Transcriptome Analysis of Goat Mammary Gland Tissue Reveals the Adaptive Strategies and Molecular Mechanisms of Lactation and Involution
Abstract
:1. Introduction
2. Results
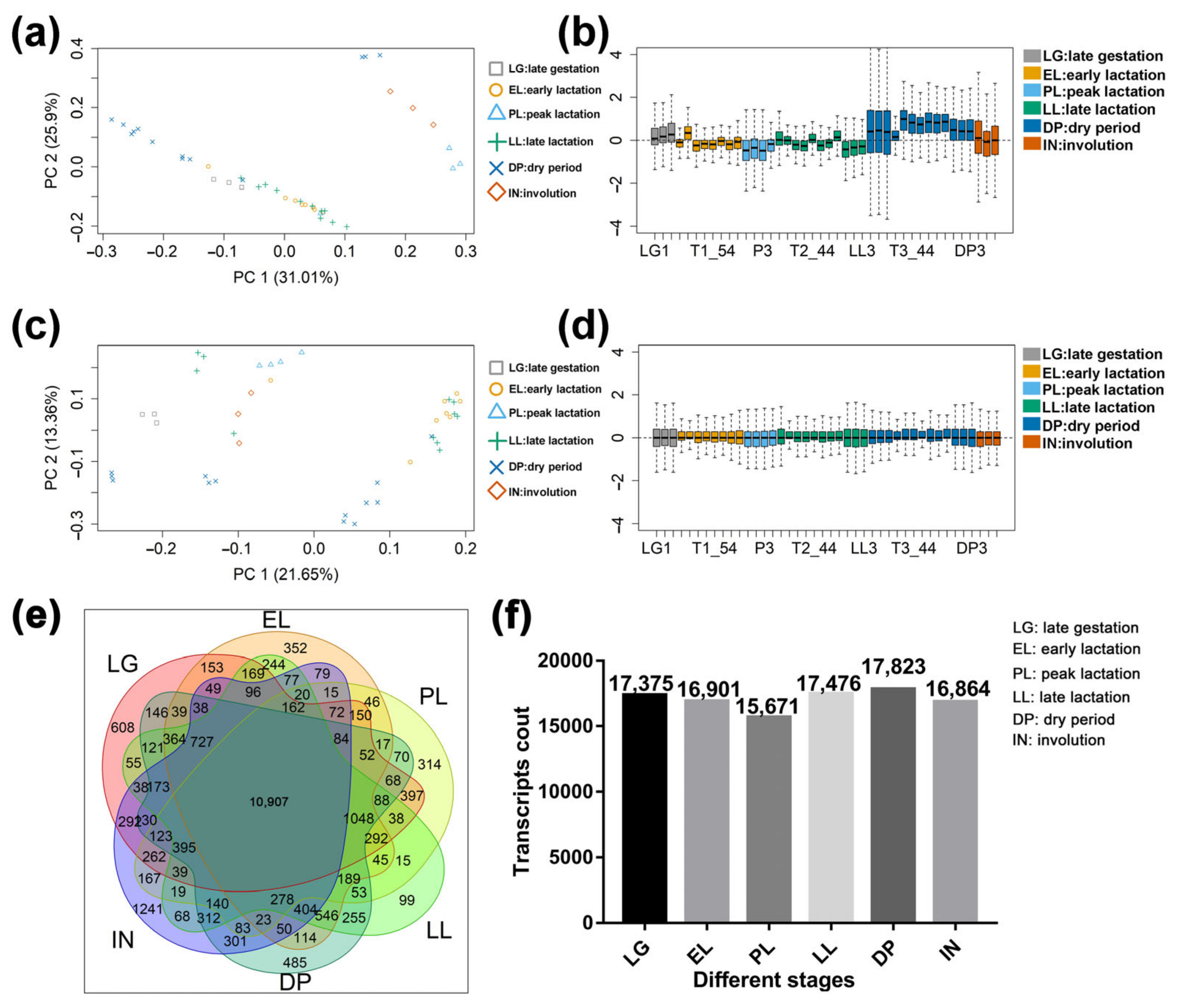
2.1. Basic Statistical Analysis Results of Sequencing Data
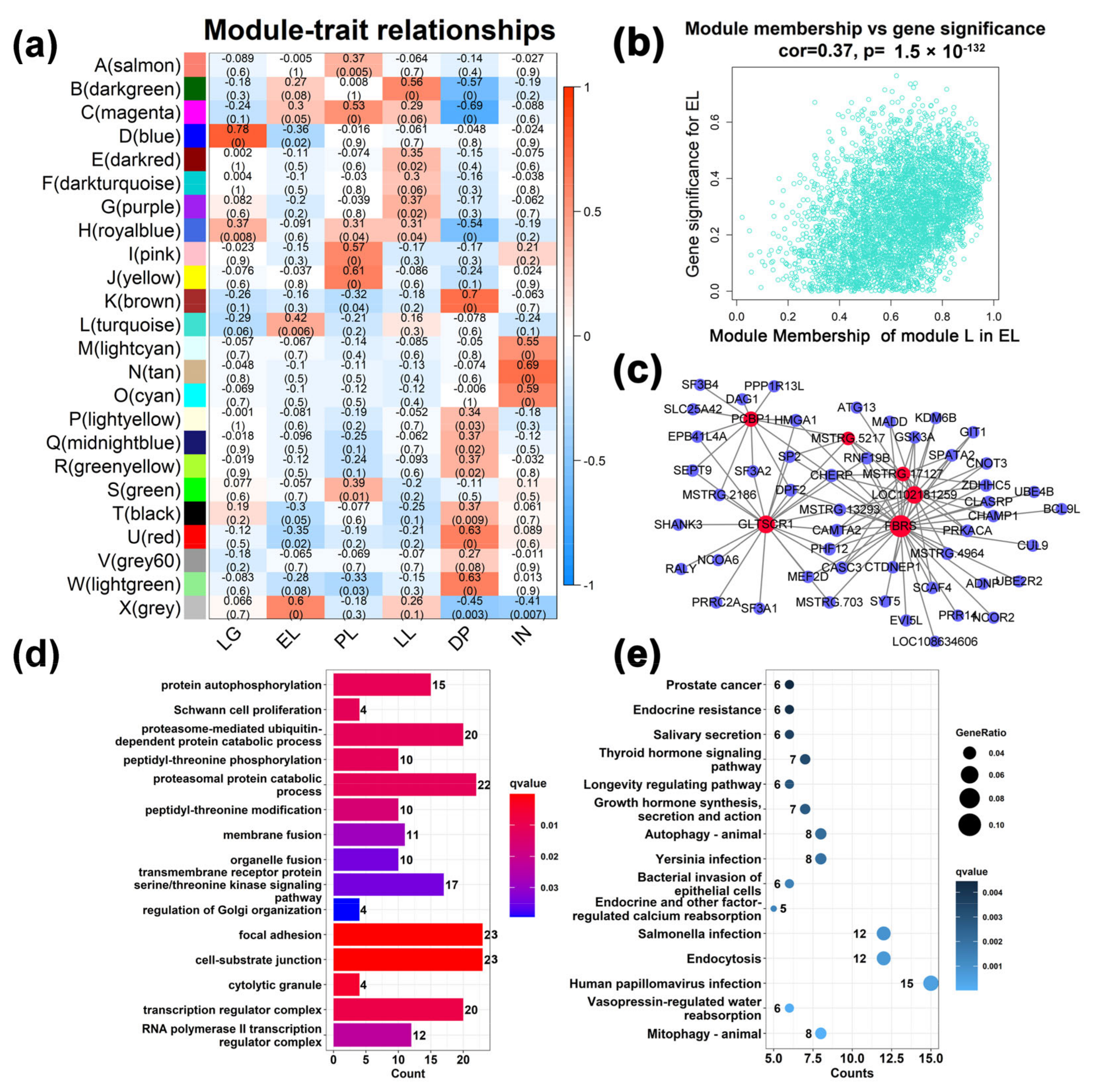
2.2. Identification of Co-Expression Modules Associated with Different Stages of Mammary Gland Development
2.3. Analysis of Gene Function in Co-Expression Modules
2.4. Differential Expression Analysis Results
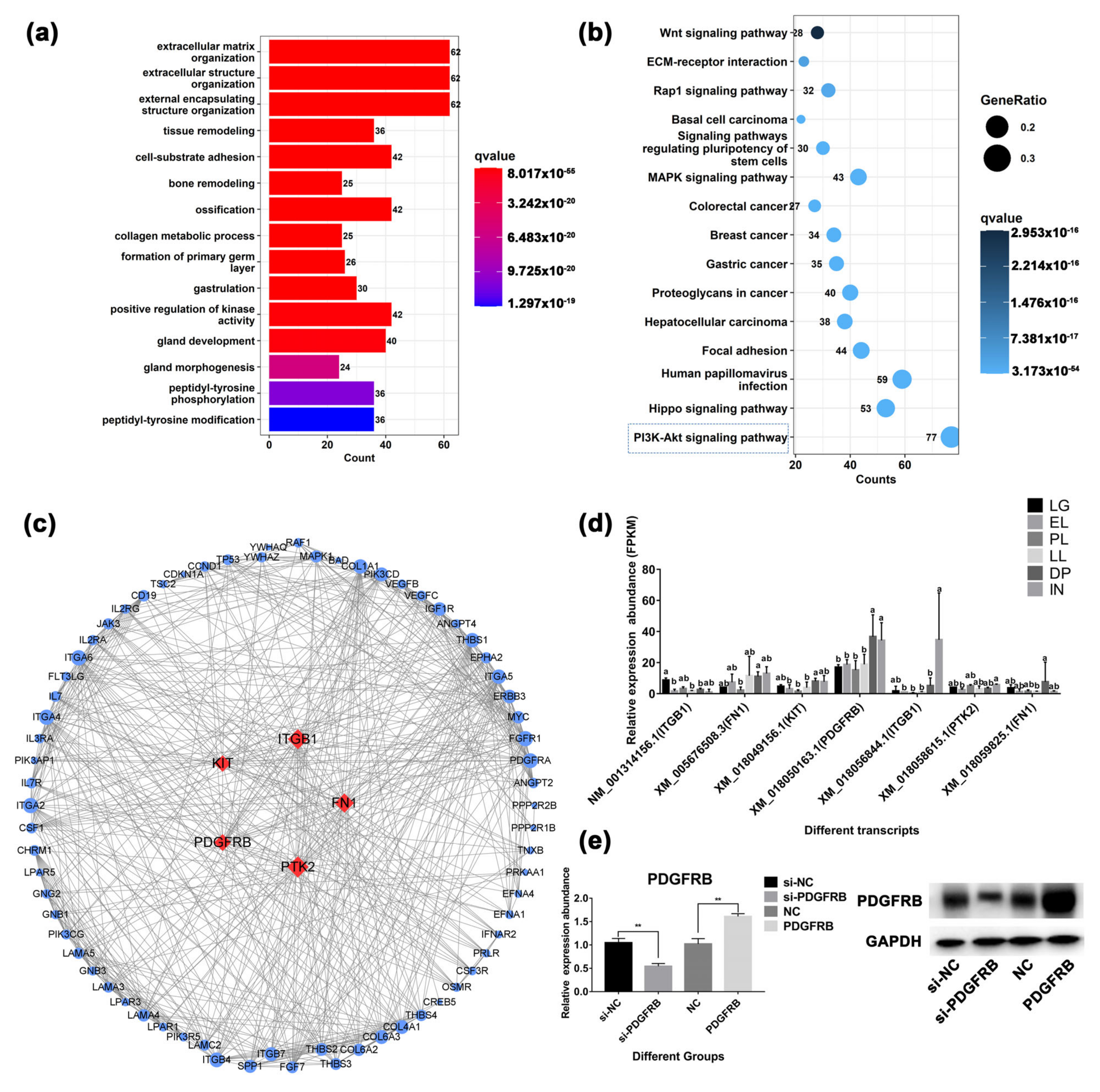
2.5. GO and KEGG Pathway Enrichment Analysis of DETs
2.6. Substance Synthesis, Transport and Metabolism Related Gene Analysis
2.7. Analysis of Immune Function Genes Expressed in Mammary Glands
2.8. Analysis of Genes Related to Cell Apoptosis and Autophagy
2.9. Analysis of Genes Related to Mammary Gland Development and Cell Growth
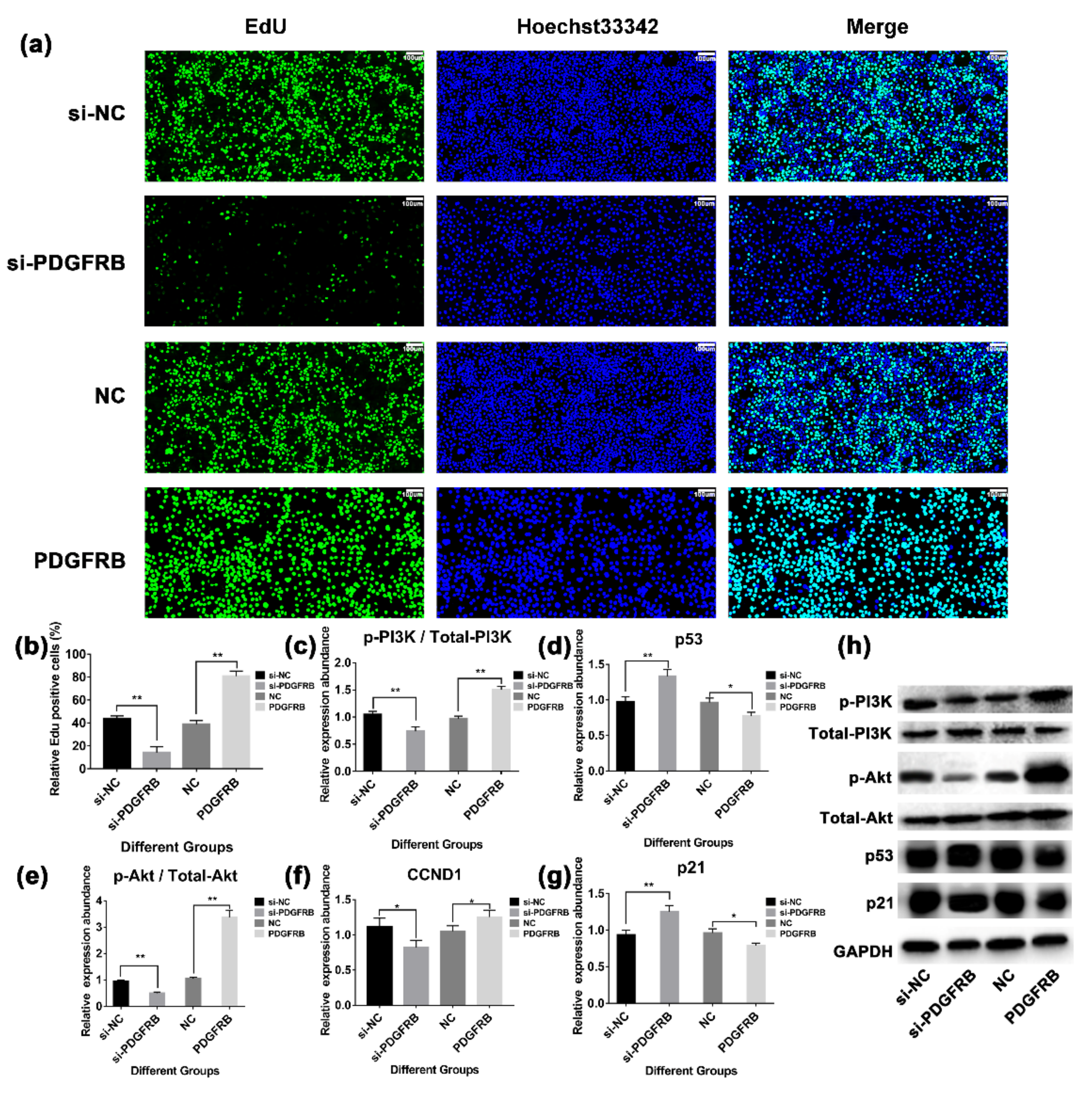
2.10. PDGFRB Overexpression In Vitro Promotes the Proliferation of Mammary Epithelial Cells by Activating PI3K-Akt Signaling Pathway
2.11. Overexpression of PDGFRB Inhibits Apoptosis of Mammary Epithelial Cells and Reduces Cell Cycle Arrest in G1/S Phase
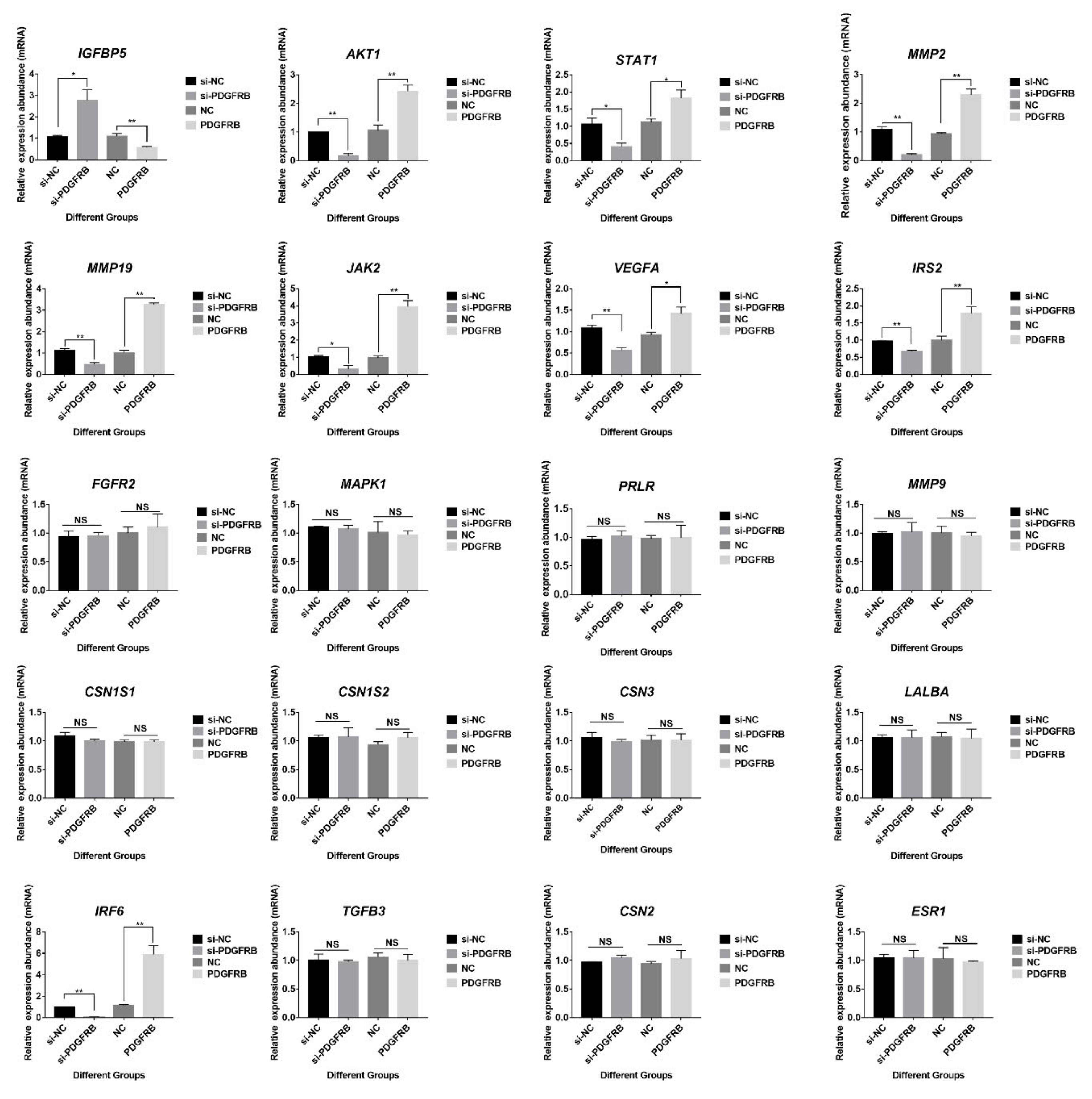
2.12. Effects of PDGFRB on Genes Related to Lactation Function and Mammary Gland Involution
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
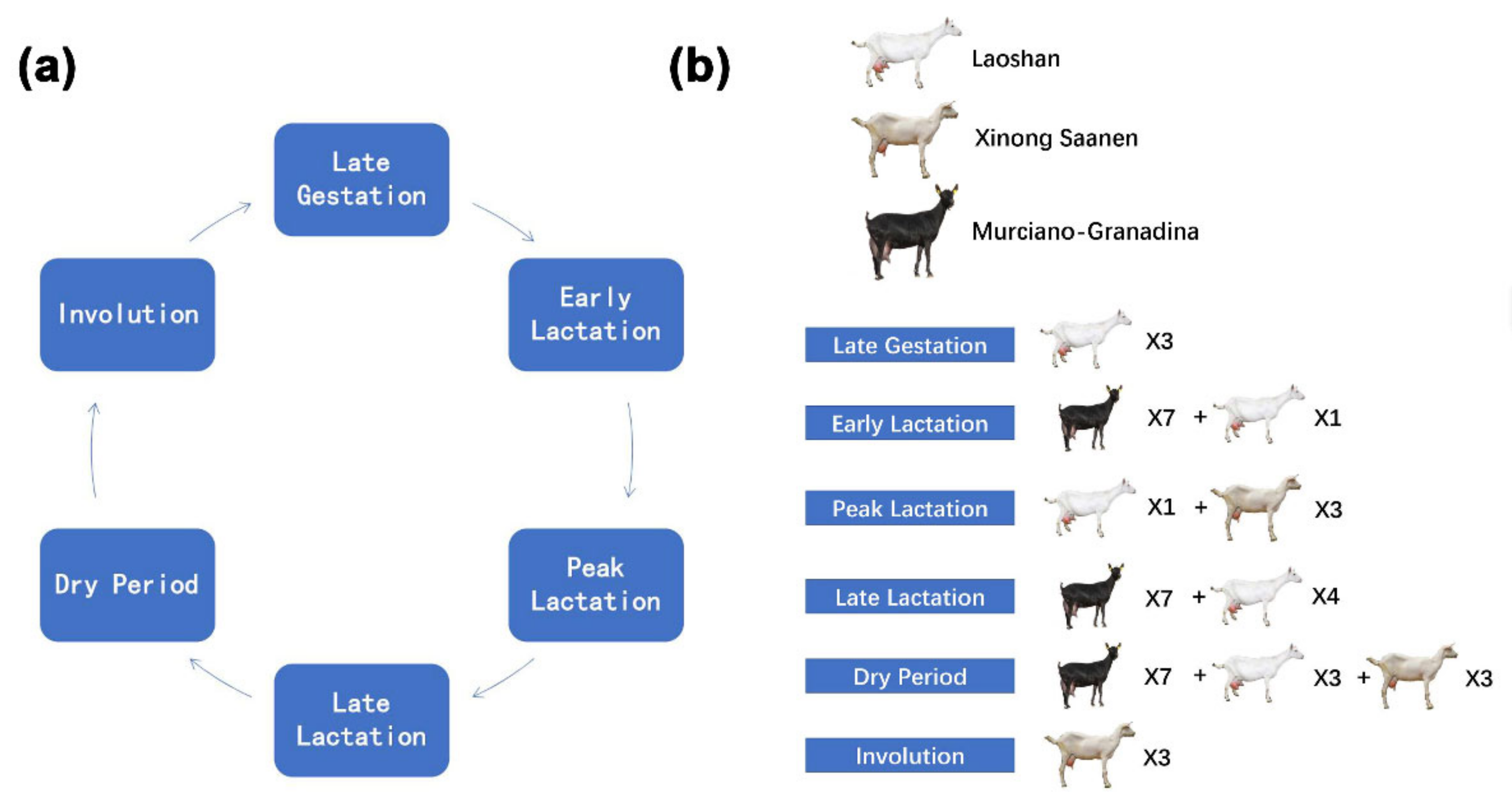
4.2. Collection of Goat Mammary Gland Samples
4.3. Sequencing Data Analysis
4.4. Weighted Gene Co-Expression Network Analysis
4.5. Differential Expression Analysis and Gene Expression Pattern Analysis
4.6. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
4.7. Goat Mammary Epithelial Cells Culture
4.8. Cell Transfection
4.9. Cell Proliferation Assay
4.10. Apoptosis and Cell Cycle Detection Assays
4.11. Western Blotting
4.12. Real-Time Quantitative Reverse-Transcription PCR Assay
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oftedal, O.T. The mammary gland and its origin during synapsid evolution. J. Mammary Gland Biol. Neoplasia 2002, 7, 225–252. [Google Scholar] [CrossRef]
- Macias, H.; Hinck, L. Mammary gland development. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 533–557. [Google Scholar] [CrossRef] [Green Version]
- Truchet, S.; Honvo-Houéto, E. Physiology of milk secretion. Best Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 367–384. [Google Scholar] [CrossRef]
- Zhao, X.; Ponchon, B.; Lanctôt, S.; Lacasse, P. Invited review: Accelerating mammary gland involution after drying-off in dairy cattle. J. Dairy Sci. 2019, 102, 6701–6717. [Google Scholar] [CrossRef]
- Hurley, W.L. Mammary gland function during involution. J. Dairy Sci. 1989, 72, 1637–1646. [Google Scholar] [CrossRef]
- Inman, J.L.; Robertson, C.; Mott, J.D.; Bissell, M.J. Mammary gland development: Cell fate specification, stem cells and the microenvironment. Development 2015, 142, 1028–1042. [Google Scholar] [CrossRef] [Green Version]
- Paten, A.M.; Duncan, E.J.; Pain, S.J.; Peterson, S.W.; Kenyon, P.R.; Blair, H.T.; Dearden, P.K. Functional development of the adult ovine mammary gland—Insights from gene expression profiling. BMC Genom. 2015, 16, 748. [Google Scholar] [CrossRef] [Green Version]
- Xuan, R.; Chao, T.; Zhao, X.; Wang, A.; Chu, Y.; Li, Q.; Zhao, Y.; Ji, Z.; Wang, J. Transcriptome profiling of the nonlactating mammary glands of dairy goats reveals the molecular genetic mechanism of mammary cell remodeling. J. Dairy Sci. 2022, 105, 5238–5260. [Google Scholar] [CrossRef]
- Ji, Z.; Chao, T.; Liu, Z.; Hou, L.; Wang, J.; Wang, A.; Zhou, J.; Xuan, R.; Wang, G.; Wang, J. Genome-wide integrated analysis demonstrates widespread functions of lncRNAs in mammary gland development and lactation in dairy goats. BMC Genom. 2020, 21, 254. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Zhu, J.; Shi, H.; Luo, J.; Zhao, W.; Shi, H.; Xu, H.; Wang, H.; Loor, J.J. Comprehensive Transcriptome Profiling of Dairy Goat Mammary Gland Identifies Genes and Networks Crucial for Lactation and Fatty Acid Metabolism. Front. Genet. 2020, 11, 878. [Google Scholar] [CrossRef]
- Shi, H.; Zhu, J.; Luo, J.; Cao, W.; Shi, H.; Yao, D.; Li, J.; Sun, Y.; Xu, H.; Yu, K.; et al. Genes regulating lipid and protein metabolism are highly expressed in mammary gland of lactating dairy goats. Funct. Integr. Genom. 2015, 15, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, H.; Hickford, J.G.H.; Hao, Z.; Shen, J.; Luo, Y.; Hu, J.; Liu, X.; Li, S. Comparison of the Transcriptome of the Ovine Mammary Gland in Lactating and Non-lactating Small-Tailed Han Sheep. Front. Genet. 2020, 11, 472. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.L.; Yang, J.X.; Li, Z.; Liu, H.Y.; Liu, J.X. Progress on the miRNA related with mammary gland development and lactation. Yi Chuan = Hereditas 2013, 35, 695–702. [Google Scholar] [CrossRef]
- Dai, W.T.; Zou, Y.X.; White, R.R.; Liu, J.X.; Liu, H.Y. Transcriptomic profiles of the bovine mammary gland during lactation and the dry period. Funct. Integr. Genom. 2018, 18, 125–140. [Google Scholar] [CrossRef]
- Palombo, V.; Loor, J.J.; D’Andrea, M.; Vailati-Riboni, M.; Shahzad, K.; Krogh, U.; Theil, P.K. Transcriptional profiling of swine mammary gland during the transition from colostrogenesis to lactogenesis using RNA sequencing. BMC Genom. 2018, 19, 322. [Google Scholar] [CrossRef] [Green Version]
- Trott, J.F.; Schennink, A.; Horigan, K.C.; Lemay, D.G.; Cohen, J.R.; Famula, T.R.; Dragon, J.A.; Hovey, R.C. Unique Transcriptomic Changes Underlie Hormonal Interactions During Mammary Histomorphogenesis in Female Pigs. Endocrinology 2022, 163, bqab256. [Google Scholar] [CrossRef]
- Pal, B.; Chen, Y.; Milevskiy, M.J.G.; Vaillant, F.; Prokopuk, L.; Dawson, C.A.; Capaldo, B.D.; Song, X.; Jackling, F.; Timpson, P.; et al. Single cell transcriptome atlas of mouse mammary epithelial cells across development. Breast Cancer Res. BCR 2021, 23, 69. [Google Scholar] [CrossRef]
- Mitra, S.D.; Ganaie, F.; Bankar, K.; Velu, D.; Mani, B.; Vasudevan, M.; Shome, R.; Rahman, H.; Kumar Ghosh, S.; Shome, B.R. Genome-wide analysis of mammary gland shows modulation of transcriptome landscape with alternative splice variants in Staphylococcus aureus mastitis in mice. Gene 2020, 735, 144278. [Google Scholar] [CrossRef]
- Farhadian, M.; Rafat, S.A.; Panahi, B.; Mayack, C. Weighted gene co-expression network analysis identifies modules and functionally enriched pathways in the lactation process. Sci. Rep. 2021, 11, 2367. [Google Scholar] [CrossRef]
- Fan, Y.; Arbab, A.A.I.; Zhang, H.; Yang, Y.; Nazar, M.; Han, Z.; Yang, Z. Lactation Associated Genes Revealed in Holstein Dairy Cows by Weighted Gene Co-Expression Network Analysis (WGCNA). Animals 2021, 11, 314. [Google Scholar] [CrossRef]
- Jiang, C.; Wu, S.; Jiang, L.; Gao, Z.; Li, X.; Duan, Y.; Li, N.; Sun, T. Network-based approach to identify biomarkers predicting response and prognosis for HER2-negative breast cancer treatment with taxane-anthracycline neoadjuvant chemotherapy. PeerJ 2019, 7, e7515. [Google Scholar] [CrossRef]
- Dado-Senn, B.; Skibiel, A.L.; Fabris, T.F.; Dahl, G.E.; Laporta, J. Dry period heat stress induces microstructural changes in the lactating mammary gland. PLoS ONE 2019, 14, e0222120. [Google Scholar] [CrossRef] [Green Version]
- Fabris, T.F.; Laporta, J.; Skibiel, A.L.; Dado-Senn, B.; Wohlgemuth, S.E.; Dahl, G.E. Effect of heat stress during the early and late dry period on mammary gland development of Holstein dairy cattle. J. Dairy Sci. 2020, 103, 8576–8586. [Google Scholar] [CrossRef]
- Kim, S.W.; Easter, R.A.; Hurley, W.L. The regression of unsuckled mammary glands during lactation in sows: The influence of lactation stage, dietary nutrients, and litter size. J. Anim. Sci. 2001, 79, 2659–2668. [Google Scholar] [CrossRef] [Green Version]
- Oliver, S.P.; Mitchell, B.A. Susceptibility of bovine mammary gland to infections during the dry period. J. Dairy Sci. 1983, 66, 1162–1166. [Google Scholar] [CrossRef]
- Gross, J.J.; Bruckmaier, R.M. Invited review: Metabolic challenges and adaptation during different functional stages of the mammary gland in dairy cows: Perspectives for sustainable milk production. J. Dairy Sci. 2019, 102, 2828–2843. [Google Scholar] [CrossRef] [Green Version]
- Heldin, C.H.; Lennartsson, J. Structural and functional properties of platelet-derived growth factor and stem cell factor receptors. Cold Spring Harb. Perspect. Biol. 2013, 5, a009100. [Google Scholar] [CrossRef] [Green Version]
- Andrae, J.; Gallini, R.; Betsholtz, C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008, 22, 1276–1312. [Google Scholar] [CrossRef] [Green Version]
- Weissmueller, S.; Manchado, E.; Saborowski, M.; Morris, J.P., IV; Wagenblast, E.; Davis, C.A.; Moon, S.H.; Pfister, N.T.; Tschaharganeh, D.F.; Kitzing, T.; et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling. Cell 2014, 157, 382–394. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Z.; Feng, T.; Lu, Z.; Wei, Y.; Meng, J.; Lin, C.P.; Zhou, B.; Liu, C.; Zhang, H. PDGFRb(+) mesenchymal cells, but not NG2(+) mural cells, contribute to cardiac fat. Cell Rep. 2021, 34, 108697. [Google Scholar] [CrossRef]
- Knight, C.H.; Peaker, M. Development of the mammary gland. J. Reprod. Fertil. 1982, 65, 521–536. [Google Scholar] [CrossRef] [Green Version]
- Ji, Z.; Chao, T.; Zhang, C.; Liu, Z.; Hou, L.; Wang, J.; Wang, A.; Wang, Y.; Zhou, J.; Xuan, R.; et al. Transcriptome Analysis of Dairy Goat Mammary Gland Tissues from Different Lactation Stages. DNA Cell Biol. 2019, 38, 129–143. [Google Scholar] [CrossRef]
- Xuan, R.; Chao, T.; Wang, A.; Zhang, F.; Sun, P.; Liu, S.; Guo, M.; Wang, G.; Ji, Z.; Wang, J.; et al. Characterization of microRNA profiles in the mammary gland tissue of dairy goats at the late lactation, dry period and late gestation stages. PLoS ONE 2020, 15, e0234427. [Google Scholar] [CrossRef]
- Lin, J.; Bao, Z.K.; Zhang, Q.; Hu, W.W.; Yu, Q.H.; Yang, Q. Transcriptome analysis of the mammary gland from GH transgenic goats during involution. Gene 2015, 565, 228–234. [Google Scholar] [CrossRef]
- Guan, D.; Landi, V.; Luigi-Sierra, M.G.; Delgado, J.V.; Such, X.; Castelló, A.; Cabrera, B.; Mármol-Sánchez, E.; Fernández-Alvarez, J.; de la Torre Casañas, J.L.R.; et al. Analyzing the genomic and transcriptomic architecture of milk traits in Murciano-Granadina goats. J. Anim. Sci. Biotechnol. 2020, 11, 35. [Google Scholar] [CrossRef] [Green Version]
- Hao, Z.; Zhou, H.; Hickford, J.G.H.; Gong, H.; Wang, J.; Hu, J.; Liu, X.; Li, S.; Zhao, M.; Luo, Y. Transcriptome Profile Analysis of Mammary Gland Tissue from Two Breeds of Lactating Sheep. Genes 2019, 10, 781. [Google Scholar] [CrossRef] [Green Version]
- Farhadian, M.; Rafat, S.A.; Panahi, B.; Ebrahimie, E. Transcriptome signature of two lactation stages in Ghezel sheep identifies using RNA-Sequencing. Anim. Biotechnol. 2022, 33, 223–233. [Google Scholar] [CrossRef]
- Li, S.; Wang, Q.; Lin, X.; Jin, X.; Liu, L.; Wang, C.; Chen, Q.; Liu, J.; Liu, H. The Use of "Omics" in Lactation Research in Dairy Cows. Int. J. Mol. Sci. 2017, 18, 983. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; Ning, C.; Zhao, P.; Feng, W.; Jin, Y.; Zhou, L.; Yu, Y.; Liu, J. Integrated analysis of long noncoding RNA and mRNA expression profiles reveals the potential role of long noncoding RNA in different bovine lactation stages. J. Dairy Sci. 2018, 101, 11061–11073. [Google Scholar] [CrossRef] [Green Version]
- Saeki, K.; Chang, G.; Kanaya, N.; Wu, X.; Wang, J.; Bernal, L.; Ha, D.; Neuhausen, S.L.; Chen, S. Mammary cell gene expression atlas links epithelial cell remodeling events to breast carcinogenesis. Commun. Biol. 2021, 4, 660. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Bakhtiarizadeh, M.R.; Mirzaei, S.; Norouzi, M.; Sheybani, N.; Vafaei Sadi, M.S. Identification of Gene Modules and Hub Genes Involved in Mastitis Development Using a Systems Biology Approach. Front. Genet. 2020, 11, 722. [Google Scholar] [CrossRef]
- Brisken, C.; Rajaram, R.D. Alveolar and lactogenic differentiation. J. Mammary Gland Biol. Neoplasia 2006, 11, 239–248. [Google Scholar] [CrossRef] [Green Version]
- Nonnecke, B.J.; Smith, K.L. Inhibition of mastitic bacteria by bovine milk apo-lactoferrin evaluated by in vitro microassay of bacterial growth. J. Dairy Sci. 1984, 67, 606–613. [Google Scholar] [CrossRef]
- Mariño, G.; Niso-Santano, M.; Baehrecke, E.H.; Kroemer, G. Self-consumption: The interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2014, 15, 81–94. [Google Scholar] [CrossRef] [Green Version]
- Ollier, S.; Robert-Granié, C.; Bernard, L.; Chilliard, Y.; Leroux, C. Mammary transcriptome analysis of food-deprived lactating goats highlights genes involved in milk secretion and programmed cell death. J. Nutr. 2007, 137, 560–567. [Google Scholar] [CrossRef] [Green Version]
- Motyl, T.; Gajkowska, B.; Zarzyńska, J.; Gajewska, M.; Lamparska-Przybysz, M. Apoptosis and autophagy in mammary gland remodeling and breast cancer chemotherapy. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2006, 57 (Suppl. S7), 17–32. [Google Scholar]
- Halaby, R. Mammary Gland Cell Death Also Involves Lysosomal Autophagy. Breast Cancer Res. 2001, 1, 4. [Google Scholar] [CrossRef] [Green Version]
- Monks, J.; Henson, P.M. Differentiation of the mammary epithelial cell during involution: Implications for breast cancer. J. Mammary Gland Biol. Neoplasia 2009, 14, 159–170. [Google Scholar] [CrossRef]
- Gajewska, M.; Gajkowska, B.; Motyl, T. Apoptosis and autophagy induced by TGF-B1 in bovine mammary epithelial BME-UV1 cells. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2005, 56 (Suppl. S3), 143–157. [Google Scholar]
- Majeski, A.E.; Dice, J.F. Mechanisms of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 2004, 36, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Bursch, W.; Hochegger, K.; Torok, L.; Marian, B.; Ellinger, A.; Hermann, R.S. Autophagic and apoptotic types of programmed cell death exhibit different fates of cytoskeletal filaments. J. Cell Sci. 2000, 113 Pt 7, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.J.; McNamara, J.P.; Wallace, C.R.; Dehoff, M.H. A review of endocrine regulation of metabolism during lactation. J. Anim. Sci. 1984, 59, 498–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, L.; He, C.; Li, Z.; Wang, Z.; Zhang, Q. FBP1 modulates cell metabolism of breast cancer cells by inhibiting the expression of HIF-1α. Neoplasma 2017, 64, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Kremer, D.M.; Nelson, B.S.; Lin, L.; Yarosz, E.L.; Halbrook, C.J.; Kerk, S.A.; Sajjakulnukit, P.; Myers, A.; Thurston, G.; Hou, S.W.; et al. GOT1 inhibition promotes pancreatic cancer cell death by ferroptosis. Nat. Commun. 2021, 12, 4860. [Google Scholar] [CrossRef] [PubMed]
- Coloff, J.L.; Murphy, J.P.; Braun, C.R.; Harris, I.S.; Shelton, L.M.; Kami, K.; Gygi, S.P.; Selfors, L.M.; Brugge, J.S. Differential Glutamate Metabolism in Proliferating and Quiescent Mammary Epithelial Cells. Cell Metab. 2016, 23, 867–880. [Google Scholar] [CrossRef]
- Zhou, X.; Curbo, S.; Li, F.; Krishnan, S.; Karlsson, A. Inhibition of glutamate oxaloacetate transaminase 1 in cancer cell lines results in altered metabolism with increased dependency of glucose. BMC Cancer 2018, 18, 559. [Google Scholar] [CrossRef]
- Jonker, J.W.; Merino, G.; Musters, S.; van Herwaarden, A.E.; Bolscher, E.; Wagenaar, E.; Mesman, E.; Dale, T.C.; Schinkel, A.H. The breast cancer resistance protein BCRP (ABCG2) concentrates drugs and carcinogenic xenotoxins into milk. Nat. Med. 2005, 11, 127–129. [Google Scholar] [CrossRef]
- Alcorn, J.; Lu, X.; Moscow, J.A.; McNamara, P.J. Transporter gene expression in lactating and nonlactating human mammary epithelial cells using real-time reverse transcription-polymerase chain reaction. J. Pharmacol. Exp. Ther. 2002, 303, 487–496. [Google Scholar] [CrossRef] [Green Version]
- Gilchrist, S.E.; Alcorn, J. Lactation stage-dependent expression of transporters in rat whole mammary gland and primary mammary epithelial organoids. Fundam. Clin. Pharmacol. 2010, 24, 205–214. [Google Scholar] [CrossRef]
- Hediger, M.A.; Romero, M.F.; Peng, J.B.; Rolfs, A.; Takanaga, H.; Bruford, E.A. The ABCs of solute carriers: Physiological, pathological and therapeutic implications of human membrane transport proteinsIntroduction. Pflug. Archiv. Eur. J. Physiol. 2004, 447, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Golabek, A.A.; Kida, E.; Walus, M.; Kaczmarski, W.; Michalewski, M.; Wisniewski, K.E. CLN3 protein regulates lysosomal pH and alters intracellular processing of Alzheimer’s amyloid-beta protein precursor and cathepsin D in human cells. Mol. Genet. Metab. 2000, 70, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Rusyn, E.; Mousallem, T.; Persaud-Sawin, D.A.; Miller, S.; Boustany, R.M. CLN3p impacts galactosylceramide transport, raft morphology, and lipid content. Pediatric Res. 2008, 63, 625–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.F.; Duan, J.J.; Wang, J.; Li, L.; Wang, D.; Liu, X.Z.; Yang, J.; Zhang, H.R.; Lv, J.; Yang, Y.J.; et al. Inhibition of the ALDH18A1-MYCN positive feedback loop attenuates MYCN-amplified neuroblastoma growth. Sci. Transl. Med. 2020, 12, eaax8694. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, C.; Delgado, P.; Busse, C.E.; Sanz-Bravo, A.; Martos-Folgado, I.; Bonzon-Kulichenko, E.; Ferrarini, A.; Gonzalez-Valdes, I.B.; Mur, S.M.; Roldán-Montero, R.; et al. ALDH4A1 is an atherosclerosis auto-antigen targeted by protective antibodies. Nature 2021, 589, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Honeth, G.; Lombardi, S.; Ginestier, C.; Hur, M.; Marlow, R.; Buchupalli, B.; Shinomiya, I.; Gazinska, P.; Bombelli, S.; Ramalingam, V.; et al. Aldehyde dehydrogenase and estrogen receptor define a hierarchy of cellular differentiation in the normal human mammary epithelium. Breast Cancer Res. BCR 2014, 16, R52. [Google Scholar] [CrossRef] [Green Version]
- Eirew, P.; Kannan, N.; Knapp, D.J.; Vaillant, F.; Emerman, J.T.; Lindeman, G.J.; Visvader, J.E.; Eaves, C.J. Aldehyde dehydrogenase activity is a biomarker of primitive normal human mammary luminal cells. Stem. Cells 2012, 30, 344–348. [Google Scholar] [CrossRef]
- Kim, R.J.; Park, J.R.; Roh, K.J.; Choi, A.R.; Kim, S.R.; Kim, P.H.; Yu, J.H.; Lee, J.W.; Ahn, S.H.; Gong, G.; et al. High aldehyde dehydrogenase activity enhances stem cell features in breast cancer cells by activating hypoxia-inducible factor-2α. Cancer Lett. 2013, 333, 18–31. [Google Scholar] [CrossRef]
- Storms, R.W.; Trujillo, A.P.; Springer, J.B.; Shah, L.; Colvin, O.M.; Ludeman, S.M.; Smith, C. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc. Natl. Acad. Sci. USA 1999, 96, 9118–9123. [Google Scholar] [CrossRef] [Green Version]
- Rudolph, M.C.; McManaman, J.L.; Phang, T.; Russell, T.; Kominsky, D.J.; Serkova, N.J.; Stein, T.; Anderson, S.M.; Neville, M.C. Metabolic regulation in the lactating mammary gland: A lipid synthesizing machine. Physiol. Genom. 2007, 28, 323–336. [Google Scholar] [CrossRef]
- Chong, B.M.; Reigan, P.; Mayle-Combs, K.D.; Orlicky, D.J.; McManaman, J.L. Determinants of adipophilin function in milk lipid formation and secretion. Trends Endocrinol. Metab. 2011, 22, 211–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, T.D.; Palmer, C.A.; Orlicky, D.J.; Fischer, A.; Rudolph, M.C.; Neville, M.C.; McManaman, J.L. Cytoplasmic lipid droplet accumulation in developing mammary epithelial cells: Roles of adipophilin and lipid metabolism. J. Lipid Res. 2007, 48, 1463–1475. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, M.C.; McManaman, J.L.; Hunter, L.; Phang, T.; Neville, M.C. Functional development of the mammary gland: Use of expression profiling and trajectory clustering to reveal changes in gene expression during pregnancy, lactation, and involution. J. Mammary Gland Biol. Neoplasia 2003, 8, 287–307. [Google Scholar] [CrossRef] [PubMed]
- Fielding, B.A.; Frayn, K.N. Lipoprotein lipase and the disposition of dietary fatty acids. Br. J. Nutr. 1998, 80, 495–502. [Google Scholar] [CrossRef] [Green Version]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [Green Version]
- Bionaz, M.; Loor, J.J. ACSL1, AGPAT6, FABP3, LPIN1, and SLC27A6 are the most abundant isoforms in bovine mammary tissue and their expression is affected by stage of lactation. J. Nutr. 2008, 138, 1019–1024. [Google Scholar] [CrossRef] [Green Version]
- Boutinaud, M.; Guinard-Flamenta, J.; Jammes, H. The number and activity of mammary epithelial cells, determining factors for milk production. Reprod. Nutr. Dev. 2004, 44, 499–508. [Google Scholar] [CrossRef] [Green Version]
- Matthews, J.R.; Clarke, A.R. p53 mediates a default programme of mammary gland involution in the absence of STAT3. Oncogene 2005, 24, 3083–3090. [Google Scholar] [CrossRef] [Green Version]
- Gatza, C.E.; Dumble, M.; Kittrell, F.; Edwards, D.G.; Dearth, R.K.; Lee, A.V.; Xu, J.; Medina, D.; Donehower, L.A. Altered mammary gland development in the p53+/m mouse, a model of accelerated aging. Dev. Biol. 2008, 313, 130–141. [Google Scholar] [CrossRef] [Green Version]
- Werb, Z.; Sympson, C.J.; Alexander, C.M.; Thomasset, N.; Lund, L.R.; MacAuley, A.; Ashkenas, J.; Bissell, M.J. Extracellular matrix remodeling and the regulation of epithelial-stromal interactions during differentiation and involution. Kidney Int. Suppl. 1996, 54, S68–S74. [Google Scholar]
- Schedin, P.; Mitrenga, T.; McDaniel, S.; Kaeck, M. Mammary ECM composition and function are altered by reproductive state. Mol. Carcinog. 2004, 41, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Ciraolo, E.; Iezzi, M.; Marone, R.; Marengo, S.; Curcio, C.; Costa, C.; Azzolino, O.; Gonella, C.; Rubinetto, C.; Wu, H.; et al. Phosphoinositide 3-kinase p110beta activity: Key role in metabolism and mammary gland cancer but not development. Sci. Signal. 2008, 1, ra3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rädler, P.D.; Wehde, B.L.; Wagner, K.U. Crosstalk between STAT5 activation and PI3K/AKT functions in normal and transformed mammary epithelial cells. Mol. Cell. Endocrinol. 2017, 451, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Xi, P.; Wang, Z.; Han, X.; Xu, Y.; Zhang, Y.; Miao, J. PI3K/Akt/mTOR signaling pathway participates in Streptococcus uberis-induced inflammation in mammary epithelial cells in concert with the classical TLRs/NF-ĸB pathway. Vet. Microbiol. 2018, 227, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Heldin, C.H. Targeting the PDGF signaling pathway in tumor treatment. Cell Commun. Signal. CCS 2013, 11, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhardwaj, B.; Klassen, J.; Cossette, N.; Sterns, E.; Tuck, A.; Deeley, R.; Sengupta, S.; Elliott, B. Localization of platelet-derived growth factor beta receptor expression in the periepithelial stroma of human breast carcinoma. Clin. Cancer. Res. 1996, 2, 773–782. [Google Scholar]
- Rodeheffer, M.S.; Birsoy, K.; Friedman, J.M. Identification of white adipocyte progenitor cells in vivo. Cell 2008, 135, 240–249. [Google Scholar] [CrossRef] [Green Version]
- Jechlinger, M.; Sommer, A.; Moriggl, R.; Seither, P.; Kraut, N.; Capodiecci, P.; Donovan, M.; Cordon-Cardo, C.; Beug, H.; Grünert, S. Autocrine PDGFR signaling promotes mammary cancer metastasis. J. Clin. Investig. 2006, 116, 1561–1570. [Google Scholar]
- Nilson, L.A.; DiMaio, D. Platelet-derived growth factor receptor can mediate tumorigenic transformation by the bovine papillomavirus E5 protein. Mol. Cell Biol. 1993, 13, 4137–4145. [Google Scholar]
- Camorani, S.; Esposito, C.L.; Rienzo, A.; Catuogno, S.; Iaboni, M.; Condorelli, G.; de Franciscis, V.; Cerchia, L. Inhibition of receptor signaling and of glioblastoma-derived tumor growth by a novel PDGFRβ aptamer. Mol. Ther. 2014, 22, 828–841. [Google Scholar]
- Allan, G.J.; Beattie, J.; Flint, D.J. The role of IGFBP-5 in mammary gland development and involution. Domest. Anim. Endocrinol. 2004, 27, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, M.T.; Nørgaard, J.V.; Theil, P.K.; Vestergaard, M.; Sejrsen, K. Cell turnover and activity in mammary tissue during lactation and the dry period in dairy cows. J. Dairy Sci. 2006, 89, 4632–4639. [Google Scholar] [CrossRef] [Green Version]
- Lee, P.P.; Hwang, J.J.; Mead, L.; Ip, M.M. Functional role of matrix metalloproteinases (MMPs) in mammary epithelial cell development. J. Cell. Physiol. 2001, 188, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Miller, N.; Delbecchi, L.; Petitclerc, D.; Wagner, G.F.; Talbot, B.G.; Lacasse, P. Effect of stage of lactation and parity on mammary gland cell renewal. J. Dairy Sci. 2006, 89, 4669–4677. [Google Scholar] [CrossRef] [Green Version]
- Hieta, N.; Impola, U.; López-Otín, C.; Saarialho-Kere, U.; Kähäri, V.M. Matrix metalloproteinase-19 expression in dermal wounds and by fibroblasts in culture. J. Investig. Dermatol. 2003, 121, 997–1004. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Gao, S.; Muegge, K.; Zhang, W.; Zhou, B. Advanced Applications of RNA Sequencing and Challenges. Bioinform. Biol. Insights 2015, 9 (Suppl. S1), 29–46. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Bhat, S.A.; Ahmad, S.M.; Mumtaz, P.T.; Malik, A.A.; Dar, M.A.; Urwat, U.; Shah, R.A.; Ganai, N.A. Long non-coding RNAs: Mechanism of action and functional utility. Non-Coding RNA Res. 2016, 1, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Martin, E.W.; Sung, M.H. Challenges of Decoding Transcription Factor Dynamics in Terms of Gene Regulation. Cells 2018, 7, 132. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. In Babraham Bioinformatics; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Wang, J.; Chen, C.; Dong, F.; Wang, G.; Ji, Z.; Qin, Z. Separation and purification method of goat mammary epithelial cells. CN103525752A; filed 21 September 2013, and issued 22 January 2014, 2013. [Google Scholar]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, Research0034. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xuan, R.; Wang, J.; Zhao, X.; Li, Q.; Wang, Y.; Du, S.; Duan, Q.; Guo, Y.; Ji, Z.; Chao, T. Transcriptome Analysis of Goat Mammary Gland Tissue Reveals the Adaptive Strategies and Molecular Mechanisms of Lactation and Involution. Int. J. Mol. Sci. 2022, 23, 14424. https://doi.org/10.3390/ijms232214424
Xuan R, Wang J, Zhao X, Li Q, Wang Y, Du S, Duan Q, Guo Y, Ji Z, Chao T. Transcriptome Analysis of Goat Mammary Gland Tissue Reveals the Adaptive Strategies and Molecular Mechanisms of Lactation and Involution. International Journal of Molecular Sciences. 2022; 23(22):14424. https://doi.org/10.3390/ijms232214424
Chicago/Turabian StyleXuan, Rong, Jianmin Wang, Xiaodong Zhao, Qing Li, Yanyan Wang, Shanfeng Du, Qingling Duan, Yanfei Guo, Zhibin Ji, and Tianle Chao. 2022. "Transcriptome Analysis of Goat Mammary Gland Tissue Reveals the Adaptive Strategies and Molecular Mechanisms of Lactation and Involution" International Journal of Molecular Sciences 23, no. 22: 14424. https://doi.org/10.3390/ijms232214424
APA StyleXuan, R., Wang, J., Zhao, X., Li, Q., Wang, Y., Du, S., Duan, Q., Guo, Y., Ji, Z., & Chao, T. (2022). Transcriptome Analysis of Goat Mammary Gland Tissue Reveals the Adaptive Strategies and Molecular Mechanisms of Lactation and Involution. International Journal of Molecular Sciences, 23(22), 14424. https://doi.org/10.3390/ijms232214424






