High Instantaneous Inhibitory Potential of Bictegravir and the New Spiro-β-Lactam BSS-730A for HIV-2 Isolates from RAL-Naïve and RAL-Failing Patients
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Susceptibility of Viral Isolates to INIs and BSS-730A
2.2. Curve Slope and Instantaneous Inhibitory Potential (IIP)
2.3. Antiviral Activity of BSS-730A and RAL Are Synergic
2.4. Analysis of Genotypic Drug Resistance
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Cells, Plasmids, and Drugs
4.3. Patient Data
4.4. Virus Stocks and Titration
4.5. Drug Susceptibility Assays
4.6. Instantaneous Inhibitory Potential (IIP)
4.7. Drug Combination Assays
4.8. DNA Extraction, PCR Amplification and Sequencing
4.9. Phylogenetic Analysis
4.10. Analysis of Genotypic Drug Resistance
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Faria, N.R.; Hodges-Mameletzis, I.; Silva, J.C.; Rodes, B.; Erasmus, S.; Paolucci, S.; Ruelle, J.; Pieniazek, D.; Taveira, N.; Trevino, A.; et al. Phylogeographical footprint of colonial history in the global dispersal of human immunodeficiency virus type 2 group A. J. Gen. Virol. 2012, 93 Pt 4, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Clavel, F.; Guetard, D.; Brun-Vezinet, F.; Chamaret, S.; Rey, M.A.; Santos-Ferreira, M.O.; Laurent, A.G.; Dauguet, C.; Katlama, C.; Rouzioux, C.; et al. Isolation of a new human retrovirus from West African patients with AIDS. Science 1986, 233, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Faria, N.R. Phylogeographic Insights into the Origins and Epidemic History of the Human Immunodeficiency Virus Type 2. In Encyclopedia of AIDS; Hope, T.J., Richman, D., Stevenson, M., Eds.; Springer: New York, NY, USA, 2013. [Google Scholar]
- Direção-Geral da Saúde, Instituto Nacional de Saúde Doutor Ricardo Jorge. Infeção VIH e SIDA em Portugal—2020; Direção-Geral da Saúde, Instituto Nacional de Saúde Doutor Ricardo Jorge: Lisboa, Portugal, 2020. [Google Scholar]
- Barin, F.; Cazein, F.; Lot, F.; Pillonel, J.; Brunet, S.; Thierry, D.; Damond, F.; Brun-Vezinet, F.; Desenclos, J.C.; Semaille, C. Prevalence of HIV-2 and HIV-1 group O infections among new HIV diagnoses in France: 2003–2006. AIDS 2007, 21, 2351–2353. [Google Scholar] [CrossRef] [PubMed]
- van Tienen, C.; van der Loeff, M.S. Epidemiology of HIV-2 infection in West Africa. In Encyclopedia of AIDS; Hope, T.J., Richman, D., Stevenson, M., Eds.; Springer: New York, NY, USA, 2015. [Google Scholar]
- Visseaux, B.; Damond, F.; Matheron, S.; Descamps, D.; Charpentier, C. Hiv-2 molecular epidemiology. Infect. Genet. Evol. 2016, 46, 233–240. [Google Scholar] [CrossRef]
- Marlink, R.; Kanki, P.; Thior, I.; Travers, K.; Eisen, G.; Siby, T.; Traore, I.; Hsieh, C.C.; Dia, M.C.; Gueye, E.H.; et al. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science 1994, 265, 1587–1590. [Google Scholar] [CrossRef]
- Tchounga, B.; Ekouevi, D.K.; Balestre, E.; Dabis, F. Mortality and survival patterns of people living with HIV-2. Curr. Opin. HIV AIDS 2016, 11, 537–544. [Google Scholar] [CrossRef] [Green Version]
- Vidya Vijayan, K.K.; Karthigeyan, K.P.; Tripathi, S.P.; Hanna, L.E. Pathophysiology of CD4+ T-Cell Depletion in HIV-1 and HIV-2 Infections. Front. Immunol. 2017, 8, 580. [Google Scholar] [CrossRef] [Green Version]
- Esbjörnsson, J.; Månsson, F.; Kvist, A.; da Silva, Z.J.; Andersson, S.; Fenyö, E.M.; Isberg, P.E.; Biague, A.J.; Lindman, J.; Palm, A.A.; et al. Long-term follow-up of HIV-2-related AIDS and mortality in Guinea-Bissau: A prospective open cohort study. Lancet HIV 2018, 6, e25–e31. [Google Scholar] [CrossRef]
- Matheron, S.; Pueyo, S.; Damond, F.; Simon, F.; Lepretre, A.; Campa, P.; Salamon, R.; Chene, G.; Brun-Vezinet, F. Factors associated with clinical progression in HIV-2 infected-patients: The French ANRS cohort. AIDS 2003, 17, 2593–2601. [Google Scholar] [CrossRef]
- Ren, J.; Bird, L.E.; Chamberlain, P.P.; Stewart-Jones, G.B.; Stuart, D.I.; Stammers, D.K. Structure of HIV-2 reverse transcriptase at 2.35-A resolution and the mechanism of resistance to non-nucleoside inhibitors. Proc. Natl. Acad. Sci. USA 2002, 99, 14410–14415. [Google Scholar] [CrossRef]
- Camacho, R.J. Special aspects of the treatment of HIV-2-infected patients. Intervirology 2012, 55, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Davenport, Y.W.; West, A.P., Jr.; Bjorkman, P.J. Structure of an HIV-2 gp120 in Complex with CD4. J. Virol. 2016, 90, 2112–2118. [Google Scholar] [CrossRef] [Green Version]
- Tie, Y.; Wang, Y.F.; Boross, P.I.; Chiu, T.Y.; Ghosh, A.K.; Tozser, J.; Louis, J.M.; Harrison, R.W.; Weber, I.T. Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors. Protein Sci. 2012, 21, 339–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witvrouw, M.; Pannecouque, C.; Switzer, W.M.; Folks, T.M.; De Clercq, E.; Heneine, W. Susceptibility of HIV-2, SIV and SHIV to various anti-HIV-1 compounds: Implications for treatment and postexposure prophylaxis. Antivir. Ther. 2004, 9, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Borrego, P.; Calado, R.; Marcelino, J.M.; Bartolo, I.; Rocha, C.; Cavaco-Silva, P.; Doroana, M.; Antunes, F.; Maltez, F.; Caixas, U.; et al. Baseline susceptibility of primary HIV-2 to entry inhibitors. Antivir. Ther. 2012, 17, 565–570. [Google Scholar] [CrossRef] [Green Version]
- Borrego, P.; Calado, R.; Marcelino, J.M.; Pereira, P.; Quintas, A.; Barroso, H.; Taveira, N. An ancestral HIV-2/simian immunodeficiency virus peptide with potent HIV-1 and HIV-2 fusion inhibitor activity. AIDS 2013, 27, 1081–1090. [Google Scholar] [CrossRef] [Green Version]
- Xiong, S.; Borrego, P.; Ding, X.; Zhu, Y.; Martins, A.; Chong, H.; Taveira, N.; He, Y. A Helical Short-Peptide Fusion Inhibitor with Highly Potent Activity against Human Immunodeficiency Virus Type 1 (HIV-1), HIV-2, and Simian Immunodeficiency Virus. J. Virol. 2017, 91, e01839-16. [Google Scholar] [CrossRef] [Green Version]
- Nowicka-Sans, B.; Gong, Y.F.; McAuliffe, B.; Dicker, I.; Ho, H.T.; Zhou, N.; Eggers, B.; Lin, P.F.; Ray, N.; Wind-Rotolo, M.; et al. In vitro antiviral characteristics of HIV-1 attachment inhibitor BMS-626529, the active component of the prodrug BMS-663068. Antimicrob. Agents Chemother. 2012, 56, 3498–3507. [Google Scholar] [CrossRef] [Green Version]
- Decker, J.M.; Bibollet-Ruche, F.; Wei, X.; Wang, S.; Levy, D.N.; Wang, W.; Delaporte, E.; Peeters, M.; Derdeyn, C.A.; Allen, S.; et al. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 2005, 201, 1407–1419. [Google Scholar] [CrossRef]
- Gaebler, C.; Nogueira, L.; Stoffel, E.; Oliveira, T.Y.; Breton, G.; Millard, K.G.; Turroja, M.; Butler, A.; Ramos, V.; Seaman, M.S.; et al. Prolonged viral suppression with anti-HIV-1 antibody therapy. Nature 2022, 606, 368–374. [Google Scholar] [CrossRef]
- Sneller, M.C.; Blazkova, J.; Justement, J.S.; Shi, V.; Kennedy, B.D.; Gittens, K.; Tolstenko, J.; McCormack, G.; Whitehead, E.J.; Schneck, R.F.; et al. Combination anti-HIV antibodies provide sustained virological suppression. Nature 2022, 606, 375–381. [Google Scholar] [CrossRef] [PubMed]
- De Mendoza, C.; Requena, R.; Caballero, E.; Cabezas, T.; Peñaranda, M.; Amengua, M.J.; Sáez, A.; Lozano, A.B.; Ramos, J.M.; Soriano, V. Antiretroviral treatment of HIV-2 infection. Future Virol. 2017, 12, 461–472. [Google Scholar] [CrossRef]
- Desbois, D.; Roquebert, B.; Peytavin, G.; Damond, F.; Collin, G.; Benard, A.; Campa, P.; Matheron, S.; Chene, G.; Brun-Vezinet, F.; et al. In vitro phenotypic susceptibility of human immunodeficiency virus type 2 clinical isolates to protease inhibitors. Antimicrob. Agents Chemother. 2008, 52, 1545–1548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, Y.; Nakata, H.; Maeda, K.; Ogata, H.; Bilcer, G.; Devasamudram, T.; Kincaid, J.F.; Boross, P.; Wang, Y.F.; Tie, Y.; et al. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 2003, 47, 3123–3129. [Google Scholar] [CrossRef] [Green Version]
- Ntemgwa, M.; Brenner, B.G.; Oliveira, M.; Moisi, D.; Wainberg, M.A. Natural polymorphisms in the human immunodeficiency virus type 2 protease can accelerate time to development of resistance to protease inhibitors. Antimicrob. Agents Chemother. 2007, 51, 604–610. [Google Scholar] [CrossRef] [Green Version]
- Ntemgwa, M.L.; d’Aquin Toni, T.; Brenner, B.G.; Camacho, R.J.; Wainberg, M.A. Antiretroviral drug resistance in human immunodeficiency virus type 2. Antimicrob. Agents Chemother. 2009, 53, 3611–3619. [Google Scholar] [CrossRef] [Green Version]
- Yoshimura, K.; Kato, R.; Kavlick, M.F.; Nguyen, A.; Maroun, V.; Maeda, K.; Hussain, K.A.; Ghosh, A.K.; Gulnik, S.V.; Erickson, J.W.; et al. A potent human immunodeficiency virus type 1 protease inhibitor, UIC-94003 (TMC-126), and selection of a novel (A28S) mutation in the protease active site. J. Virol. 2002, 76, 1349–1358. [Google Scholar] [CrossRef] [Green Version]
- Raugi, D.N.; Ba, S.; Cisse, O.; Diallo, K.; Tamba, I.T.; Ndour, C.; Badiane, N.M.D.; Fortes, L.; Diallo, M.B.; Faye, D.; et al. Long-term Experience and Outcomes of Programmatic Antiretroviral Therapy for Human Immunodeficiency Virus Type 2 Infection in Senegal, West Africa. Clin. Infect. Dis. 2021, 72, 369–378. [Google Scholar] [CrossRef]
- Wittkop, L.; Arsandaux, J.; Trevino, A.; Schim van der Loeff, M.; Anderson, J.; van Sighem, A.; Böni, J.; Brun-Vezinet, F.; Soriano, V.; Boufassa, F.; et al. CD4 cell count response to first-line combination ART in HIV-2+ patients compared with HIV-1+ patients: A multinational, multicohort European study. J. Antimicrob. Chemother. 2017, 72, 2869–2878. [Google Scholar] [CrossRef] [Green Version]
- Ntemgwa, M.L.; Toni, T.; Brenner, B.G.; Oliveira, M.; Asahchop, E.L.; Moisi, D.; Wainberg, M.A. Nucleoside and nucleotide analogs select in culture for different patterns of drug resistance in human immunodeficiency virus types 1 and 2. Antimicrob. Agents Chemother. 2009, 53, 708–715. [Google Scholar] [CrossRef]
- Roquebert, B.; Damond, F.; Collin, G.; Matheron, S.; Peytavin, G.; Benard, A.; Campa, P.; Chene, G.; Brun-Vezinet, F.; Descamps, D.; et al. HIV-2 integrase gene polymorphism and phenotypic susceptibility of HIV-2 clinical isolates to the integrase inhibitors raltegravir and elvitegravir in vitro. J. Antimicrob. Chemother. 2008, 62, 914–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, K.; Ruelle, J.; Vekemans, M.; Siegal, F.P.; Deayton, J.R.; Colebunders, R. The role of raltegravir in the treatment of HIV-2 infections: Evidence from a case series. Antivir. Ther. 2012, 17, 1097–1100. [Google Scholar] [CrossRef] [Green Version]
- Requena, S.; Trevino, A.; Cabezas, T.; Garcia-Delgado, R.; Amengual, M.J.; Lozano, A.B.; Penaranda, M.; Fernandez, J.M.; Soriano, V.; de Mendoza, C.; et al. Drug resistance mutations in HIV-2 patients failing raltegravir and influence on dolutegravir response. J. Antimicrob. Chemother. 2017, 72, 2083–2088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trevino, A.; Cabezas, T.; Lozano, A.B.; Garcia-Delgado, R.; Force, L.; Fernandez-Montero, J.M.; Mendoza, C.; Caballero, E.; Soriano, V. Dolutegravir for the treatment of HIV-2 infection. J. Clin. Virol. 2015, 64, 12–15. [Google Scholar] [CrossRef]
- Zheng, Y.; Lambert, C.; Arendt, V.; Seguin-Devaux, C. Virological and immunological outcomes of elvitegravir-based regimen in a treatment-naive HIV-2-infected patient. AIDS 2014, 28, 2329–2331. [Google Scholar] [CrossRef] [PubMed]
- Andreatta, K.; Miller, M.D.; White, K.L. HIV-2 antiviral potency and selection of drug resistance mutations by the integrase strand transfer inhibitor elvitegravir and NRTIs emtricitabine and tenofovir in vitro. J. Acquir. Immune Defic. Syndr. 2013, 62, 367–374. [Google Scholar] [CrossRef]
- Marinello, J.; Marchand, C.; Mott, B.T.; Bain, A.; Thomas, C.J.; Pommier, Y. Comparison of raltegravir and elvitegravir on HIV-1 integrase catalytic reactions and on a series of drug-resistant integrase mutants. Biochemistry 2008, 47, 9345–9354. [Google Scholar] [CrossRef] [Green Version]
- Shimura, K.; Kodama, E.; Sakagami, Y.; Matsuzaki, Y.; Watanabe, W.; Yamataka, K.; Watanabe, Y.; Ohata, Y.; Doi, S.; Sato, M.; et al. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137). J. Virol. 2008, 82, 764–774. [Google Scholar] [CrossRef] [Green Version]
- Smith, R.A.; Raugi, D.N.; Pan, C.; Coyne, M.; Hernandez, A.; Church, B.; Parker, K.; Mullins, J.I.; Sow, P.S.; Gottlieb, G.S.; et al. Three main mutational pathways in HIV-2 lead to high-level raltegravir and elvitegravir resistance: Implications for emerging HIV-2 treatment regimens. PLoS ONE 2012, 7, e45372. [Google Scholar] [CrossRef]
- Charpentier, C.; Larrouy, L.; Collin, G.; Damond, F.; Matheron, S.; Chene, G.; Nie, T.; Schinazi, R.; Brun-Vezinet, F.; Descamps, D.; et al. In-vitro phenotypic susceptibility of HIV-2 clinical isolates to the integrase inhibitor S/GSK1349572. AIDS 2010, 24, 2753–2755. [Google Scholar] [CrossRef]
- Smith, R.A.; Raugi, D.N.; Pan, C.; Sow, P.S.; Seydi, M.; Mullins, J.I.; Gottlieb, G.S.; University of Washington-Dakar, H.I.V.S.G. In vitro activity of dolutegravir against wild-type and integrase inhibitor-resistant HIV-2. Retrovirology 2015, 12, 10. [Google Scholar] [CrossRef] [Green Version]
- Smith, R.A.; Wu, V.H.; Zavala, C.G.; Raugi, D.N.; Ba, S.; Seydi, M.; Gottlieb, G.S. In Vitro Antiviral Activity of Cabotegravir against HIV-2. Antimicrob. Agents Chemother. 2018, 62, e01299-18. [Google Scholar] [CrossRef] [Green Version]
- Smith, R.A.; Raugi, D.N.; Wu, V.H.; Zavala, C.G.; Song, J.; Diallo, K.M.; Seydi, M.; Gottlieb, G.S.; University of Washington-Dakar HIV-2 Study Group. Comparison of the Antiviral Activity of Bictegravir against HIV-1 and HIV-2 Isolates and Integrase Inhibitor-Resistant HIV-2 Mutants. Antimicrob. Agents Chemother. 2019, 63, e00014-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Hingrat, Q.; Collin, G.; Le, M.; Peytavin, G.; Visseaux, B.; Bertine, M.; Tubiana, R.; Karmochkine, M.; Valin, N.; Collin, F.; et al. A New Mechanism of Resistance of Human Immunodeficiency Virus Type 2 to Integrase Inhibitors: A 5-Amino-Acid Insertion in the Integrase C-Terminal Domain. Clin. Infect. Dis. 2019, 69, 657–667. [Google Scholar] [CrossRef]
- Descamps, D.; Peytavin, G.; Visseaux, B.; Tubiana, R.; Damond, F.; Campa, P.; Charpentier, C.; Khuong-Josses, M.A.; Duvivier, C.; Karmochkine, M.; et al. Dolutegravir in HIV-2-Infected Patients With Resistant Virus to First-line Integrase Inhibitors From the French Named Patient Program. Clin. Infect. Dis. 2015, 60, 1521–1527. [Google Scholar] [CrossRef] [Green Version]
- Requena, S.; Lozano, A.B.; Caballero, E.; Garcia, F.; Nieto, M.C.; Tellez, R.; Fernandez, J.M.; Trigo, M.; Rodriguez-Avial, I.; Martin-Carbonero, L.; et al. Clinical experience with integrase inhibitors in HIV-2-infected individuals in Spain. J. Antimicrob. Chemother. 2019, 74, 1357–1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berzow, D.; Descamps, D.; Obermeier, M.; Charpentier, C.; Kaiser, R.; Guertler, L.; Eberle, J.; Wensing, A.; Sierra, S.; Ruelle, J.; et al. Human Immunodeficiency Virus-2 (HIV-2): A Summary of the Present Standard of Care and Treatment Options for Individuals Living with HIV-2 in Western Europe. Clin. Infect. Dis. 2021, 72, 503–509. [Google Scholar] [CrossRef]
- Tzou, P.L.; Descamps, D.; Rhee, S.Y.; Raugi, D.N.; Charpentier, C.; Taveira, N.; Smith, R.A.; Soriano, V.; de Mendoza, C.; Holmes, S.P.; et al. Expanded Spectrum of Antiretroviral-Selected Mutations in Human Immunodeficiency Virus Type 2. J. Infect. Dis. 2020, 221, 1962–1972. [Google Scholar] [CrossRef] [PubMed]
- Northrop, A.J.; Pomeroy, L.W. Forecasting Prevalence of HIV-1 Integrase Strand Transfer Inhibitor (INSTI) Drug Resistance: A Modeling Study. J. Acquir. Immune Defic. Syndr. 2020, 83, 65–71. [Google Scholar] [CrossRef]
- McGee, K.S.; Okeke, N.L.; Hurt, C.B.; McKellar, M.S. Canary in the Coal Mine? Transmitted Mutations Conferring Resistance to All Integrase Strand Transfer Inhibitors in a Treatment-Naive Patient. Open Forum Infect. Dis. 2018, 5, ofy294. [Google Scholar] [CrossRef]
- Alves, A.J.S.; Alves, N.G.; Caratao, C.C.; Esteves, M.I.M.; Fontinha, D.; Bartolo, I.; Soares, M.I.L.; Lopes, S.M.M.; Prudencio, M.; Taveira, N.; et al. Spiro-Lactams as Novel Antimicrobial Agents. Curr. Top. Med. Chem. 2020, 20, 140–152. [Google Scholar] [CrossRef] [PubMed]
- Alves, N.G.; Bartolo, I.; Alves, A.J.S.; Fontinha, D.; Francisco, D.; Lopes, S.M.M.; Soares, M.I.L.; Simoes, C.J.V.; Prudencio, M.; Taveira, N.; et al. Synthesis and structure-activity relationships of new chiral spiro-beta-lactams highly active against HIV-1 and Plasmodium. Eur. J. Med. Chem. 2021, 219, 113439. [Google Scholar] [CrossRef] [PubMed]
- Bartolo, I.; Santos, B.S.; Fontinha, D.; Machado, M.; Francisco, D.; Sepodes, B.; Rocha, J.; Mota-Filipe, H.; Pinto, R.; Figueira, M.E.; et al. Spiro-beta-lactam BSS-730A Displays Potent Activity against HIV and Plasmodium. ACS Infect. Dis. 2021, 7, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Rocha, C.; Calado, R.; Borrego, P.; Marcelino, J.M.; Bartolo, I.; Rosado, L.; Cavaco-Silva, P.; Gomes, P.; Familia, C.; Quintas, A.; et al. Evolution of the human immunodeficiency virus type 2 envelope in the first years of infection is associated with the dynamics of the neutralizing antibody response. Retrovirology 2013, 10, 110. [Google Scholar] [CrossRef] [Green Version]
- Shafer, R.W. Rationale and Uses of a Public HIV Drug-Resistance Database. J. Infect. Dis. 2006, 194 Suppl. 1, S51–S58. [Google Scholar] [CrossRef] [Green Version]
- Ni, X.J.; Delelis, O.; Charpentier, C.; Storto, A.; Collin, G.; Damond, F.; Descamps, D.; Mouscadet, J.F. G140S/Q148R and N155H mutations render HIV-2 Integrase resistant to raltegravir whereas Y143C does not. Retrovirology 2011, 8, 68. [Google Scholar] [CrossRef] [Green Version]
- Smith, R.A.; Wu, V.H.; Song, J.; Raugi, D.N.; Mbaye, K.D.; Seydi, M.; Gottlieb, G.S.; University of Washington-Senegal, H.I.V.S.G. Spectrum of Activity of Raltegravir and Dolutegravir Against Novel Treatment-Associated Mutations in HIV-2 Integrase: A Phenotypic Analysis Using An Expanded Panel of Site-Directed Mutants. J. Infect. Dis. 2022, 226, 497–509. [Google Scholar] [CrossRef]
- Cavaco-Silva, J.; Abecasis, A.; Miranda, A.C.; Pocas, J.; Narciso, J.; Aguas, M.J.; Maltez, F.; Almeida, I.; Germano, I.; Diniz, A.; et al. HIV-2 integrase polymorphisms and longitudinal genotypic analysis of HIV-2 infected patients failing a raltegravir-containing regimen. PLoS ONE 2014, 9, e92747. [Google Scholar] [CrossRef] [Green Version]
- Charpentier, C.; Roquebert, B.; Delelis, O.; Larrouy, L.; Matheron, S.; Tubiana, R.; Karmochkine, M.; Duval, X.; Chene, G.; Storto, A.; et al. Hot spots of integrase genotypic changes leading to HIV-2 resistance to raltegravir. Antimicrob. Agents Chemother. 2011, 55, 1293–1295. [Google Scholar] [CrossRef] [Green Version]
- Salgado, M.; Toro, C.; Simon, A.; Garrido, C.; Blanco, F.; Soriano, V.; Rodes, B. Mutation N155H in HIV-2 integrase confers high phenotypic resistance to raltegravir and impairs replication capacity. J. Clin. Virol. 2009, 46, 173–175. [Google Scholar] [CrossRef]
- Shen, L.; Peterson, S.; Sedaghat, A.R.; McMahon, M.A.; Callender, M.; Zhang, H.; Zhou, Y.; Pitt, E.; Anderson, K.S.; Acosta, E.P.; et al. Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat. Med. 2008, 14, 762–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, L.; Rabi, S.A.; Siliciano, R.F. A novel method for determining the inhibitory potential of anti-HIV drugs. Trends Pharmacol. Sci. 2009, 30, 610–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampah, M.E.; Shen, L.; Jilek, B.L.; Siliciano, R.F. Dose-response curve slope is a missing dimension in the analysis of HIV-1 drug resistance. Proc. Natl. Acad. Sci. USA 2011, 108, 7613–7618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavaco-Silva, P.; Taveira, N.C.; Rosado, L.; Lourenco, M.H.; Moniz-Pereira, J.; Douglas, N.W.; Daniels, R.S.; Santos-Ferreira, M.O. Virological and molecular demonstration of human immunodeficiency virus type 2 vertical transmission. J. Virol. 1998, 72, 3418–3422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.Y.; Shingai, M.; Welbourn, S.; Martin, M.A.; Borrego, P.; Taveira, N.; Strebel, K. Antagonism of BST-2/Tetherin Is a Conserved Function of the Env Glycoprotein of Primary HIV-2 Isolates. J. Virol. 2016, 90, 11062–11074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcelino, J.M.; Borrego, P.; Nilsson, C.; Familia, C.; Barroso, H.; Maltez, F.; Doroana, M.; Antunes, F.; Quintas, A.; Taveira, N. Resistance to antibody neutralization in HIV-2 infection occurs in late stage disease and is associated with X4 tropism. AIDS 2012, 26, 2275–2284. [Google Scholar] [CrossRef] [Green Version]
- Bartolo, I.; Borrego, P.; Gomes, P.; Goncalves, F.; Caixas, U.; Pinto, I.V.; Taveira, N. In vitro evaluation of novel reverse transcriptase inhibitors TAF (tenofovir alafenamide) and OBP-601 (2,3-didehydro-3-deoxy-4-ethynylthymidine) against multi-drug resistant primary isolates of HIV-2. Antivir. Res. 2019, 161, 85–89. [Google Scholar] [CrossRef]
- Podany, A.T.; Scarsi, K.K.; Pham, M.M.; Fletcher, C.V. Comparative Clinical Pharmacokinetics and Pharmacodynamics of HIV-1 Integrase Strand Transfer Inhibitors: An Updated Review. Clin. Pharmacokinet. 2020, 59, 1085–1107. [Google Scholar] [CrossRef]
- Scarsi, K.K.; Havens, J.P.; Podany, A.T.; Avedissian, S.N.; Fletcher, C.V. HIV-1 Integrase Inhibitors: A Comparative Review of Efficacy and Safety. Drugs 2020, 80, 1649–1676. [Google Scholar] [CrossRef]
- Chou, T.C.; Talalay, P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]
- Chou, T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [PubMed]

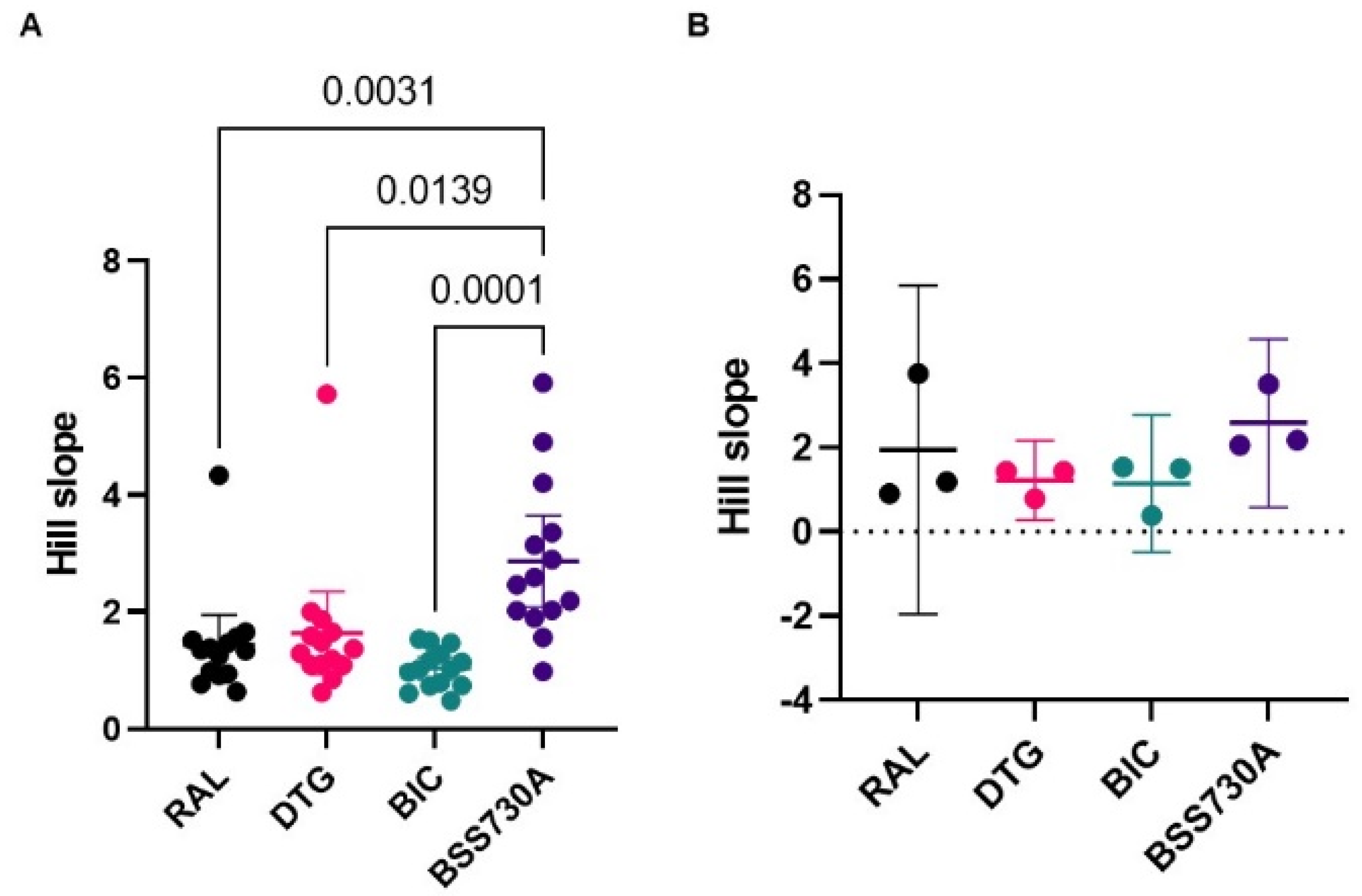
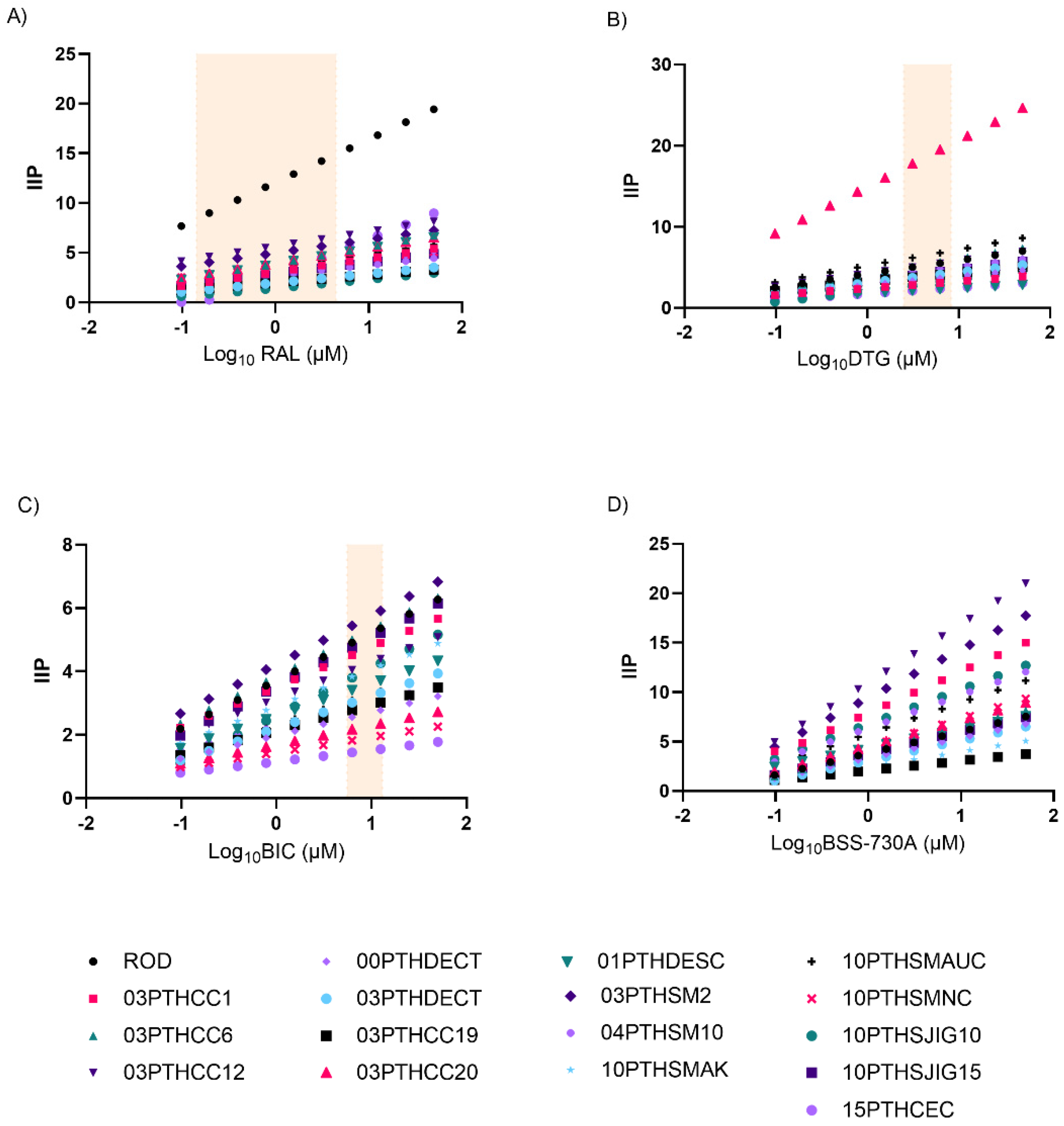


| Parameter a | RAL-Naive Isolates [Mean Values (95% CI)] (n = 14) | RAL-Experienced Isolates [Mean Values (95% CI)] (n = 3) | p-Value b |
|---|---|---|---|
| RALTEGRAVIR | |||
| IC50 (nM) | 2.692 (1.351, 4.032) | 78.178 (−195.1, 351.5) | 0.0294 |
| Fold-change IC50 | 1.681 (0.803, 2.559) | 29.043 (−72.491, 130.578) | 0.1393 |
| IC90 (nM) | 19.998 (7.463, 32.533) | 236.169 (−242.727, 715,065) | 0.0324 |
| Fold-change IC90 | 1.066 (0.401, 1.731) | 11.809 (−12.138, 35.757) | 0.0393 |
| Hill slope | 1.44 (0.922, 1.952) | 1.95 (−1.952, 5.846) | 0.9529 |
| DOLUTEGRAVIR | |||
| IC50 (nM) | 2.778 (2.037, 3.518) | 14.467 (−24.96, 53.89) | 0.0059 |
| Fold-change IC50 | 0.970 (0.688, 1.253) | 5.208 (−8.985, 19.401) | 0.0071 |
| IC90 (nM) | 16.264 (11.307, 21.221) | 89.460 (−70.462, 249,383) | 0.0132 |
| Fold-change IC90 | 1.607 (1.099, 2.116) | 5.501 (−4.332, 15.334) | 0.1464 |
| Hill slope | 1.64 (0.925, 2.352) | 1.22 (0.276, 2.162) | 0.6765 |
| BICTEGRAVIR | |||
| IC50 (nM) | 2.595 (1.707, 3.483) | 8.322 (−14.37, 31.01) | 0.3618 |
| Fold-change IC50 | 0.735 (0.458, 1.013) | 3.207 (−5.537, 11.951) | 0.3536 |
| IC90 (nM) | 30.493 (17.248, 43.738) | 156.100 (−312.135, 624.335) | 0.1485 |
| Fold-change IC90 | 2.375 (1.322, 3.427) | 5.119 (−10.236, 20.474) | 0.7821 |
| Hill slope | 1.03 (0.840, 1.228) | 1.14 (−0.488, 2.773) | 0.5912 |
| BSS-730A | |||
| IC50 (nM) | 18.101 (13.37, 22.84) | 15.667 (7.61, 23.65) | 0.6044 |
| Fold-change IC50 | 0.993 (0.710, 1.277) | 0.865 (0.425, 1.306) | 0.8161 |
| IC90 (nM) | 46.549 (31.850, 61.247) | 42.974 (−0.975, 86.924) | 0.9529 |
| Fold-change IC90 | 0.957 (0.607, 1.308) | 0.903 (−0.021, 1.827) | >0.9999 |
| Hill slope | 2.87 (2.087, 3.651) | 2.58 (0.581, 4.579) | >0.9999 |
| Drug Combination (Combination Ratio) | CI Values at Inhibition of 1: | CIwt-Values 3 | |||
|---|---|---|---|---|---|
| 50% | 75% | 90% | 95% | ||
| RAL + BSS-730A (1:1) | 0.24265 | 0.17127 | 0.18068 | 0.19165 | 0.19 |
| ++++ 2 | ++++ | ++++ | ++++ | ++++ | |
| RAL + BSS-730A (1:3) | 0.17775 | 0.16790 | 0.18613 | 0.20118 | 0.15 |
| ++++ | ++++ | ++++ | ++++ | ++++ | |
| RAL + BSS-730A (3:1) | 0.09422 | 0.05240 | 0.06646 | 0.08334 | 0.07 |
| +++++ | +++++ | +++++ | +++++ | +++++ | |
| Patient (Isolate) | Major Resistance Mutations | Accessory Resistance Mutations | Predicted Susceptibility * | ||
|---|---|---|---|---|---|
| RAL | DTG | BIC ** | |||
| 10PTHSJIG | E92Q | T97A | Resistant | Intermediate | Sensitive |
| 15PTHSJIG | None | I84V | Sensitive | Sensitive | Sensitive |
| 15PTHCEC | E92A, Q148K | I84V | Resistant | Resistant | Sensitive |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bártolo, I.; Moranguinho, I.; Gonçalves, P.; Diniz, A.R.; Borrego, P.; Martin, F.; Figueiredo, I.; Gomes, P.; Gonçalves, F.; Alves, A.J.S.; et al. High Instantaneous Inhibitory Potential of Bictegravir and the New Spiro-β-Lactam BSS-730A for HIV-2 Isolates from RAL-Naïve and RAL-Failing Patients. Int. J. Mol. Sci. 2022, 23, 14300. https://doi.org/10.3390/ijms232214300
Bártolo I, Moranguinho I, Gonçalves P, Diniz AR, Borrego P, Martin F, Figueiredo I, Gomes P, Gonçalves F, Alves AJS, et al. High Instantaneous Inhibitory Potential of Bictegravir and the New Spiro-β-Lactam BSS-730A for HIV-2 Isolates from RAL-Naïve and RAL-Failing Patients. International Journal of Molecular Sciences. 2022; 23(22):14300. https://doi.org/10.3390/ijms232214300
Chicago/Turabian StyleBártolo, Inês, Inês Moranguinho, Paloma Gonçalves, Ana Rita Diniz, Pedro Borrego, Francisco Martin, Inês Figueiredo, Perpétua Gomes, Fátima Gonçalves, Américo J. S. Alves, and et al. 2022. "High Instantaneous Inhibitory Potential of Bictegravir and the New Spiro-β-Lactam BSS-730A for HIV-2 Isolates from RAL-Naïve and RAL-Failing Patients" International Journal of Molecular Sciences 23, no. 22: 14300. https://doi.org/10.3390/ijms232214300







