis. Biol. Reprod. 1980, 22, 73–106. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Pando, J.M.; Alcocer-Gómez, E.; Castejón-Vega, B.; Navarro-Villarán, E.; Condés-Hervás, M.; Mundi-Roldan, M.; Muntané, J.; Pérez-Pulido, A.J.; Bullon, P.; Wang, C.; et al. Inhibition of the NLRP3 inflammasome prevents ovarian aging. Sci. Adv. 2021, 7, eabc7409. [Google Scholar] [CrossRef] [PubMed]
- Liddicoat, B.J.; Chalk, A.M.; Walkley, C.R. ADAR1, inosine and the immune sensing system: Distinguishing self from non-self. Wiley Interdiscip. Rev. RNA 2016, 7, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Bajad, P.; Ebner, F.; Amman, F.; Szabó, B.; Kapoor, U.; Manjali, G.; Hildebrandt, A.; Janisiw, M.P.; Jantsch, M.F. An internal deletion of ADAR rescued by MAVS deficiency leads to a minute phenotype. Nucleic Acids Res. 2020, 48, 3286–3303. [Google Scholar] [CrossRef]
- John, S.P.; Sun, J.; Carlson, R.J.; Cao, B.; Bradfield, C.J.; Song, J.; Smelkinson, M.; Fraser, I.D.C. IFIT1 Exerts Opposing Regulatory Effects on the Inflammatory and Interferon Gene Programs in LPS-Activated Human Macrophages. Cell Rep. 2018, 25, 95–106.e106. [Google Scholar] [CrossRef]
- Johnson, B.; VanBlargan, L.A.; Xu, W.; White, J.P.; Shan, C.; Shi, P.Y.; Zhang, R.; Adhikari, J.; Gross, M.L.; Leung, D.W.; et al. Human IFIT3 Modulates IFIT1 RNA Binding Specificity and Protein Stability. Immunity 2018, 48, 487–499.e485. [Google Scholar] [CrossRef]
- Lee, D.L.; Kim, S.H.; Kim, E.; Chun, S.Y.; Kim, T.S. Interferon-alpha is involved in the luteinizing hormone-induced differentiation of rat preovulatory granulosa cells. J. Interferon Cytokine Res. 2009, 29, 801–808. [Google Scholar] [CrossRef]
- Jang, Y.J.; Park, J.I.; Moon, W.J.; Dam, P.T.; Cho, M.K.; Chun, S.Y. Cumulus cell-expressed type I interferons induce cumulus expansion in mice. Biol. Reprod. 2015, 92, 20. [Google Scholar] [CrossRef][Green Version]
- DeDiego, M.L.; Nogales, A.; Martinez-Sobrido, L.; Topham, D.J. Interferon-Induced Protein 44 Interacts with Cellular FK506-Binding Protein 5, Negatively Regulates Host Antiviral Responses, and Supports Virus Replication. mBio 2019, 10, e01839-19. [Google Scholar] [CrossRef]
- Briley, S.M.; Jasti, S.; McCracken, J.M.; Hornick, J.E.; Fegley, B.; Pritchard, M.T.; Duncan, F.E. Reproductive age-associated fibrosis in the stroma of the mammalian ovary. Reproduction 2016, 152, 245–260. [Google Scholar] [CrossRef]
- Aust, G.; Simchen, C.; Heider, U.; Hmeidan, F.A.; Blumenauer, V.; Spanel-Borowski, K. Eosinophils in the human corpus luteum: The role of RANTES and eotaxin in eosinophil attraction into periovulatory structures. Mol. Hum. Reprod. 2000, 6, 1085–1091. [Google Scholar] [CrossRef] [PubMed]
- Hovnanian, A.; Rebouillat, D.; Levy, E.R.; Mattei, M.G.; Hovanessian, A.G. The human 2’,5’-oligoadenylate synthetase-like gene (OASL) encoding the interferon-induced 56-kDa protein maps to chromosome 12q24.2 in the proximity of the 2’,5’-OAS locus. Genomics 1999, 56, 362–363. [Google Scholar] [CrossRef] [PubMed]
- Hovnanian, A.; Rebouillat, D.; Mattei, M.G.; Levy, E.R.; Marié, I.; Monaco, A.P.; Hovanessian, A.G. The human 2’,5’-oligoadenylate synthetase locus is composed of three distinct genes clustered on chromosome 12q24.2 encoding the 100-, 69-, and 40-kDa forms. Genomics 1998, 52, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Kakuta, S.; Shibata, S.; Iwakura, Y. Genomic structure of the mouse 2’,5’-oligoadenylate synthetase gene family. J. Interferon Cytokine Res. 2002, 22, 981–993. [Google Scholar] [CrossRef]
- Mashimo, T.; Glaser, P.; Lucas, M.; Simon-Chazottes, D.; Ceccaldi, P.E.; Montagutelli, X.; Desprès, P.; Guénet, J.-L. Structural and functional genomics and evolutionary relationships in the cluster of genes encoding murine 2′,5′-oligoadenylate synthetases. Genomics 2003, 82, 537–552. [Google Scholar] [CrossRef]
- Yan, W.; Ma, L.; Stein, P.; Pangas, S.A.; Burns, K.H.; Bai, Y.; Schultz, R.M.; Matzuk, M.M. Mice deficient in oocyte-specific oligoadenylate synthetase-like protein OAS1D display reduced fertility. Mol. Cell Biol. 2005, 25, 4615–4624. [Google Scholar] [CrossRef]
- Hunter, R.H. Have the Fallopian tubes a vital rôle in promoting fertility? Acta Obstet. Gynecol. Scand. 1998, 77, 475–486. [Google Scholar]
- Algarra, B.; Han, L.; Soriano-Úbeda, C.; Avilés, M.; Coy, P.; Jovine, L.; Jiménez-Movilla, M. The C-terminal region of OVGP1 remodels the zona pellucida and modifies fertility parameters. Sci. Rep. 2016, 6, 32556. [Google Scholar] [CrossRef]
- Choudhary, S.; Kumaresan, A.; Kumar, M.; Chhillar, S.; Malik, H.; Kumar, S.; Kaushik, J.K.; Datta, T.K.; Mohanty, A.K. Effect of recombinant and native buffalo OVGP1 on sperm functions and in vitro embryo development: A comparative study. J. Anim. Sci. Biotechnol. 2017, 8, 69. [Google Scholar] [CrossRef]
- Hartner, J.C.; Walkley, C.R.; Lu, J.; Orkin, S.H. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 2009, 10, 109–115. [Google Scholar] [CrossRef]
- Fan, H.-Y.; Shimada, M.; Liu, Z.; Cahill, N.; Noma, N.; Wu, Y.; Gossen, J.; Richards, J.S. Selective expression of KrasG12D in granulosa cells of the mouse ovary causes defects in follicle development and ovulation. Development 2008, 135, 2127–2137. [Google Scholar] [CrossRef] [PubMed]
- McLean, A.C.; Valenzuela, N.; Fai, S.; Bennett, S.A. Performing vaginal lavage, crystal violet staining, and vaginal cytological evaluation for mouse estrous cycle staging identification. J. Vis. Exp. 2012, 67, e4389. [Google Scholar] [CrossRef] [PubMed]
- Masters, W.G., 3rd; Wheeler, M.B. Timing of induced ovulation in C.B-17/Icr-scid/scid and B6SJLF1 mice. Lab. Anim. Sci. 1996, 46, 663–666. [Google Scholar] [PubMed]
- Puchtler, H.; Waldrop, F.S.; Valentine, L.S. Polarization microscopic studies of connective tissue stained with picro-sirius red FBA. Beitr. Pathol. 1973, 150, 174–187. [Google Scholar] [CrossRef]
- Junqueira, L.C.; Bignolas, G.; Brentani, R.R. Picrosirius staining plus polarization microscopy, a specific method for collagen detection in tissue sections. Histochem. J. 1979, 11, 447–455. [Google Scholar] [CrossRef] [PubMed]

| Symbol | Reference mRNA | Forward Primer and Reverse Primers | Amplicon |
|---|---|---|---|
| Ifi44 | NM_001370771.1 | F: CCAACTGACTGCTCGCAATA R: AATAGGACCCAGCAGCAGAA | 190 bp |
| Ifit1 | NM_008331.3 | F:ATGGGAGAGAATGCTGATGG R: AGGAACTGGACCTGCTCTGA | 137 bp |
| Ifit3b | NM_001005858.3 | F: CCATGTTCCGCCTAGAAGAA R: TCTCCCATCCTCAGCAGTTT | 130 bp |
| Oas1g | NM_011852.3 | F:GCTGGGAGACCCAGGAAG R: GCACCTTGGAAGCATCTCTC | 180 bp |
| Ovgp1 | NM_007696.2 | F: TTGTGGCCAAGAATCTGCAG R: GCAAGTGTGGAGAGCATAGC | 148 bp |
| Rn18s | NR_046237.1 | F:GCAATTATTCCCCATGAACG R: GGCCTCACTAAACCATCCAA | 123 bp |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nelson, R.N.; Chakravarthi, V.P.; Ratri, A.; Hong, X.; Gossen, J.A.; Christenson, L.K. Granulosa Cell Specific Loss of Adar in Mice Delays Ovulation, Oocyte Maturation and Leads to Infertility. Int. J. Mol. Sci. 2022, 23, 14001. https://doi.org/10.3390/ijms232214001
Nelson RN, Chakravarthi VP, Ratri A, Hong X, Gossen JA, Christenson LK. Granulosa Cell Specific Loss of Adar in Mice Delays Ovulation, Oocyte Maturation and Leads to Infertility. International Journal of Molecular Sciences. 2022; 23(22):14001. https://doi.org/10.3390/ijms232214001
Chicago/Turabian StyleNelson, Rikki N., V. Praveen Chakravarthi, Anamika Ratri, Xiaoman Hong, Jan A. Gossen, and Lane K. Christenson. 2022. "Granulosa Cell Specific Loss of Adar in Mice Delays Ovulation, Oocyte Maturation and Leads to Infertility" International Journal of Molecular Sciences 23, no. 22: 14001. https://doi.org/10.3390/ijms232214001
APA StyleNelson, R. N., Chakravarthi, V. P., Ratri, A., Hong, X., Gossen, J. A., & Christenson, L. K. (2022). Granulosa Cell Specific Loss of Adar in Mice Delays Ovulation, Oocyte Maturation and Leads to Infertility. International Journal of Molecular Sciences, 23(22), 14001. https://doi.org/10.3390/ijms232214001









