Integrating Single-Cell Transcriptome and Network Analysis to Characterize the Therapeutic Response of Chronic Myeloid Leukemia
Abstract
:1. Introduction
2. Results
2.1. CP-CML Stem Cells
2.2. Abnormal Gene Expression in Different Types of CP-CML Stem Cells
2.3. Inferring Cellular Regulatory Networks
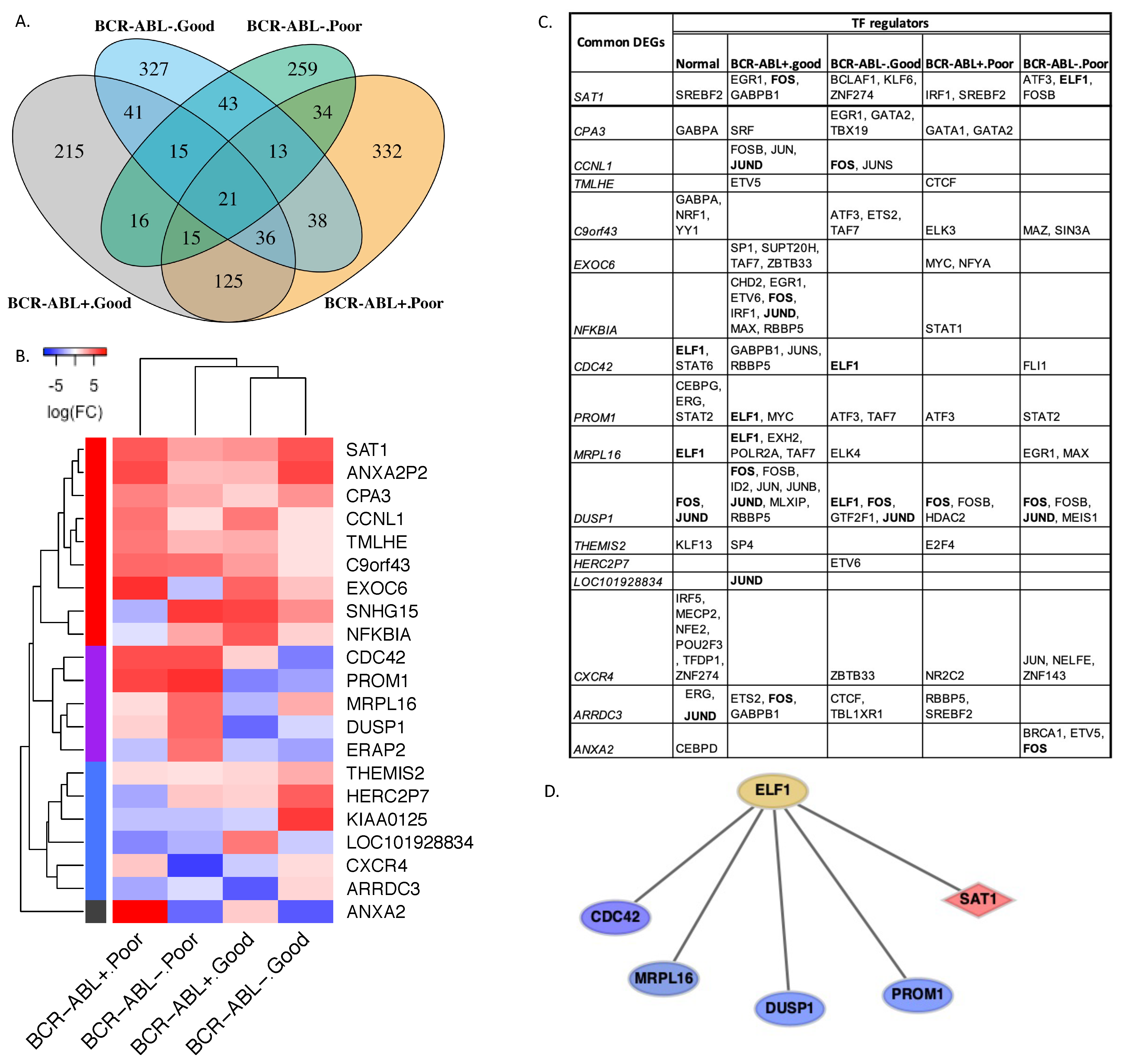
2.4. CP-CML Stem Cells with Different TKI Response
2.5. Protein–Protein Interaction Networks Reveal Putative Drugs for Response Predictive Markers
3. Materials and Methods
3.1. Data Process and Visualization
3.2. Transcription Network Inference
3.3. Protein Interaction Network
3.4. Cancer Drug Database
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gunnarsson, N.; Sandin, F.; Höglund, M.; Stenke, L.; Björkholm, M.; Lambe, M.; Olsson-Strömberg, U.; Richter, J.; Själander, A. Population-based assessment of chronic myeloid leukemia in Sweden: Striking increase in survival and prevalence. Eur. J. Haematol. 2016, 97, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Bennour, A.; Ouahchi, I.; Moez, M.; Elloumi, M.; Khelif, A.; Saad, A.; Sennana, H. Comprehensive analysis of BCR/ABL variants in chronic myeloid leukemia patients using multiplex RT-PCR. Clin. Lab. 2012, 58, 433. [Google Scholar] [PubMed]
- Salesse, S.; Verfaillie, C.M. BCR/ABL: From molecular mechanisms of leukemia induction to treatment of chronic myelogenous leukemia. Oncogene 2002, 21, 8547–8559. [Google Scholar] [CrossRef] [Green Version]
- Copland, M. Is there a role for dose modification of TKI therapy in CML? Curr. Hematol. Malig. Rep. 2019, 14, 337–345. [Google Scholar] [CrossRef] [Green Version]
- Jabbour, E.J.; Hughes, T.P.; Cortes, J.E.; Kantarjian, H.M.; Hochhaus, A. Potential mechanisms of disease progression and management of advanced-phase chronic myeloid leukemia. Leuk. Lymphoma 2014, 55, 1451–1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umeh, C.A.; Garcia-Gonzalez, P.; Tremblay, D.; Laing, R. The survival of patients enrolled in a global direct-to-patient cancer medicine donation program: The Glivec International Patient Assistance Program (GIPAP). EClinicalMedicine 2020, 19, 100257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochhaus, A.; Larson, R.A.; Guilhot, F.; Radich, J.P.; Branford, S.; Hughes, T.P.; Baccarani, M.; Deininger, M.W.; Cervantes, F.; Fujihara, S.; et al. Long-term outcomes of imatinib treatment for chronic myeloid leukemia. N. Engl. J. Med. 2017, 376, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.P.; Eide, C.A.; Druker, B.J. Response and resistance to BCR-ABL1-targeted therapies. Cancer Cell 2020, 37, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Talati, C.; Pinilla-Ibarz, J. Resistance in chronic myeloid leukemia: Definitions and novel therapeutic agents. Curr. Opin. Hematol. 2018, 25, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Branford, S.; Wang, P.; Yeung, D.T.; Thomson, D.; Purins, A.; Wadham, C.; Shahrin, N.H.; Marum, J.E.; Nataren, N.; Parker, W.T.; et al. Integrative genomic analysis reveals cancer-associated mutations at diagnosis of CML in patients with high-risk disease. Blood J. Am. Soc. Hematol. 2018, 132, 948–961. [Google Scholar] [CrossRef] [PubMed]
- Pisco, A.O.; Huang, S. Non-genetic cancer cell plasticity and therapy-induced stemness in tumour relapse: ‘What does not kill me strengthens me’. Br. J. Cancer 2015, 112, 1725–1732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biddle, A.; Gammon, L.; Liang, X.; Costea, D.E.; Mackenzie, I.C. Phenotypic plasticity determines cancer stem cell therapeutic resistance in oral squamous cell carcinoma. EBioMedicine 2016, 4, 138–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayob, A.Z.; Ramasamy, T.S. Cancer stem cells as key drivers of tumour progression. J. Biomed. Sci. 2018, 25, 20. [Google Scholar] [CrossRef] [Green Version]
- Tehranchi, R.; Woll, P.S.; Anderson, K.; Buza-Vidas, N.; Mizukami, T.; Mead, A.J.; Åstrand-Grundström, I.; Strömbeck, B.; Horvat, A.; Ferry, H.; et al. Persistent malignant stem cells in del (5q) myelodysplasia in remission. N. Engl. J. Med. 2010, 363, 1025–1037. [Google Scholar] [CrossRef]
- Woll, P.S.; Kjällquist, U.; Chowdhury, O.; Doolittle, H.; Wedge, D.C.; Thongjuea, S.; Erlandsson, R.; Ngara, M.; Anderson, K.; Deng, Q.; et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell 2014, 25, 794–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magee, J.A.; Piskounova, E.; Morrison, S.J. Cancer stem cells: Impact, heterogeneity, and uncertainty. Cancer Cell 2012, 21, 283–296. [Google Scholar] [CrossRef] [Green Version]
- Holyoake, T.L.; Vetrie, D. The chronic myeloid leukemia stem cell: Stemming the tide of persistence. Blood J. Am. Soc. Hematol. 2017, 129, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; McDonald, T.; Lin, A.; Chakraborty, S.; Huang, Q.; Snyder, D.S.; Bhatia, R. Persistence of leukemia stem cells in chronic myelogenous leukemia patients in prolonged remission with imatinib treatment. Blood J. Am. Soc. Hematol. 2011, 118, 5565–5572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallipoli, P.; Abraham, S.A.; Holyoake, T.L. Hurdles toward a cure for CML: The CML stem cell. Hematol. Clin. 2011, 25, 951–966. [Google Scholar] [CrossRef]
- Mahon, F.X.; Réa, D.; Guilhot, J.; Guilhot, F.; Huguet, F.; Nicolini, F.; Legros, L.; Charbonnier, A.; Guerci, A.; Varet, B.; et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: The prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010, 11, 1029–1035. [Google Scholar] [CrossRef]
- Welner, R.S.; Amabile, G.; Bararia, D.; Czibere, A.; Yang, H.; Zhang, H.; Pontes, L.L.D.F.; Ye, M.; Levantini, E.; Di Ruscio, A.; et al. Treatment of chronic myelogenous leukemia by blocking cytokine alterations found in normal stem and progenitor cells. Cancer Cell 2015, 27, 671–681. [Google Scholar] [CrossRef] [Green Version]
- Reynaud, D.; Pietras, E.; Barry-Holson, K.; Mir, A.; Binnewies, M.; Jeanne, M.; Sala-Torra, O.; Radich, J.P.; Passegué, E. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer Cell 2011, 20, 661–673. [Google Scholar] [CrossRef] [Green Version]
- Giustacchini, A.; Thongjuea, S.; Barkas, N.; Woll, P.S.; Povinelli, B.J.; Booth, C.A.; Sopp, P.; Norfo, R.; Rodriguez-Meira, A.; Ashley, N.; et al. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat. Med. 2017, 23, 692–702. [Google Scholar] [CrossRef]
- Loscocco, F.; Visani, G.; Galimberti, S.; Curti, A.; Isidori, A. BCR-ABL independent mechanisms of resistance in chronic myeloid leukemia. Front. Oncol. 2019, 9, 939. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Zhang, Y.; Qu, X. Effects of Cdc42 overexpression on the estrogen-enhanced multidrug resistance in breast cancer cells. Zhonghua Zhong Liu Za Zhi [Chin. J. Oncol.] 2011, 33, 489–493. [Google Scholar]
- Saha, S.K.; Islam, S.; Kwak, K.S.; Rahman, M.; Cho, S.G. PROM1 and PROM2 expression differentially modulates clinical prognosis of cancer: A multiomics analysis. Cancer Gene Ther. 2020, 27, 147–167. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Gore, A.J.; Wilson, J.L.; Korc, M. DUSP1 is a novel target for enhancing pancreatic cancer cell sensitivity to gemcitabine. PLoS ONE 2014, 9, e84982. [Google Scholar] [CrossRef]
- Teng, F.; Xu, Z.; Chen, J.; Zheng, G.; Zheng, G.; Lv, H.; Wang, Y.; Wang, L.; Cheng, X. DUSP1 induces apatinib resistance by activating the MAPK pathway in gastric cancer. Oncol. Rep. 2018, 40, 1203–1222. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, Y.; Yu, H.; Shen, B.; Liang, Y.; Jin, R.; Liu, X.; Shi, L.; Cai, X. Role of DUSP1/MKP1 in tumorigenesis, tumor progression and therapy. Cancer Med. 2016, 5, 2061–2068. [Google Scholar] [CrossRef] [Green Version]
- Budka, J.A.; Ferris, M.W.; Capone, M.J.; Hollenhorst, P.C. Common ELF1 deletion in prostate cancer bolsters oncogenic ETS function, inhibits senescence and promotes docetaxel resistance. Genes Cancer 2018, 9, 198. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, S.; Yamayoshi, Y.; Nishimuta, A.; Tanigawara, Y. S100A10 protein expression is associated with oxaliplatin sensitivity in human colorectal cancer cells. Proteome Sci. 2011, 9, 76. [Google Scholar] [CrossRef]
- Florea, A.M.; Varghese, E.; McCallum, J.E.; Mahgoub, S.; Helmy, I.; Varghese, S.; Gopinath, N.; Sass, S.; Theis, F.J.; Reifenberger, G.; et al. Calcium-regulatory proteins as modulators of chemotherapy in human neuroblastoma. Oncotarget 2017, 8, 22876. [Google Scholar] [CrossRef] [Green Version]
- Johansson, H.J.; Sanchez, B.C.; Forshed, J.; Stål, O.; Fohlin, H.; Lewensohn, R.; Hall, P.; Bergh, J.; Lehtiö, J.; Linderholm, B.K. Proteomics profiling identify CAPS as a potential predictive marker of tamoxifen resistance in estrogen receptor positive breast cancer. Clin. Proteom. 2015, 12, 8. [Google Scholar] [CrossRef] [Green Version]
- Nymoen, D.A.; Falkenthal, T.E.H.; Holth, A.; Ow, G.S.; Ivshina, A.V.; Tropé, C.G.; Kuznetsov, V.A.; Staff, A.C.; Davidson, B. Expression and clinical role of chemoresponse-associated genes in ovarian serous carcinoma. Gynecol. Oncol. 2015, 139, 30–39. [Google Scholar] [CrossRef]
- Pantziarka, P.; Capistrano, R.; De Potter, A.; Vandeborne, L.; Bouche, G. An Open Access Database of Licensed Cancer Drugs. Front. Pharmacol. 2021, 12, 236. [Google Scholar] [CrossRef]
- The AACR Project GENIE Consortium; André, F.; Arnedos, M.; Baras, A.S.; Baselga, J.; Bedard, P.L.; Berger, M.F.; Bierkens, M.; Calvo, F.; Cerami, E.; et al. AACR Project GENIE: Powering precision medicine through an international consortium. Cancer Discov. 2017, 7, 818–831. [Google Scholar]
- Kim, Y.S.; Gupta Vallur, P.; Phaëton, R.; Mythreye, K.; Hempel, N. Insights into the Dichotomous Regulation of SOD2 in Cancer. Antioxidants 2017, 6, 86. [Google Scholar] [CrossRef] [Green Version]
- Madureira, P.A.; O’Connell, P.A.; Surette, A.P.; Miller, V.A.; Waisman, D.M. The biochemistry and regulation of S100A10: A multifunctional plasminogen receptor involved in oncogenesis. J. Biomed. Biotechnol. 2012, 2012, 353687. [Google Scholar] [CrossRef] [Green Version]
- Coutinho, A.E.; Chapman, K.E. The anti-inflammatory and immunosuppressive effects of glucocorticoids, recent developments and mechanistic insights. Mol. Cell. Endocrinol. 2011, 335, 2–13. [Google Scholar] [CrossRef]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the p53 tumor suppressor gene: Important milestones at the various steps of tumorigenesis. Genes Cancer 2011, 2, 466–474. [Google Scholar] [CrossRef] [Green Version]
- Walker, K.; Boyd, N.H.; Anderson, J.C.; Willey, C.D.; Hjelmeland, A.B. Kinomic profiling of glioblastoma cells reveals PLCG1 as a target in restricted glucose. Biomark. Res. 2018, 6, 1–9. [Google Scholar] [CrossRef]
- Cremer, A.; Ellegast, J.M.; Alexe, G.; Frank, E.S.; Ross, L.; Chu, S.H.; Pikman, Y.; Robichaud, A.; Goodale, A.; Häupl, B.; et al. Resistance mechanisms to SYK inhibition in acute myeloid leukemia. Cancer Discov. 2020, 10, 214–231. [Google Scholar] [CrossRef]
- Brown, D.P.; Chin-Sinex, H.; Nie, B.; Mendonca, M.S.; Wang, M. Targeting superoxide dismutase 1 to overcome cisplatin resistance in human ovarian cancer. Cancer Chemother. Pharmacol. 2009, 63, 723–730. [Google Scholar] [CrossRef]
- Gene Expression Omnibus. Available online: https://www.ncbi.nlm.nih.gov/geo/ (accessed on 10 October 2020).
- Ramsköld, D.; Wang, E.T.; Burge, C.B.; Sandberg, R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009, 5, e1000598. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Van de Sande, B.; Flerin, C.; Davie, K.; De Waegeneer, M.; Hulselmans, G.; Aibar, S.; Seurinck, R.; Saelens, W.; Cannoodt, R.; Rouchon, Q.; et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 2020, 15, 2247–2276. [Google Scholar] [CrossRef]
- Moerman, T.; Aibar Santos, S.; Bravo, G.C.; Simm, J.; Moreau, Y.; Aerts, J.; Aerts, S. GRNBoost2 and Arboreto: Efficient and scalable inference of gene regulatory networks. Bioinformatics. 2019, 35, 2159–2161. [Google Scholar] [CrossRef]
- STRING: Functional Protein Association Networks. Available online: https://string-db.org/ (accessed on 15 January 2022).
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Assouline, S.; Lipton, J.H. Monitoring response and resistance to treatment in chronic myeloid leukemia. Curr. Oncol. 2011, 18, 71–83. [Google Scholar] [CrossRef] [Green Version]
- Apperley, J.F. Part I: Mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007, 8, 1018–1029. [Google Scholar] [CrossRef]
- Svenningsson, P.; Greengard, P. p11 (S100A10)—An inducible adaptor protein that modulates neuronal functions. Curr. Opin. Pharmacol. 2007, 7, 27–32. [Google Scholar] [CrossRef]
- Lou, Y.; Han, M.; Liu, H.; Niu, Y.; Liang, Y.; Guo, J.; Zhang, W.; Wang, H. Essential roles of S100A10 in Toll-like receptor signaling and immunity to infection. Cell. Mol. Immunol. 2020, 17, 1053–1062. [Google Scholar] [CrossRef]
- Shchebliakov, D.; Logunov, Y.; Tukhvatulin, A.; Shmarov, M.; Naroditsky, B.; Ginzburg, A. Toll-like receptors (TLRs): The role in tumor progression. Acta Naturae. 2010, 3, 21–29. [Google Scholar] [CrossRef] [Green Version]
- Deschamps, M.; Laval, G.; Fagny, M.; Itan, Y.; Abel, L.; Casanova, J.L.; Patin, E.; Quintana-Murci, L. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am. J. Hum. Genet. 2016, 98, 5–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potaczek, D.; Sanak, M.; Szczeklik, A. Additive association between FCER1A and FCER1B genetic polymorphisms and total serum IgE levels. Allergy 2007, 62, 1095–1096. [Google Scholar] [CrossRef]
- Palikhe, N.S.; Kim, S.H.; Cho, B.Y.; Ye, Y.M.; Hur, G.Y.; Park, H.S. Association of three sets of high-affinity IgE receptor (FcepsilonR1) polymorphisms with aspirin-intolerant asthma. Respir. Med. 2008, 102, 1132–1139. [Google Scholar] [CrossRef] [Green Version]
- Roshanizadeh, Z.; Ghandil, P.; Khodadadi, A.; Tavakold, H.; Angali, K.A.; Ghadiri, A. Genetic association study of CTLA4 and FCεRIα polymorphisms in asthmatic patients in the southwestern region of Iran. Nucleosides Nucleotides Nucleic Acids 2021, 40, 914–925. [Google Scholar] [CrossRef]
- Lee, J.Y.; Park, A.K.; Lee, K.M.; Park, S.K.; Han, S.; Han, W.; Noh, D.Y.; Yoo, K.Y.; Kim, H.; Chanock, S.J.; et al. Candidate gene approach evaluates association between innate immunity genes and breast cancer risk in Korean women. Carcinogenesis 2009, 30, 1528–1531. [Google Scholar] [CrossRef] [Green Version]
- Jiang, P.; Zhang, P.; Mukthavaram, R.; Nomura, N.; Pingle, S.C.; Teng, D.; Chien, S.; Guo, F.; Kesari, S. Anti-cancer effects of nitrogen-containing bisphosphonates on human cancer cells. Oncotarget 2016, 7, 57932. [Google Scholar] [CrossRef] [Green Version]
- Subramani, D.; Alahari, S.K. Integrin-mediated function of Rab GTPases in cancer progression. Mol. Cancer 2010, 9, 312. [Google Scholar] [CrossRef] [Green Version]
- American Cancer Society. Cancer Facts & Figures 2022. Available online: https://www.cancer.org/cancer/chronic-myeloid-leukemia/about/statistics.html (accessed on 26 October 2022).
- Pemmaraju, N.; Kantarjian, H.; Shan, J.; Jabbour, E.; Quintas-Cardama, A.; Verstovsek, S.; Ravandi, F.; Wierda, W.; O’Brien, S.; Cortes, J. Analysis of outcomes in adolescents and young adults with chronic myelogenous leukemia treated with upfront tyrosine kinase inhibitor therapy. Haematologica 2012, 97, 1029–1035. [Google Scholar] [CrossRef]


| BCR-ABL | TKI Response | |
|---|---|---|
| Good | Poor | |
| Positive | 255 | 181 |
| Negative | 188 | 138 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Pettit, N.; Talburt, J.; Wang, S.; Weissman, S.M.; Yang, M.Q. Integrating Single-Cell Transcriptome and Network Analysis to Characterize the Therapeutic Response of Chronic Myeloid Leukemia. Int. J. Mol. Sci. 2022, 23, 14335. https://doi.org/10.3390/ijms232214335
Ma J, Pettit N, Talburt J, Wang S, Weissman SM, Yang MQ. Integrating Single-Cell Transcriptome and Network Analysis to Characterize the Therapeutic Response of Chronic Myeloid Leukemia. International Journal of Molecular Sciences. 2022; 23(22):14335. https://doi.org/10.3390/ijms232214335
Chicago/Turabian StyleMa, Jialu, Nathan Pettit, John Talburt, Shanzhi Wang, Sherman M. Weissman, and Mary Qu Yang. 2022. "Integrating Single-Cell Transcriptome and Network Analysis to Characterize the Therapeutic Response of Chronic Myeloid Leukemia" International Journal of Molecular Sciences 23, no. 22: 14335. https://doi.org/10.3390/ijms232214335
APA StyleMa, J., Pettit, N., Talburt, J., Wang, S., Weissman, S. M., & Yang, M. Q. (2022). Integrating Single-Cell Transcriptome and Network Analysis to Characterize the Therapeutic Response of Chronic Myeloid Leukemia. International Journal of Molecular Sciences, 23(22), 14335. https://doi.org/10.3390/ijms232214335






