Dynamic Change of R-Loop Implicates in the Regulation of Zygotic Genome Activation in Mouse
Abstract
1. Introduction
2. Results
2.1. Optimization of R-Loop Immunofluorescence (IF) in Mouse Preimplantation Embryos
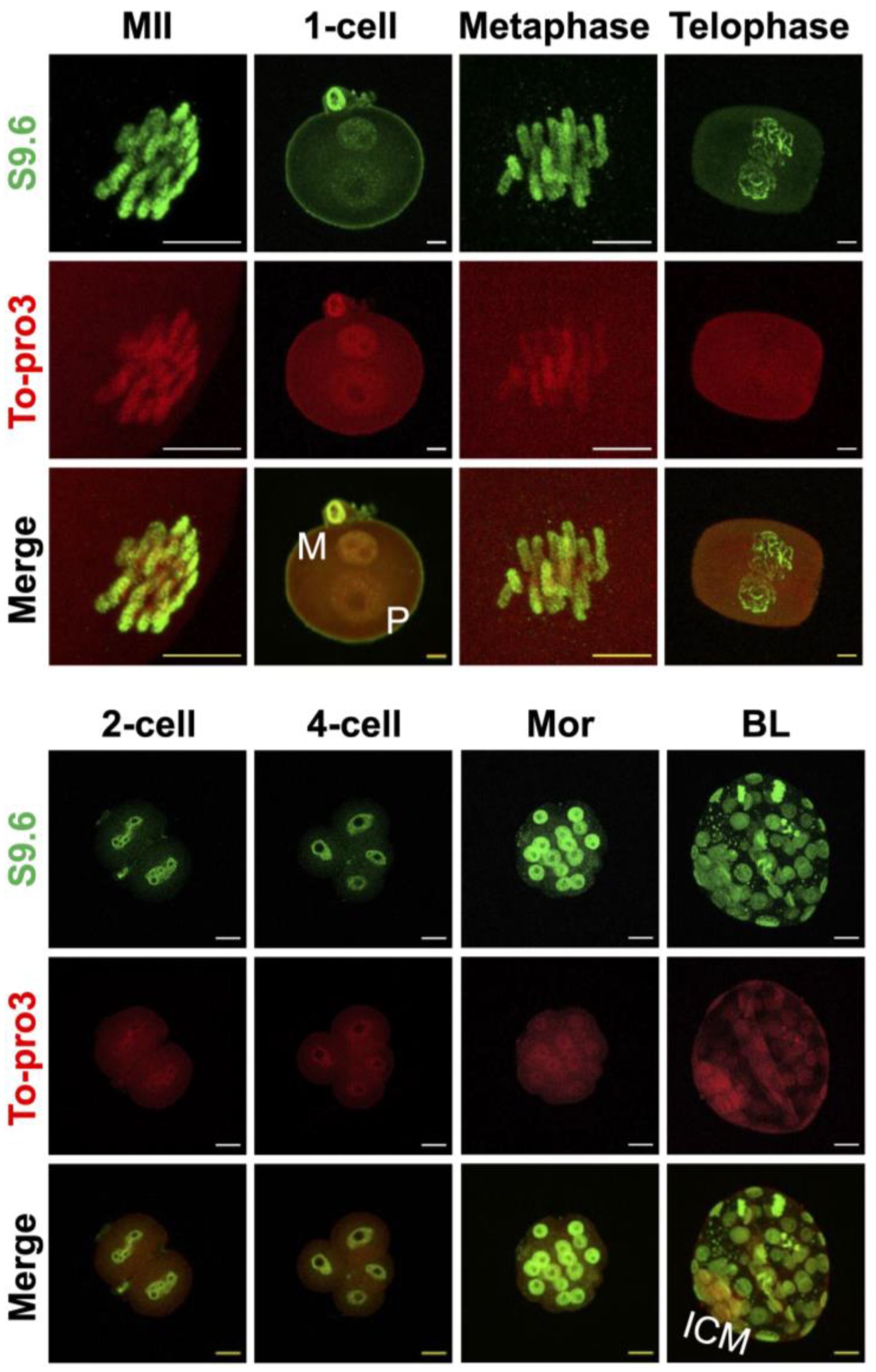
2.2. R-Loops Are Detected throughout Preimplantation Stage Embryos and All Cell Cycles
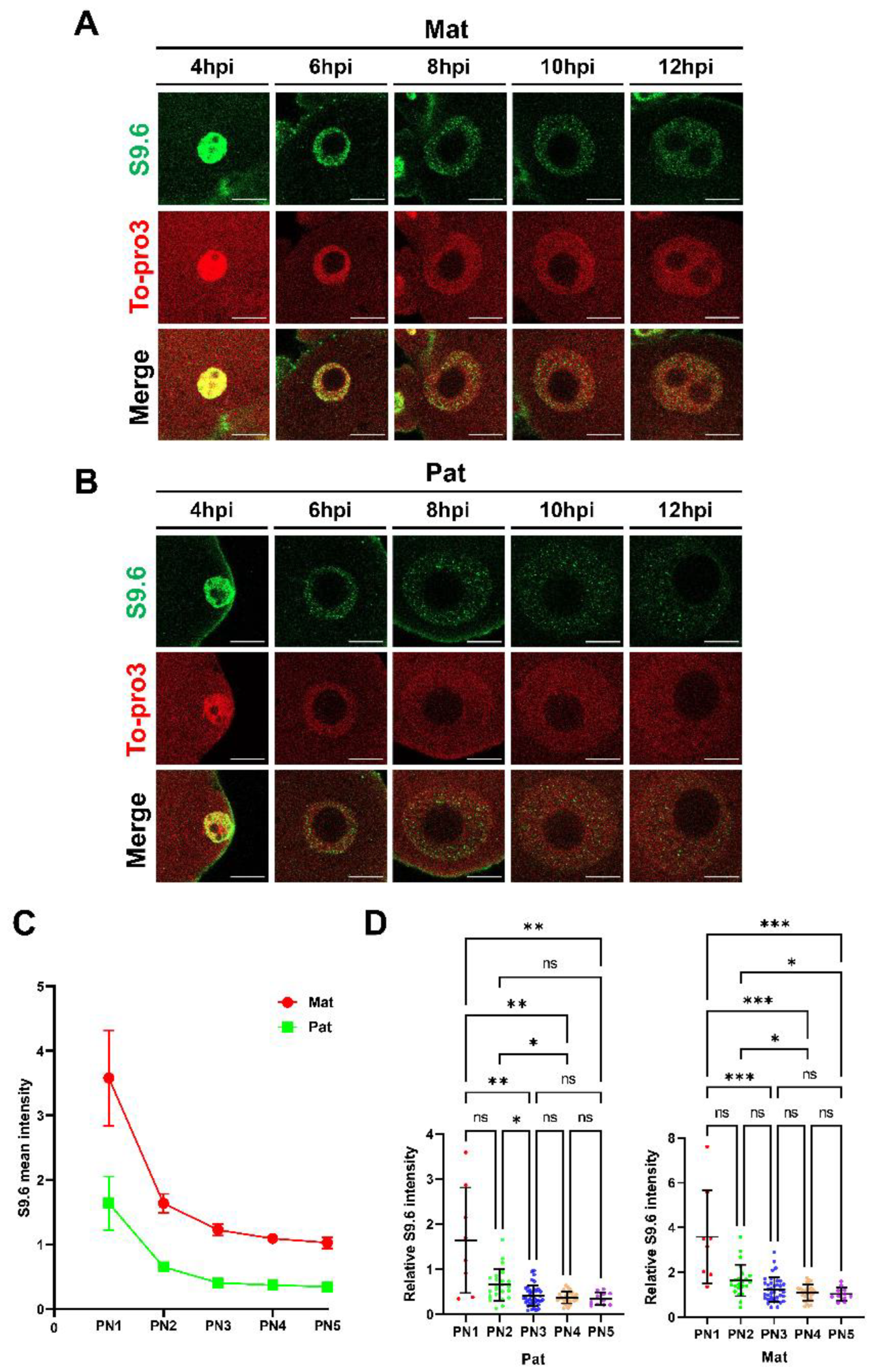
2.3. R-Loop Level Is Dynamically Changed in One-Cell Embryo
2.4. R-Loop Homeostasis Is Associated with Transcription and DNA Replication
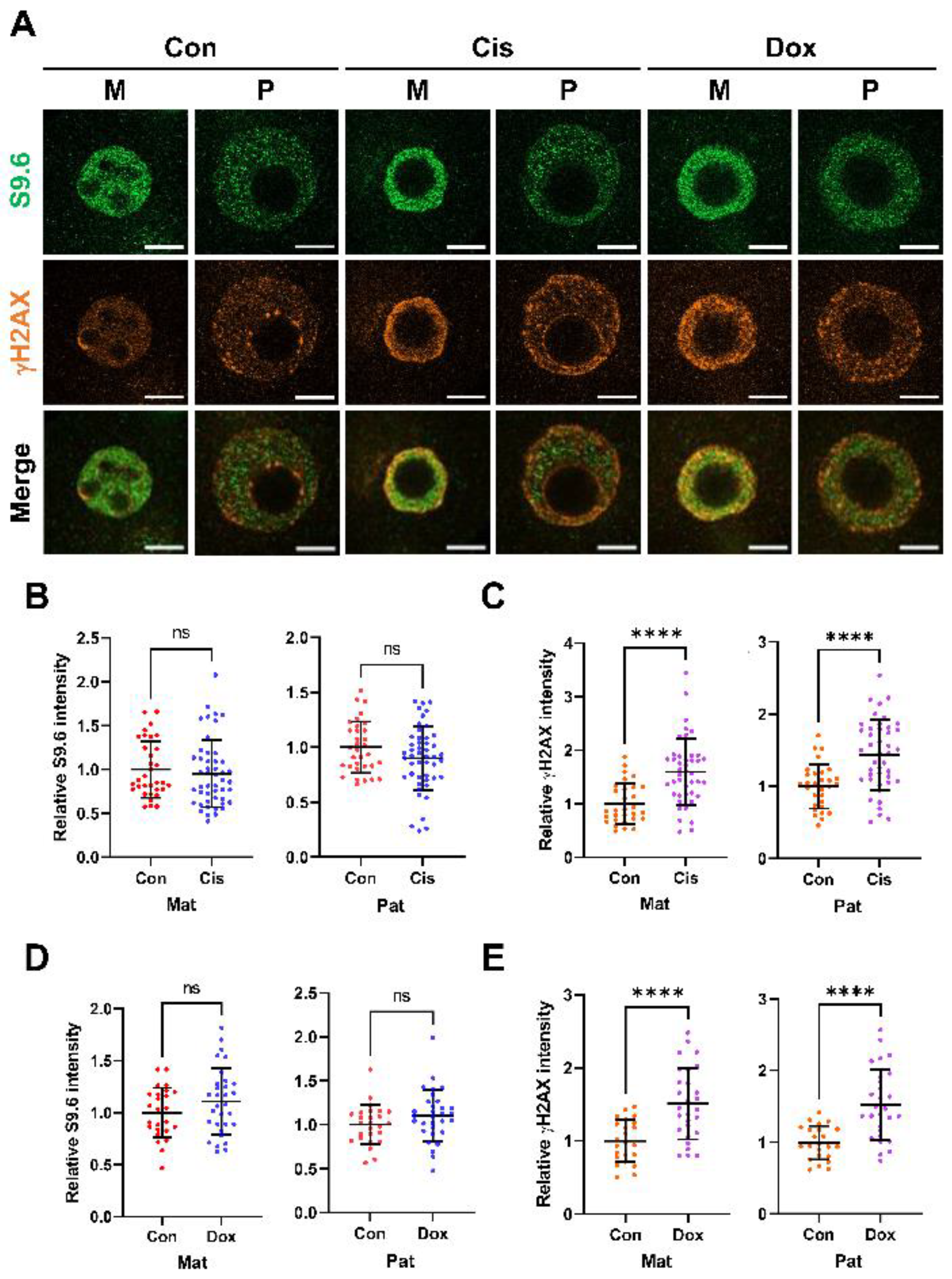
2.5. Anticancer Agents Do Not Alter Zygotic R-Loop Levels Globally
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Metaphase II Oocyte Collection and In Vitro Fertilization
4.3. Immunofluorescence and Quantification
4.4. Treatment of Inhibitors
4.5. Rnase H Treatment
4.6. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ram, P.T.; Schultz, R.M. Reporter gene expression in G2 of the 1-cell mouse embryo. Dev. Biol. 1993, 156, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.-i.; Funaya, S.; Tsukioka, D.; Kawamura, M.; Suzuki, Y.; Schultz, R.M.; Aoki, F. Minor zygotic gene activation is essential for mouse preimplantation development. Proc. Natl. Acad. Sci. USA 2018, 115, E6780–E6788. [Google Scholar] [CrossRef]
- Svoboda, P. Mammalian zygotic genome activation. Semin. Cell Dev. Biol. 2018, 84, 118–126. [Google Scholar] [CrossRef]
- Warner, C.M.; Versteegh, L.R. In vivo and in vitro effect of alpha-amanitin on preimplantation mouse embryo RNA polymerase. Nature 1974, 248, 678–680. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.-i.; Yamamoto, R.; Franke, V.; Cao, M.; Suzuki, Y.; Suzuki, M.G.; Vlahovicek, K.; Svoboda, P.; Schultz, R.M.; Aoki, F. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3’ processing. EMBO J. 2015, 34, 1523–1537. [Google Scholar] [CrossRef]
- Jukam, D.; Shariati, S.A.M.; Skotheim, J.M. Zygotic Genome Activation in Vertebrates. Dev. Cell 2017, 42, 316–332. [Google Scholar] [CrossRef] [PubMed]
- Schulz, K.N.; Harrison, M.M. Mechanisms regulating zygotic genome activation. Nat. Rev. Genet. 2019, 20, 221–234. [Google Scholar] [CrossRef]
- Schultz, R.M. Regulation of zygotic gene activation in the mouse. BioEssays News Rev. Mol. Cell. Dev. Biol. 1993, 15, 531–538. [Google Scholar] [CrossRef]
- Wu, D.; Dean, J. Maternal factors regulating preimplantation development in mice. Curr. Top. Dev. Biol. 2020, 140, 317–340. [Google Scholar] [CrossRef]
- Vastenhouw, N.L.; Cao, W.X.; Lipshitz, H.D. The maternal-to-zygotic transition revisited. Development 2019, 146, dev161471. [Google Scholar] [CrossRef]
- Latham, K.E.; Solter, D.; Schultz, R.M. Acquisition of a transcriptionally permissive state during the 1-cell stage of mouse embryogenesis. Dev. Biol. 1992, 149, 457–462. [Google Scholar] [CrossRef]
- Aoki, F. Zygotic gene activation in mice: Profile and regulation. J. Reprod. Dev. 2022, 68, 79–84. [Google Scholar] [CrossRef]
- Vassena, R.; Boué, S.; Gonzalez-Roca, E.; Aran, B.; Auer, H.; Veiga, A.; Belmonte, J.C.I. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development 2011, 138, 3699–3709. [Google Scholar] [CrossRef]
- Graf, A.; Krebs, S.; Zakhartchenko, V.; Schwalb, B.; Blum, H.; Wolf, E. Fine mapping of genome activation in bovine embryos by RNA sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, 4139–4144. [Google Scholar] [CrossRef]
- Halstead, M.M.; Ma, X.; Zhou, C.; Schultz, R.M.; Ross, P.J. Chromatin remodeling in bovine embryos indicates species-specific regulation of genome activation. Nat. Commun. 2020, 11, 4654. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Xu, Q.; Wang, Q.; Feng, S.; Lai, F.; Wang, P.; Zheng, F.; Xiang, Y.; Wu, J.; Nie, J.; et al. The landscape of RNA Pol II binding reveals a stepwise transition during ZGA. Nature 2020, 587, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, M.; Li, Y.; Zhang, Y. Profiling and functional characterization of maternal mRNA translation during mouse maternal-to-zygotic transition. Sci. Adv. 2022, 8, eabj3967. [Google Scholar] [CrossRef]
- Merrikh, H.; Machón, C.; Grainger, W.H.; Grossman, A.D.; Soultanas, P. Co-directional replication-transcription conflicts lead to replication restart. Nature 2011, 470, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Sankar, T.S.; Wastuwidyaningtyas, B.D.; Dong, Y.; Lewis, S.A.; Wang, J.D. The nature of mutations induced by replication–transcription collisions. Nature 2016, 535, 178–181. [Google Scholar] [CrossRef]
- Hamperl, S.; Bocek, M.J.; Saldivar, J.C.; Swigut, T.; Cimprich, K.A. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 2017, 170, 774–786.e19. [Google Scholar] [CrossRef]
- Munden, A.; Benton, M.L.; Capra, J.A.; Nordman, J.T. R-loop Mapping and Characterization During Drosophila Embryogenesis Reveals Developmental Plasticity in R-loop Signatures. J. Mol. Biol. 2022, 434, 167645. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; García-Muse, T. R loops: From transcription byproducts to threats to genome stability. Mol. Cell 2012, 46, 115–124. [Google Scholar] [CrossRef]
- White, R.L.; Hogness, D.S. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell 1977, 10, 177–192. [Google Scholar] [CrossRef]
- Drolet, M.; Bi, X.; Liu, L.F. Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J. Biol. Chem. 1994, 269, 2068–2074. [Google Scholar] [CrossRef]
- Wahba, L.; Costantino, L.; Tan, F.J.; Zimmer, A.; Koshland, D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016, 30, 1327–1338. [Google Scholar] [CrossRef]
- Xu, W.; Xu, H.; Li, K.; Fan, Y.; Liu, Y.; Yang, X.; Sun, Q. The R-loop is a common chromatin feature of the Arabidopsis genome. Nat. Plants 2017, 3, 704–714. [Google Scholar] [CrossRef]
- Sanz, L.A.; Hartono, S.R.; Lim, Y.W.; Steyaert, S.; Rajpurkar, A.; Ginno, P.A.; Xu, X.; Chédin, F. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol. Cell 2016, 63, 167–178. [Google Scholar] [CrossRef]
- Manzo, S.G.; Hartono, S.R.; Sanz, L.A.; Marinello, J.; Biasi, S.D.; Cossarizza, A.; Capranico, G.; Chedin, F. DNA Topoisomerase I differentially modulates R-loops across the human genome. Genome Biol. 2018, 19, 100. [Google Scholar] [CrossRef] [PubMed]
- Kuzminov, A. When DNA Topology Turns Deadly—RNA Polymerases Dig in Their R-Loops to Stand Their Ground: New Positive and Negative (Super)Twists in the Replication-Transcription Conflict. Trends Genet. TIG 2018, 34, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Manley, J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 2005, 122, 365–378. [Google Scholar] [CrossRef]
- Gómez-González, B.; García-Rubio, M.; Bermejo, R.; Gaillard, H.; Shirahige, K.; Marín, A.; Foiani, M.; Aguilera, A. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 2011, 30, 3106–3119. [Google Scholar] [CrossRef]
- Promonet, A.; Padioleau, I.; Liu, Y.; Sanz, L.; Biernacka, A.; Schmitz, A.-L.; Skrzypczak, M.; Sarrazin, A.; Mettling, C.; Rowicka, M.; et al. Topoisomerase 1 prevents replication stress at R-loop-enriched transcription termination sites. Nat. Commun. 2020, 11, 3940. [Google Scholar] [CrossRef]
- Skourti-Stathaki, K.; Proudfoot, N.J.; Gromak, N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 2011, 42, 794–805. [Google Scholar] [CrossRef]
- Cerritelli, S.M.; Crouch, R.J. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009, 276, 1494–1505. [Google Scholar] [CrossRef] [PubMed]
- Mersaoui, S.Y.; Yu, Z.; Coulombe, Y.; Karam, M.; Busatto, F.F.; Masson, J.-Y.; Richard, S. Arginine methylation of the DDX5 helicase RGG/RG motif by PRMT5 regulates resolution of RNA:DNA hybrids. EMBO J. 2019, 38, e100986. [Google Scholar] [CrossRef]
- Aoki, F.; Schultz, R.M. DNA replication in the 1-cell mouse embryo: Stimulatory effect of histone acetylation. Zygote 1999, 7, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Wossidlo, M.; Nakamura, T.; Lepikhov, K.; Marques, C.J.; Zakhartchenko, V.; Boiani, M.; Arand, J.; Nakano, T.; Reik, W.; Walter, J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2011, 2, 241. [Google Scholar] [CrossRef] [PubMed]
- Vanoosthuyse, V. Strengths and Weaknesses of the Current Strategies to Map and Characterize R-Loops. Non-Coding RNA 2018, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.-L.; Sun, S.-C.; Han, J.; Kwak, Y.-C.; Kim, N.-H.; Cui, X.-S. Doxorubicin induces early embryo apoptosis by inhibiting poly(ADP ribose) polymerase. In Vivo 2012, 26, 827–834. [Google Scholar] [PubMed]
- Hostetter, A.A.; Chapman, E.G.; DeRose, V.J. Rapid cross-linking of an RNA internal loop by the anticancer drug cisplatin. J. Am. Chem. Soc. 2009, 131, 9250–9257. [Google Scholar] [CrossRef]
- Al-Hadid, Q.; Yang, Y. R-loop: An emerging regulator of chromatin dynamics. Acta Biochim. Et Biophys. Sin. 2016, 48, 623–631. [Google Scholar] [CrossRef]
- Smolka, J.A.; Sanz, L.A.; Hartono, S.R.; Chédin, F. Recognition of RNA by the S9.6 antibody creates pervasive artifacts when imaging RNA:DNA hybrids. J. Cell Biol. 2021, 220, e202004079. [Google Scholar] [CrossRef] [PubMed]
- Hartono, S.R.; Malapert, A.; Legros, P.; Bernard, P.; Chédin, F.; Vanoosthuyse, V. The Affinity of the S9.6 Antibody for Double-Stranded RNAs Impacts the Accurate Mapping of R-Loops in Fission Yeast. J. Mol. Biol. 2018, 430, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Boguslawski, S.J.; Smith, D.E.; Michalak, M.A.; Mickelson, K.E.; Yehle, C.O.; Patterson, W.L.; Carrico, R.J. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods 1986, 89, 123–130. [Google Scholar] [CrossRef]
- Bou-Nader, C.; Bothra, A.; Garboczi, D.N.; Leppla, S.H.; Zhang, J. Structural basis of R-loop recognition by the S9.6 monoclonal antibody. Nat. Commun. 2022, 13, 1641. [Google Scholar] [CrossRef] [PubMed]
- König, F.; Schubert, T.; Längst, G. The monoclonal S9.6 antibody exhibits highly variable binding affinities towards different R-loop sequences. PLoS ONE 2017, 12, e0178875. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.C.; Coonen, E.; Ramaekers, F.C.S.; Delhanty, D.A.J.; Handyside, A.H.; Winston, R.M.L.; Hopman, A.H.N. Identification of the sex of human preimplantation embryos in two hours using an improved spreading method and fluorescent in-situ hybridization (FISH) using directly labelled probes. Hum. Reprod. 1994, 9, 721–724. [Google Scholar] [CrossRef]
- Barton, S.C.; Arney, K.L.; Niveleau, W.S.A.; Fundele, R.; Surani, M.A.; Haaf, T. Genome-wide methylation patterns in normal and uniparental early mouse embryos. Hum. Mol. Genet. 2001, 10, 2983–2987. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, J.J.; Grifo, J.A.; Krey, L.C. DNA methylation patterns in human tripronucleate zygotes. Mol. Hum. Reprod. 2005, 11, 167–171. [Google Scholar] [CrossRef]
- Santos, F.; Hendrich, B.; Reik, W.; Dean, W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2002, 241, 172–182. [Google Scholar] [CrossRef]
- Yamakawa, K.; Nakagomi, O. Improved detection of rotavirus RNA in dot-blot hybridization assay by chromatographic extraction and acid denaturation of double-stranded RNA. Mol. Cell. Probes 1990, 4, 415–418. [Google Scholar] [CrossRef]
- Graf, M.; Bonetti, D.; Lockhart, A.; Serhal, K.; Kellner, V.; Maicher, A.; Jolivet, P.; Teixeira, M.T.; Luke, B. Telomere Length Determines TERRA and R-Loop Regulation through the Cell Cycle. Cell 2017, 170, 72–85.e14. [Google Scholar] [CrossRef]
- Mishra, P.K.; Chakraborty, A.; Yeh, E.; Feng, W.; Bloom, K.S.; Basrai, M.A. R-loops at centromeric chromatin contribute to defects in kinetochore integrity and chromosomal instability in budding yeast. Mol. Biol. Cell 2021, 32, 74–89. [Google Scholar] [CrossRef] [PubMed]
- Wahba, L.; Gore, S.K.; Koshland, D. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. eLife 2013, 2, e00505. [Google Scholar] [CrossRef]
- Westover, K.D.; Bushnell, D.A.; Kornberg, R.D. Structural basis of transcription: Nucleotide selection by rotation in the RNA polymerase II active center. Cell 2004, 119, 481–489. [Google Scholar] [CrossRef]
- Aoki, F.; Worrad, D.M.; Schultz, R.M. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 1997, 181, 296–307. [Google Scholar] [CrossRef]
- Krokan, H.; Wist, E.; Krokan, R.H. Aphidicolin inhibits DNA synthesis by DNA polymerase alpha and isolated nuclei by a similar mechanism. Nucleic Acids Res. 1981, 9, 4709–4719. [Google Scholar] [CrossRef] [PubMed]
- Nascakova, Z.; Boleslavska, B.; Urban, V.; Oravetzova, A.; Vlachova, E.; Janscak, P.; Dobrovolna, J. RAD51 Inhibition Induces R-Loop Formation in Early G1 Phase of the Cell Cycle. Int. J. Mol. Sci. 2021, 22, 3740. [Google Scholar] [CrossRef] [PubMed]
- Martin-Alonso, M.S.; Soler-Oliva, M.E.; García-Rubio, M.; García-Muse, T.; Aguilera, A. Harmful R-loops are prevented via different cell cycle-specific mechanisms. Nat. Commun. 2021, 12, 4451. [Google Scholar] [CrossRef]
- Wossidlo, M.; Arand, J.; Sebastiano, V.; Lepikhov, K.; Boiani, M.; Reinhardt, R.; Schöler, H.; Walter, J. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 2010, 29, 1877–1888. [Google Scholar] [CrossRef]
- Rinaldi, C.; Pizzul, P.; Longhese, M.P.; Bonetti, D. Sensing R-Loop-Associated DNA Damage to Safeguard Genome Stability. Front. Cell Dev. Biol. 2021, 8, 618157. [Google Scholar] [CrossRef] [PubMed]
- Derijck, A.; Heijden, G.v.d.; Giele, M.; Philippens, M.; Boer, P.d. DNA double-strand break repair in parental chromatin of mouse zygotes, the first cell cycle as an origin of de novo mutation. Hum. Mol. Genet. 2008, 17, 1922–1937. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.; You, S.-Y.; Han, D.W.; La, H.; Park, C.; Yoo, S.; Kang, K.; Kang, M.-H.; Choi, Y.; Hong, K. Dynamic Change of R-Loop Implicates in the Regulation of Zygotic Genome Activation in Mouse. Int. J. Mol. Sci. 2022, 23, 14345. https://doi.org/10.3390/ijms232214345
Lee H, You S-Y, Han DW, La H, Park C, Yoo S, Kang K, Kang M-H, Choi Y, Hong K. Dynamic Change of R-Loop Implicates in the Regulation of Zygotic Genome Activation in Mouse. International Journal of Molecular Sciences. 2022; 23(22):14345. https://doi.org/10.3390/ijms232214345
Chicago/Turabian StyleLee, Hyeonji, Seong-Yeob You, Dong Wook Han, Hyeonwoo La, Chanhyeok Park, Seonho Yoo, Kiye Kang, Min-Hee Kang, Youngsok Choi, and Kwonho Hong. 2022. "Dynamic Change of R-Loop Implicates in the Regulation of Zygotic Genome Activation in Mouse" International Journal of Molecular Sciences 23, no. 22: 14345. https://doi.org/10.3390/ijms232214345
APA StyleLee, H., You, S.-Y., Han, D. W., La, H., Park, C., Yoo, S., Kang, K., Kang, M.-H., Choi, Y., & Hong, K. (2022). Dynamic Change of R-Loop Implicates in the Regulation of Zygotic Genome Activation in Mouse. International Journal of Molecular Sciences, 23(22), 14345. https://doi.org/10.3390/ijms232214345







