Integrated Transcriptomic, Metabolomic, and Physiological Analyses Reveal New Insights into Fragrance Formation in the Heartwood of Phoebe hui
Abstract
1. Introduction
2. Results
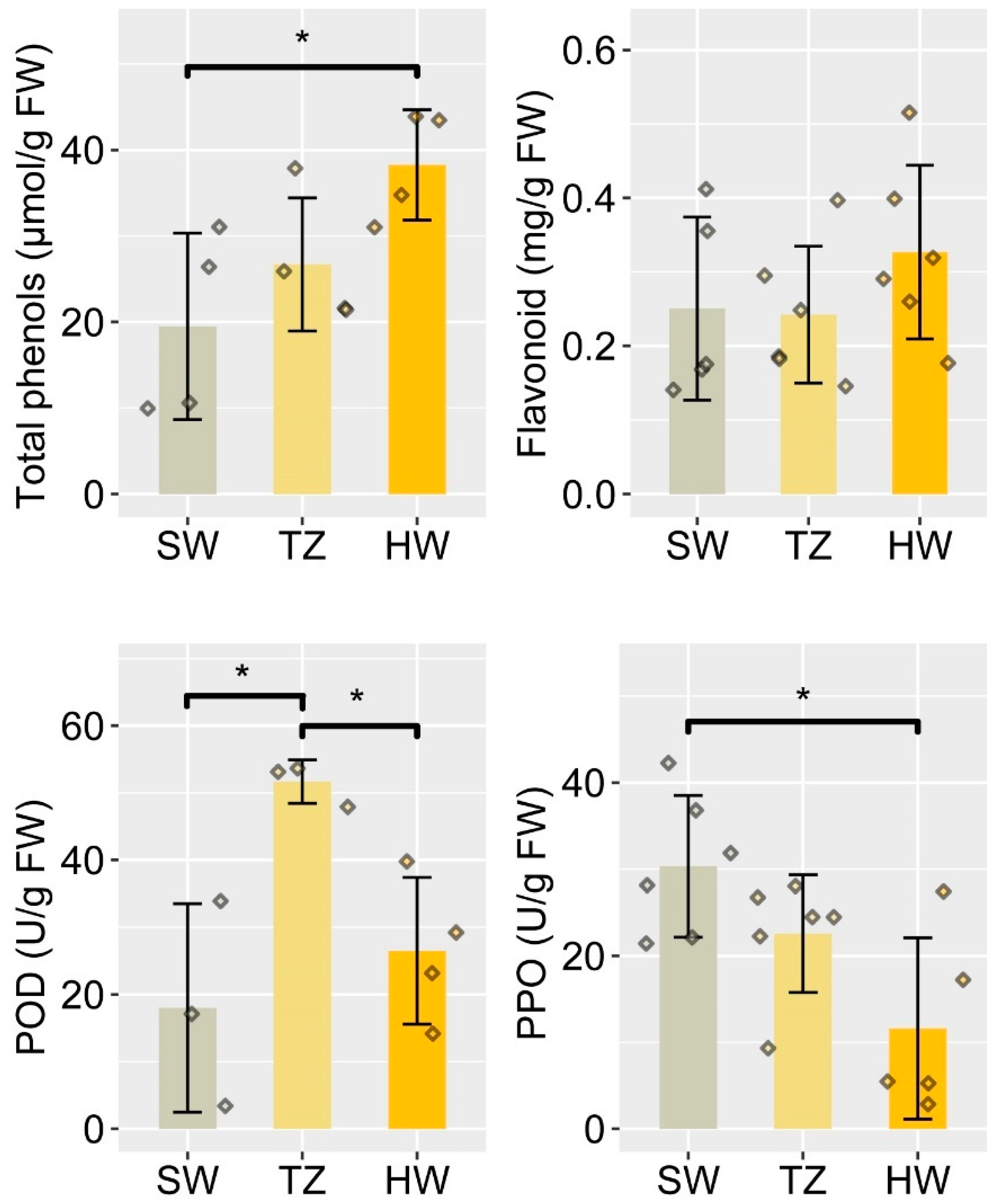
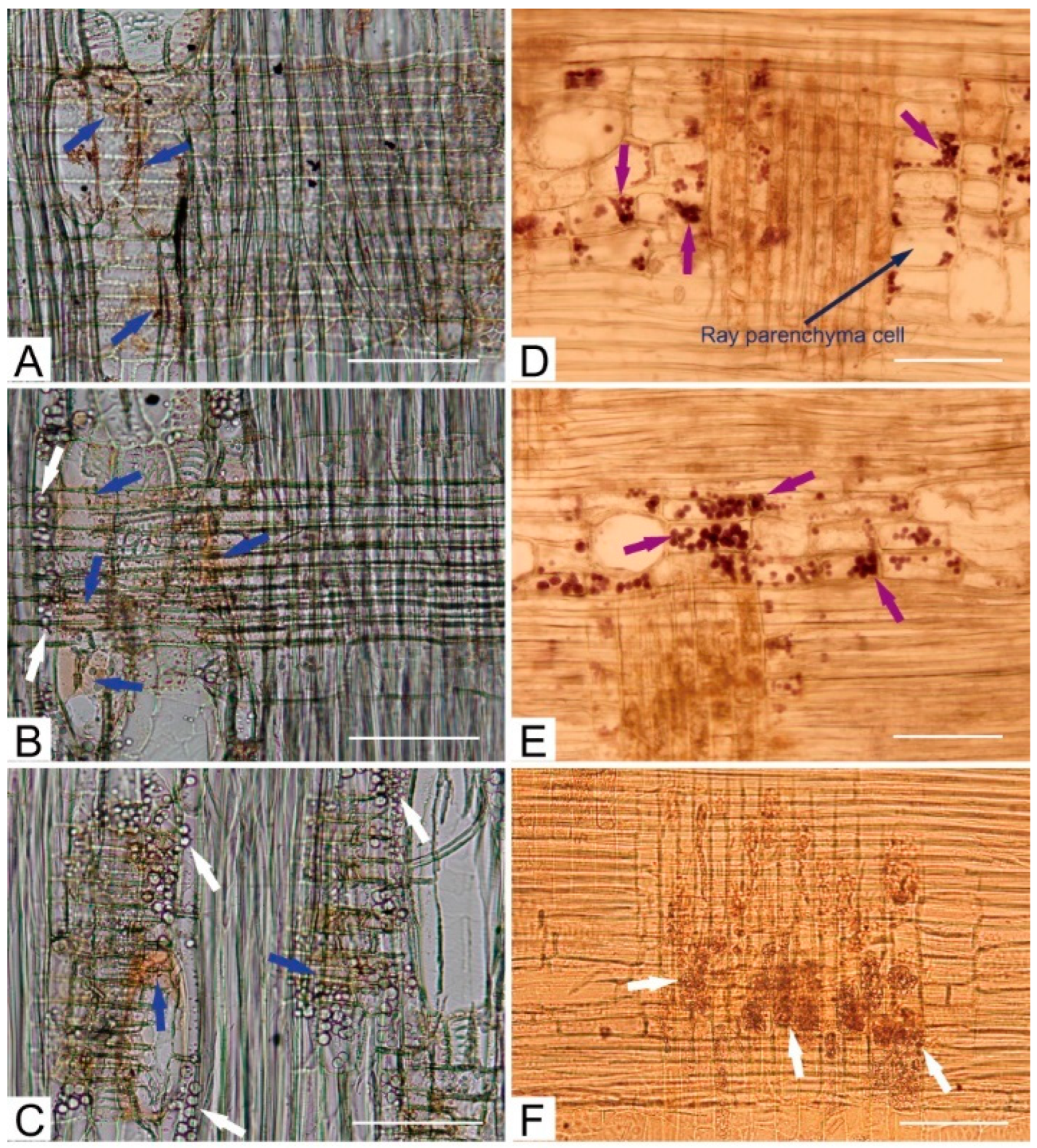
2.1. Radial Distribution of Bioactive Compounds and Enzyme Activities
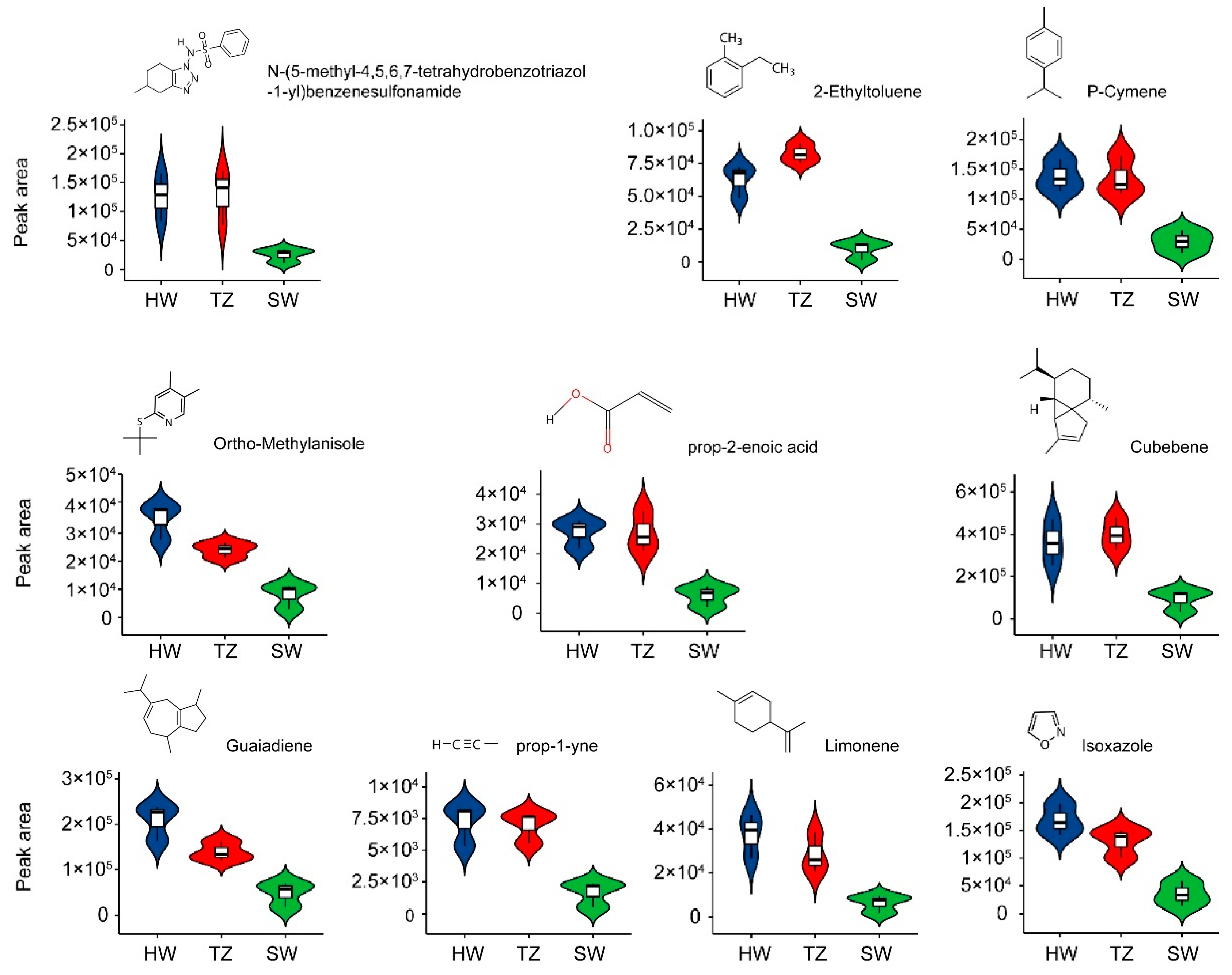
2.2. The Detection of Volatile Metabolites Related to Fragrance
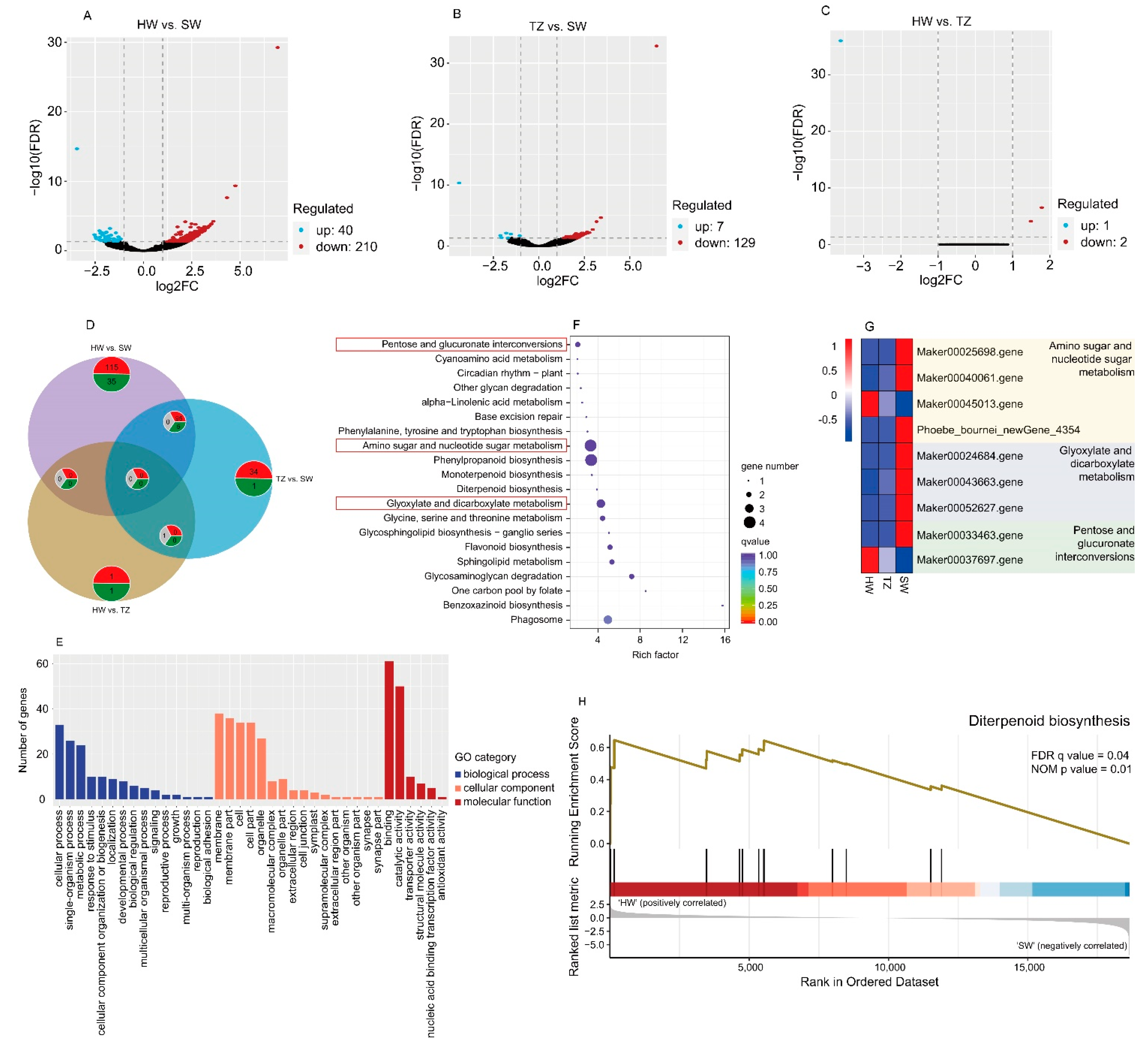
2.3. Transcriptome Analysis and DEGs among HW, TZ, and SW
2.4. Analysis of the Key Pathways Involved in Terpenoid Biosynthesis in P. hui Heartwood
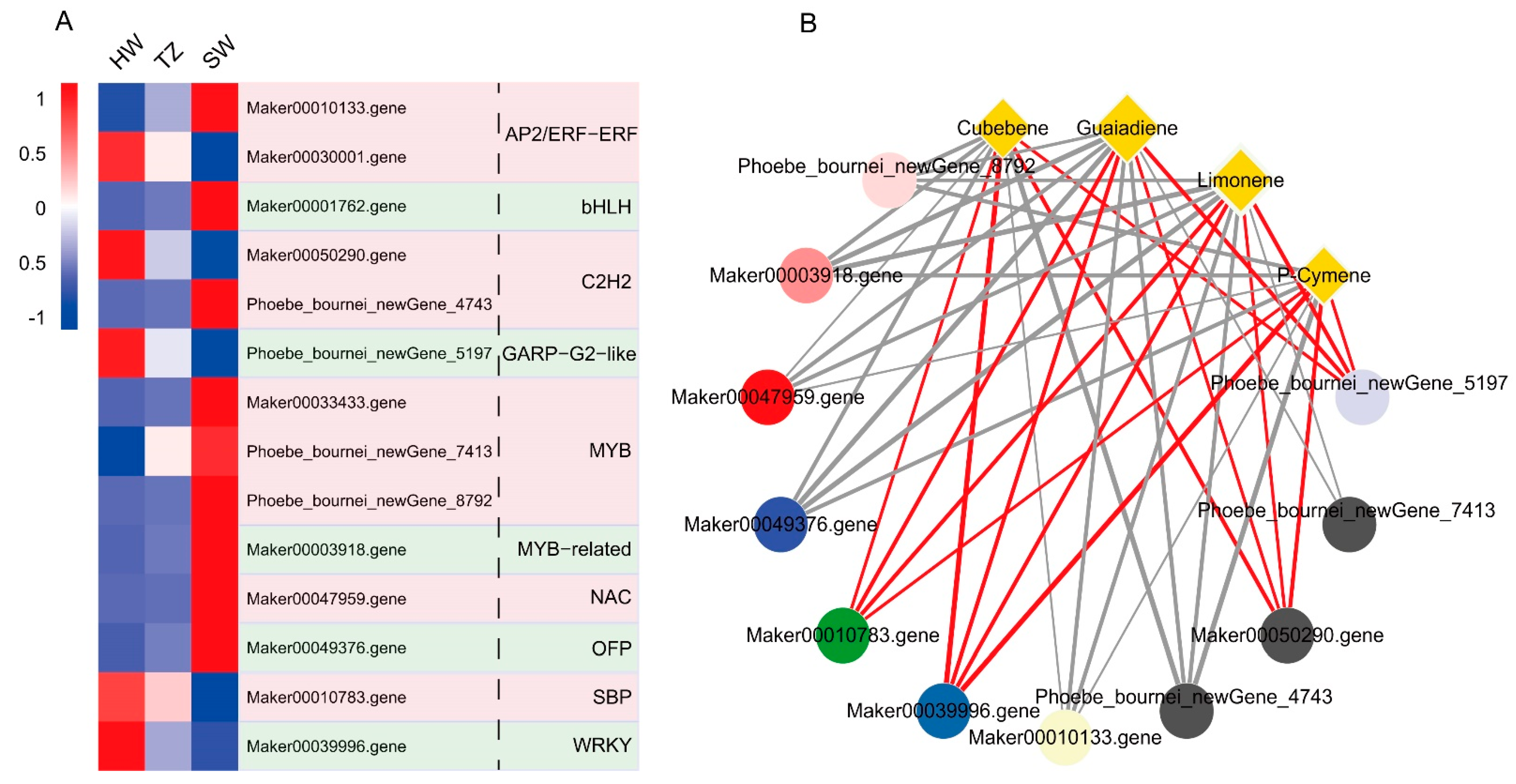
2.5. Transcription Factors Related to Terpenoid Synthesis
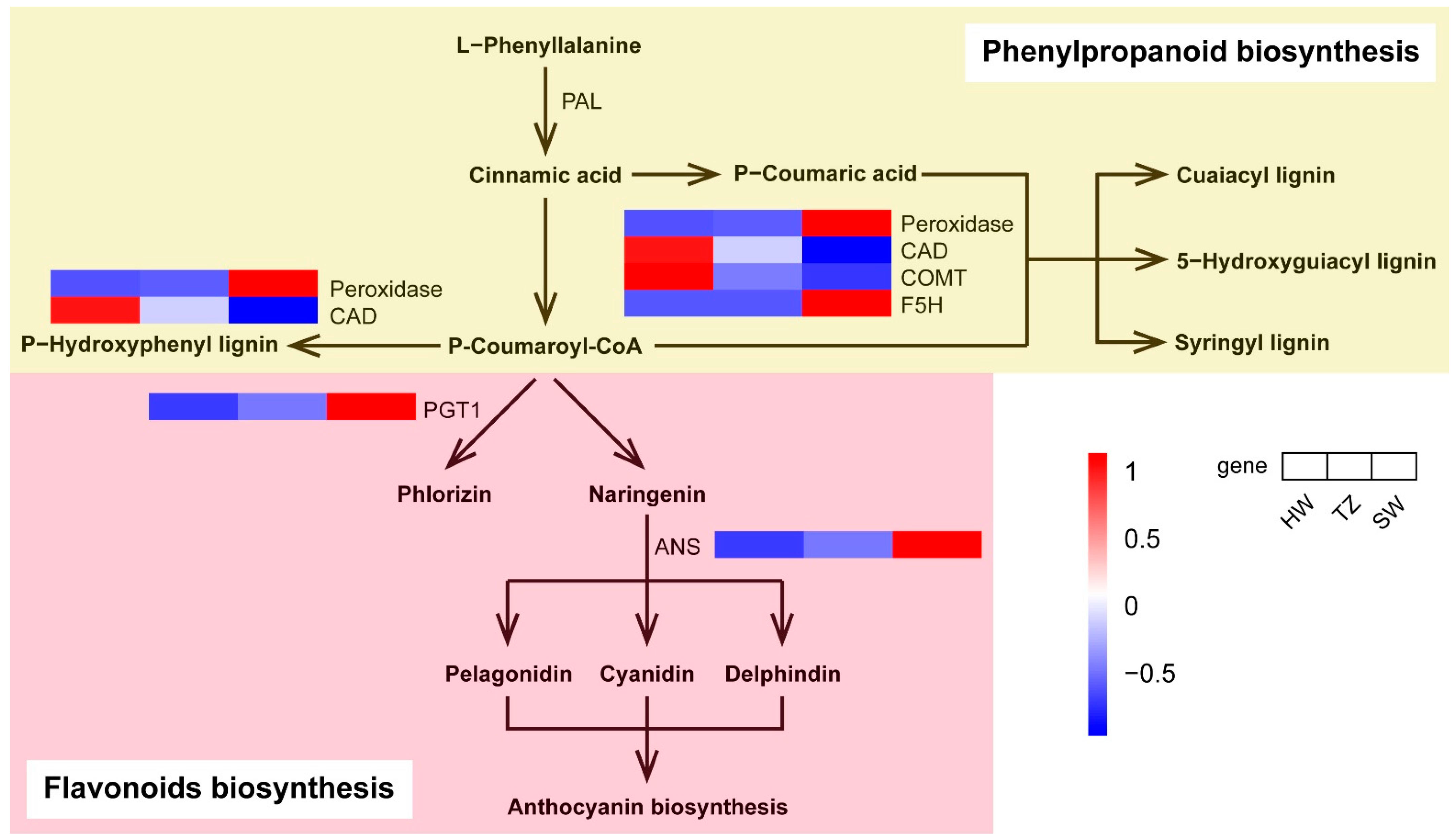
2.6. Phenylpropanoid and Flavonoid Biosynthesis Pathways
3. Discussion
4. Materials and Methods
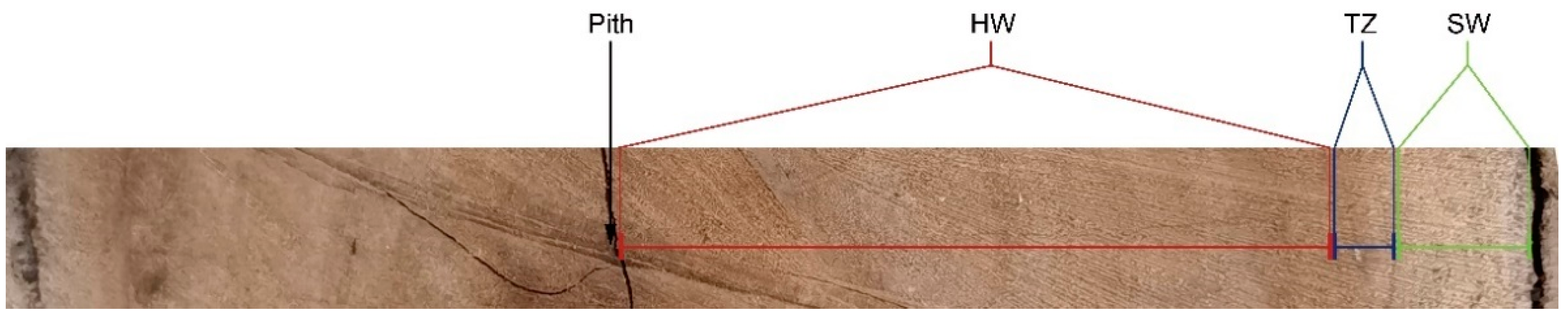
4.1. Plant Material and Sampling
4.2. Microscopy Analyses and Physiological Tests
4.3. GC-MS Analysis
4.4. RNA-seq Analysis
4.5. Correlation Analysis of Metabolites and Transcription Factors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Celedon, J.M.; Bohlmann, J. An extended model of heartwood secondary metabolism informed by functional genomics. Tree Physiol. 2018, 38, 311–319. [Google Scholar] [CrossRef]
- Ding, X.; Xiao, J.H.; Li, L.; Conran, J.G.; Li, J. Congruent species delimitation of two controversial gold-thread nanmu tree species based on morphological and restriction site-associated DNA sequencing data. J. Syst. Evol. 2019, 57, 234–246. [Google Scholar] [CrossRef]
- Yu, D.; Peng, Q.; Li, X.; Xie, K.; Qi, D. Phoebe hui hole tray seedling technique. For. Sci. Technol. 2012, 12, 39–40. [Google Scholar]
- Jiao, L.; Lu, Y.; Zhang, M.; Chen, Y.; Wang, Z.; Guo, Y.; Xu, C.; Guo, J.; He, T.; Ma, L.; et al. Ancient plastid genomes solve the tree species mystery of the imperial wood “Nanmu” in the Forbidden City, the largest existing wooden palace complex in the world. Plants People Planet 2022. [Google Scholar] [CrossRef]
- Ding, W.; Ning, L.; Xiong, Y.; Shi, H.; Wang, T.; An, R. Essential oils extracted from Phoebe hui Cheng ex Yang: Chemical constituents, antitumor and antibacterial activities, and potential use as a species identifier. J. Wood Chem. Technol. 2017, 37, 201–210. [Google Scholar] [CrossRef]
- Kampe, A.; Magel, E. New Insights into Heartwood and Heartwood Formation. In Cellular Aspects of Wood Formation; Fromm, J., Ed.; Plant Cell Monographs; Springer: Berlin/Heidelberg, Germany, 2013; Volume 20. [Google Scholar] [CrossRef]
- Lim, K.-J.; Paasela, T.; Harju, A.; Venäläinen, M.; Paulin, L.; Auvinen, P.; Kärkkäinen, K.; Teeri, T.H. Developmental changes in scots pine transcriptome during heartwood formation. Plant Physiol. 2016, 172, 1403–1417. [Google Scholar] [CrossRef]
- Ma, R.; Liu, H.; Fu, Y.; Li, Y.; Wei, P.; Liu, Z. Variation of chemical components in sapwood, transition zone, and heartwood of Dalbergia odorifera and its relationship with heartwood formation. Forests 2021, 12, 577. [Google Scholar] [CrossRef]
- Spicer, R. Senescence in secondary xylem: Heartwood formation as an active developmental program. In Vascular Transport in Plants; Elsevier: Amsterdam, The Netherlands, 2005; pp. 457–475. [Google Scholar]
- Dehon, L.; Macheix, J.J.; Durand, M. Involvement of peroxidases in the formation of the brown coloration of heartwood in Juglans Nigra. J. Exp. Bot. 2002, 53, 303–311. [Google Scholar] [CrossRef]
- Chen, S.-Y.; Yen, P.-L.; Chang, T.-C.; Chang, S.-T.; Huang, S.-K.; Yeh, T.-F. Distribution of living ray parenchyma cells and major bioactive compounds during the heartwood formation of Taiwania cryptomerioides hayata. J. Wood Chem. Technol. 2018, 38, 84–95. [Google Scholar] [CrossRef]
- Yang, J.; Kamdem, D.P.; Keathley, D.E.; Han, K.-H. Seasonal changes in gene expression at the sapwood—heartwood transition zone of black locust (Robinia pseudoacacia) revealed by cDNA microarray analysis. Tree Physiol. 2004, 24, 461–474. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, X.; Liu, Y.; Shi, Z.; Li, L.; Xie, S.; Zhu, G.; Zhao, P. Comparative metabolomics analysis reveals the color variation between heartwood and sapwood of Chinese fir (Cunninghamia lanceolata (Lamb.) Hook. Ind. Crops Prod. 2021, 169, 113656. [Google Scholar] [CrossRef]
- Chang, S.-T.; Wang, S.-Y.; Wu, C.-L.; Chen, P.-F.; Kuo, Y.-H. Comparison of the antifungal activity of cadinane skeletal sesquiterpenoids from Taiwania (Taiwania cryptomerioides Hayata) heartwood. Holzforschung 2000, 54, 241–245. [Google Scholar] [CrossRef]
- Celedon, J.M.; Chiang, A.; Yuen, M.M.S.; Diaz-Chavez, M.L.; Madilao, L.L.; Finnegan, P.M.; Barbour, E.L.; Bohlmann, J. Heartwood-specific transcriptome and metabolite signatures of tropical sandalwood (Santalum album) reveal the final step of (Z)-santalol fragrance biosynthesis. Plant J. 2016, 86, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wang, J.; Yang, F.; Lei, C. Comparative analysis of essential oil components of two Cryptomeria species from China. Ind. Crops Prod. 2011, 34, 1226–1230. [Google Scholar] [CrossRef]
- Ghadiriasli, R.; Mahmoud, M.A.; Wagenstaller, M.; Van de Kuilen, J.-W.; Buettner, A. Molecular and sensory characterization of odorants in Cembran pine (Pinus cembra L.) from different geographic regions. Talanta 2020, 220, 121380. [Google Scholar] [CrossRef]
- Schreiner, L.; Bauer, P.; Buettner, A. Resolving the smell of wood-identification of odour-active compounds in Scots pine (Pinus sylvestris L.). Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Wang, Q.; Shen, J.; Zeng, B.; Wang, H. Research on VOCs and odor from heartwood and sapwood of paper mulberry (Broussonetia papyrifera (L.) Vent.) with different moisture content. Wood Sci. Technol. 2021, 55, 1153–1170. [Google Scholar] [CrossRef]
- Joshi, S.C.; Padalia, R.C.; Bisht, D.S.; Mathela, C.S. Terpenoid diversity in the leaf essential oils of Himalayan Lauraceae species. Chem. Biodivers. 2009, 6, 1364–1373. [Google Scholar] [CrossRef]
- Xie, J.; Qi, J.; Huang, X.; Zhou, N.; Hu, Y. Comparative analysis of modern and ancient buried Phoebe zhennan wood: Surface color, chemical components, infrared spectroscopy, and essential oil composition. J. For. Res. 2015, 26, 501–507. [Google Scholar] [CrossRef]
- Ding, W.; Liping, N.; Xing, H.; Wei, Z.; Zhoua, Q.; Nong, R.; Chen, J. Essential oil extracted from leaf of Phoebe bournei (Hemsl.) yang: Chemical constituents, antitumor, antibacterial, hypoglycemic activities. Nat. Prod. Res. 2020, 34, 2524–2527. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhang, J.; Han, S.; Chong, S.L.; Meng, G.; Song, M.; Wang, Y.; Zhou, S.; Liu, C.; Lou, L.; et al. The chromosome-scale genome of Phoebe bournei reveals contrasting fates of terpene synthase (TPS)-a and TPS-b subfamilies. Plant Commun. 2022, 3, 100410. [Google Scholar] [CrossRef] [PubMed]
- Yeh, T.F.; Chu, J.H.; Liu, L.Y.; Chen, S.Y. Differential gene profiling of the heartwood formation process in Taiwania cryptomerioides Hayata Xylem tissues. Int. J. Mol. Sci. 2020, 21, 960. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Deng, H.; Zhao, Y.; Zhang, Z.; Tian, Y.; Sun, Y.; Li, Y.; Zheng, H. Metabolite profiling and transcriptome analysis unveil the mechanisms of red-heart Chinese fir [Cunninghamia lanceolata (Lamb.) Hook] heartwood coloration. Front. Plant Sci. 2022, 13, 854716. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Moniodis, J.; Jones, C.G.; Barbour, E.L.; Plummer, J.A.; Ghisalberti, E.L.; Bohlmann, J. The transcriptome of sesquiterpenoid biosynthesis in heartwood xylem of Western Australian sandalwood (Santalum spicatum). Phytochemistry 2015, 113, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ma, Y.; Zeng, J.; Duan, L.; Xue, X.; Wang, H.; Lin, T.; Liu, Z.; Zeng, K.; Zhong, Y. Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nat. Plants 2016, 2, 1–8. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, W.; Ling, Z.; Li, J.; Bai, H.; Li, H.; Shi, L. Advances in transcription factors regulating plant terpenoids biosynthesis. Chin. Bull. Bot. 2020, 55, 340. [Google Scholar]
- Rouet-Mayer, M.-A.; Ralambosoa, J.; Philippon, J. Roles of o-quinones and their polymers in the enzymic browning of apples. Phytochemistry 1990, 29, 435–440. [Google Scholar] [CrossRef]
- Shah, J.; Baqui, S.; Pandalai, R.; Patel, K.R. Histochemical changes in Acacia nilotica L. during transition from sapwood to heartwood. IAWA J. 1981, 2, 31–36. [Google Scholar] [CrossRef]
- Liu, X.; Hao, N.; Feng, R.; Meng, Z.; Li, Y.; Zhao, Z. Transcriptome and metabolite profiling analyses provide insight into volatile compounds of the apple cultivar ‘Ruixue’and its parents during fruit development. BMC Plant Biol. 2021, 21, 1–16. [Google Scholar] [CrossRef]
- Li, N.; Dong, Y.; Lv, M.; Qian, L.; Sun, X.; Liu, L.; Cai, Y.; Fan, H. Combined analysis of volatile terpenoid metabolism and transcriptome reveals transcription factors related to terpene synthase in two cultivars of Dendrobium officinale flowers. Front. Genet. 2021, 450. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, M.; Wen, H.; Wang, Z.; Chen, J.; Fang, L.; Zhang, H.; Xie, Z.; Jiang, D.; Cheng, Y. Transcriptomic and metabolomic analyses provide insight into the volatile compounds of citrus leaves and flowers. BMC Plant Biol. 2020, 20, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Wan, S.; Lin, C.; Zhou, C.; Hu, L.; Deng, C.; Zhang, L. Integration of metabolome and transcriptome reveals the relationship of benzenoid–phenylpropanoid pigment and aroma in purple tea flowers. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, X.; Chen, Z.; Ye, J.; Liao, Y.; Zhang, W.; Chang, J.; Xu, F. De novo assembly and comparative transcriptome analysis: Novel insights into terpenoid biosynthesis in Chamaemelum nobile L. Plant Cell Rep. 2019, 38, 101–116. [Google Scholar] [CrossRef]
- Pichersky, E.; Raguso, R.A. Why do plants produce so many terpenoid compounds? New Phytol. 2018, 220, 692–702. [Google Scholar] [CrossRef]
- Yao, S.; Tang, C.-P.; Ke, C.-Q.; Ye, Y. Abietane diterpenoids from the bark of Cryptomeria fortunei. J. Nat. Prod. 2008, 71, 1242–1246. [Google Scholar] [CrossRef]
- Wu, J.; Zhao, W.-M. New sesquiterpene and triterpene from the fruits of Cryptomeria fortunei. J. Asian Nat. Prod. Res. 2010, 12, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cui, J.; Hu, H.; Xue, J.; Yang, J.; Xu, J. Integrated four comparative-omics reveals the mechanism of the terpenoid biosynthesis in two different overwintering Cryptomeria fortunei phenotypes. Front. Plant Sci. 2021, 12, 740755. [Google Scholar] [CrossRef]
- Beekwilder, J.; van Houwelingen, A.; Cankar, K.; van Dijk, A.D.J.; de Jong, R.M.; Stoopen, G.; Bouwmeester, H.; Achkar, J.; Sonke, T.; Bosch, D. Valencene synthase from the heartwood of Nootka cypress (Callitropsis nootkatensis) for biotechnological production of valencene. Plant Biotechnol. J. 2014, 12, 174–182. [Google Scholar] [CrossRef]
- Cankar, K.; van Houwelingen, A.; Goedbloed, M.; Renirie, R.; de Jong, R.M.; Bouwmeester, H.; Bosch, D.; Sonke, T.; Beekwilder, J. Valencene oxidase CYP706M1 from Alaska cedar (Callitropsis nootkatensis). FEBS Lett. 2014, 588, 1001–1007. [Google Scholar] [CrossRef]
- Jones, C.G.; Keeling, C.I.; Ghisalberti, E.L.; Barbour, E.L.; Plummer, J.A.; Bohlmann, J. Isolation of cDNAs and functional characterisation of two multi-product terpene synthase enzymes from sandalwood, Santalum album L. Arch. Biochem. Biophys. 2008, 477, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Nakada, R.; Fukatsu, E. Seasonal variation of heartwood formation in Larix kaempferi. Tree Physiol. 2012, 32, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhang, J.; Brown, B.; Li, R.; Rodríguez-Romero, J.; Berasategui, A.; Liu, B.; Xu, M.; Luo, D.; Pan, Z. Inferring roles in defense from metabolic allocation of rice diterpenoids. Plant Cell 2018, 30, 1119–1131. [Google Scholar] [CrossRef]
- Mao, L.; Kawaide, H.; Higuchi, T.; Chen, M.; Miyamoto, K.; Hirata, Y.; Kimura, H.; Miyazaki, S.; Teruya, M.; Fujiwara, K. Genomic evidence for convergent evolution of gene clusters for momilactone biosynthesis in land plants. Proc. Natl. Acad. Sci. USA 2020, 117, 12472–12480. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.A.; Wildung, M.R.; Williams, D.C.; Hyatt, D.C.; Croteau, R. cDNA isolation, functional expression, and characterization of (+)-α-pinene synthase and (−)-α-pinene synthase from loblolly pine (Pinus taeda): Stereocontrol in pinene biosynthesis. Arch. Biochem. Biophys. 2003, 411, 267–276. [Google Scholar] [CrossRef]
- Martin, D.M.; Bohlmann, J. Identification of Vitis vinifera (−)-α-terpineol synthase by in silico screening of full-length cDNA ESTs and functional characterization of recombinant terpene synthase. Phytochemistry 2004, 65, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q. Lignification: Flexibility, Biosynthesis and Regulation. Trends Plant Sci. 2016, 21, 713–721. [Google Scholar] [CrossRef]
- Dong, N.-Q.; Lin, H.-X. Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Xie, M.; Zhang, J.; Tschaplinski, T.J.; Tuskan, G.A.; Chen, J.-G.; Muchero, W. Regulation of lignin biosynthesis and its role in growth-defense tradeoffs. Front. Plant Sci. 2018, 9, 1427. [Google Scholar] [CrossRef]
- Cesarino, I. Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 2019, 56, 209–214. [Google Scholar] [CrossRef]
- Huang, G.; Liao, X.; Han, Q.; Zhou, Z.; Liang, K.; Li, G.; Yang, G.; Tembrock, L.R.; Wang, X.; Wu, Z. Integrated metabolome and transcriptome analyses reveal dissimilarities in the anthocyanin synthesis pathway between different developmental leaf color transitions in Hopea hainanensis (Dipterocarpaceae). Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Latchman, D.S. Transcription factors: An overview. Int. J. Biochem. Cell Biol. 1997, 29, 1305–1312. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Bevan, M. The regulation of transcription factor activity in plants. Trends Plant Sci. 1998, 3, 378–383. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, C.; Xu, C.; Sun, J.; Grierson, D.; Zhang, B.; Chen, K. Integration of metabolite profiling and transcriptome analysis reveals genes related to volatile terpenoid metabolism in finger citron (C. medica var. sarcodactylis). Molecules 2019, 24, 2564. [Google Scholar] [CrossRef]
- Ye, W.; Wu, H.; He, X.; Wang, L.; Zhang, W.; Li, H.; Fan, Y.; Tan, G.; Liu, T.; Gao, X. Transcriptome sequencing of chemically induced Aquilaria sinensis to identify genes related to agarwood formation. PLoS ONE 2016, 11, e0155505. [Google Scholar] [CrossRef]
- Spyropoulou, E.A.; Haring, M.A.; Schuurink, R.C. RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genom. 2014, 15, 1–16. [Google Scholar] [CrossRef]
- He, X.; Wang, H.; Yang, J.; Deng, K.; Wang, T. RNA sequencing on Amomum villosum Lour. induced by MeJA identifies the genes of WRKY and terpene synthases involved in terpene biosynthesis. Genome 2018, 61, 91–102. [Google Scholar] [CrossRef]
- Ma, D.; Pu, G.; Lei, C.; Ma, L.; Wang, H.; Guo, Y.; Chen, J.; Du, Z.; Wang, H.; Li, G. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4, 11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 2009, 50, 2146–2161. [Google Scholar] [CrossRef]
- Suttipanta, N.; Pattanaik, S.; Kulshrestha, M.; Patra, B.; Singh, S.K.; Yuan, L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011, 157, 2081–2093. [Google Scholar] [CrossRef]
- Xiang, N.; Hu, J.; Zhang, B.; Cheng, Y.; Wang, S.; Guo, X. Effect of light qualities on volatiles metabolism in maize (Zea mays L.) sprouts. Food Res. Int. 2022, 156, 111340. [Google Scholar] [CrossRef]
- Sharma, B.; Seth, R.; Thakur, S.; Parmar, R.; Masand, M.; Devi, A.; Singh, G.; Dhyani, P.; Choudhary, S.; Sharma, R.K. Genome-wide transcriptional analysis unveils the molecular basis of organ-specific expression of isosteroidal alkaloids biosynthesis in critically endangered Fritillaria roylei Hook. Phytochemistry 2021, 187, 112772. [Google Scholar] [CrossRef] [PubMed]
- Nieuwenhuizen, N.J.; Chen, X.; Wang, M.Y.; Matich, A.J.; Perez, R.L.; Allan, A.C.; Green, S.A.; Atkinson, R.G. Natural variation in monoterpene synthesis in kiwifruit: Transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-like transcription factors. Plant Physiol. 2015, 167, 1243–1258. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, Y.; Shen, S.; Yin, X.; Klee, H.; Zhang, B.; Chen, K. Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E-geraniol in sweet orange fruit. J. Exp. Bot. 2017, 68, 4929–4938. [Google Scholar] [CrossRef]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S. HMDB: The human metabolome database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.; Russell, D.W. Lmsd: Lipid maps structure database. Nucleic Acids Res. 2007, 35, D527–D532. [Google Scholar] [CrossRef]
- Abdelrazig, S.; Safo, L.; Rance, G.A.; Fay, M.W.; Theodosiou, E.; Topham, P.D.; Kim, D.-H.; Fernández-Castané, A. Metabolic characterisation of Magnetospirillum gryphiswaldense MSR-1 using LC-MS-based metabolite profiling. RSC Adv. 2020, 10, 32548–32560. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 2011, 6, 743–760. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-P.; Sun, W.-H.; Xiong, Y.-F.; Jiang, Y.-T.; Liu, X.-D.; Liao, X.-Y.; Zhang, D.-Y.; Jiang, S.-Z.; Li, Y.; Liu, B.; et al. The Phoebe genome sheds light on the evolution of magnoliids. Hortic. Res. 2020, 7, 146. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2-and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; An, W.; Wang, F.; Gu, Y.; Guo, H.; Jiang, Y.; Peng, J.; Liu, M.; Chen, L.; Zhang, F.; et al. Integrated Transcriptomic, Metabolomic, and Physiological Analyses Reveal New Insights into Fragrance Formation in the Heartwood of Phoebe hui. Int. J. Mol. Sci. 2022, 23, 14044. https://doi.org/10.3390/ijms232214044
Yang H, An W, Wang F, Gu Y, Guo H, Jiang Y, Peng J, Liu M, Chen L, Zhang F, et al. Integrated Transcriptomic, Metabolomic, and Physiological Analyses Reveal New Insights into Fragrance Formation in the Heartwood of Phoebe hui. International Journal of Molecular Sciences. 2022; 23(22):14044. https://doi.org/10.3390/ijms232214044
Chicago/Turabian StyleYang, Hanbo, Wenna An, Fang Wang, Yunjie Gu, Hongying Guo, Yongze Jiang, Jian Peng, Minhao Liu, Lianghua Chen, Fan Zhang, and et al. 2022. "Integrated Transcriptomic, Metabolomic, and Physiological Analyses Reveal New Insights into Fragrance Formation in the Heartwood of Phoebe hui" International Journal of Molecular Sciences 23, no. 22: 14044. https://doi.org/10.3390/ijms232214044
APA StyleYang, H., An, W., Wang, F., Gu, Y., Guo, H., Jiang, Y., Peng, J., Liu, M., Chen, L., Zhang, F., Zhu, P., Huang, X., & Wan, X. (2022). Integrated Transcriptomic, Metabolomic, and Physiological Analyses Reveal New Insights into Fragrance Formation in the Heartwood of Phoebe hui. International Journal of Molecular Sciences, 23(22), 14044. https://doi.org/10.3390/ijms232214044





