Loss of Runx1 Induces Granulosa Cell Defects and Development of Ovarian Tumors in the Mouse
Abstract
1. Introduction
2. Results
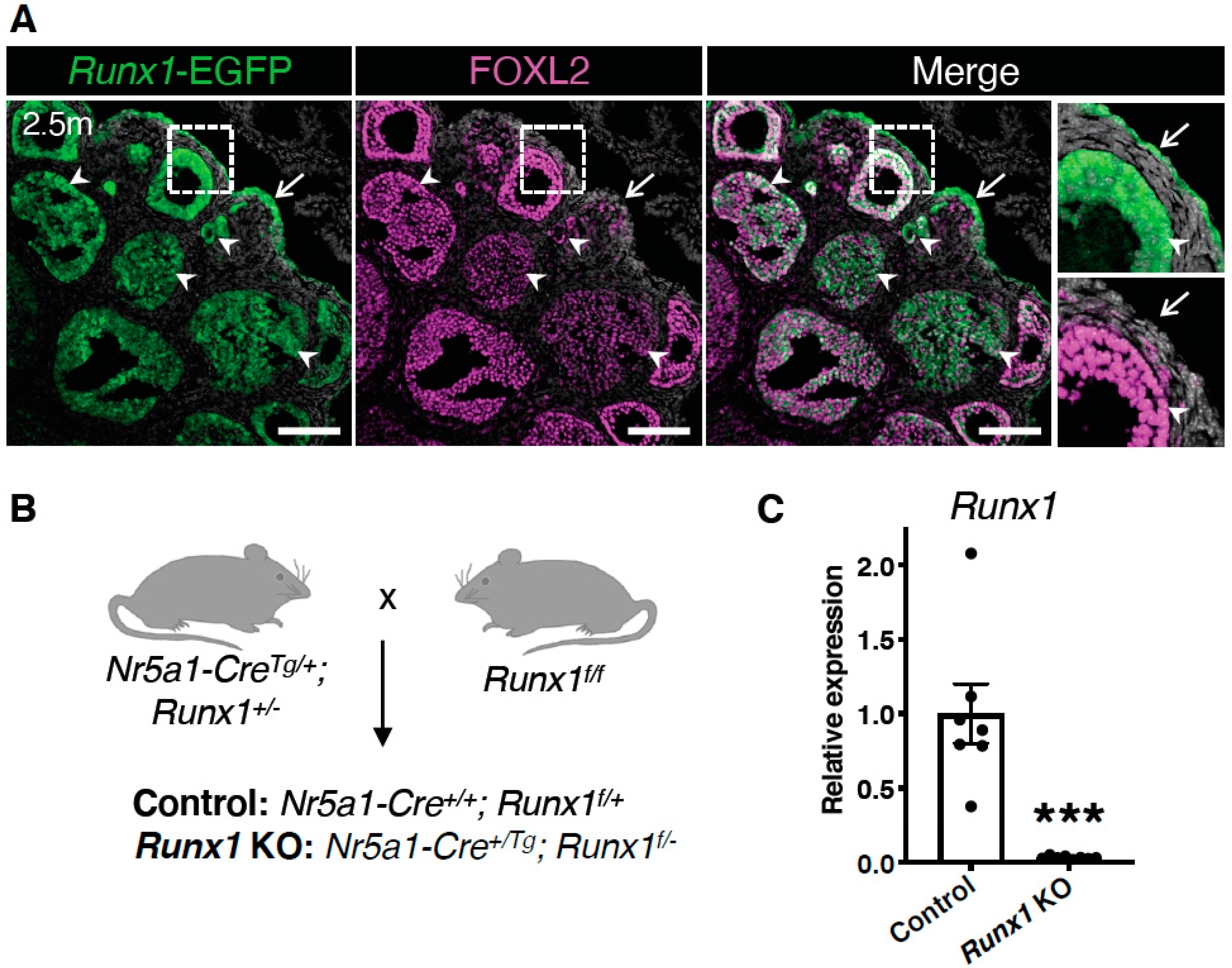
2.1. Runx1 Is Expressed in Granulosa Cells and the Surface Epithelium of Adult Mouse Ovaries
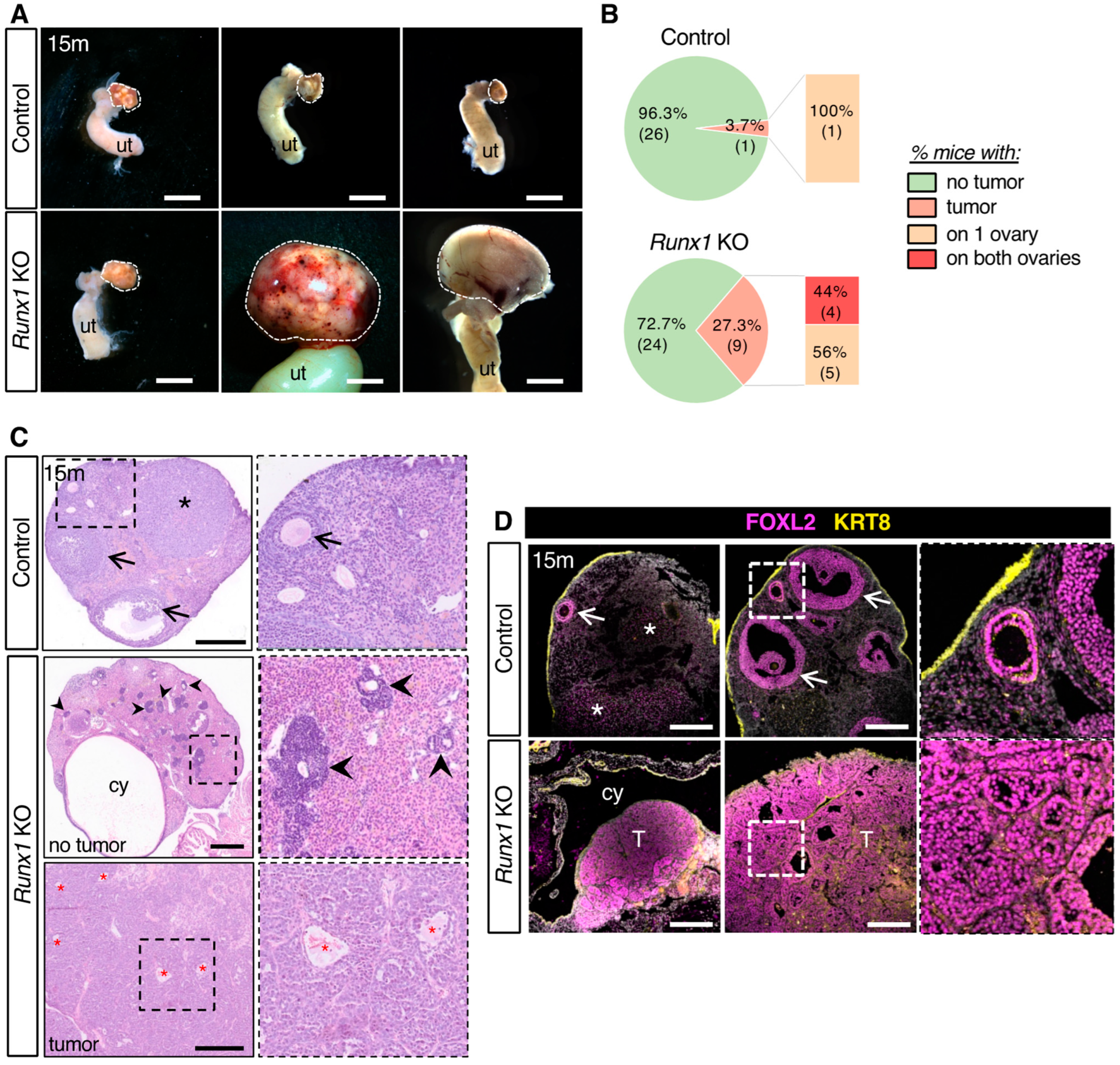
2.2. Loss of Runx1 Leads to Increased Prevalence of Ovarian Tumors
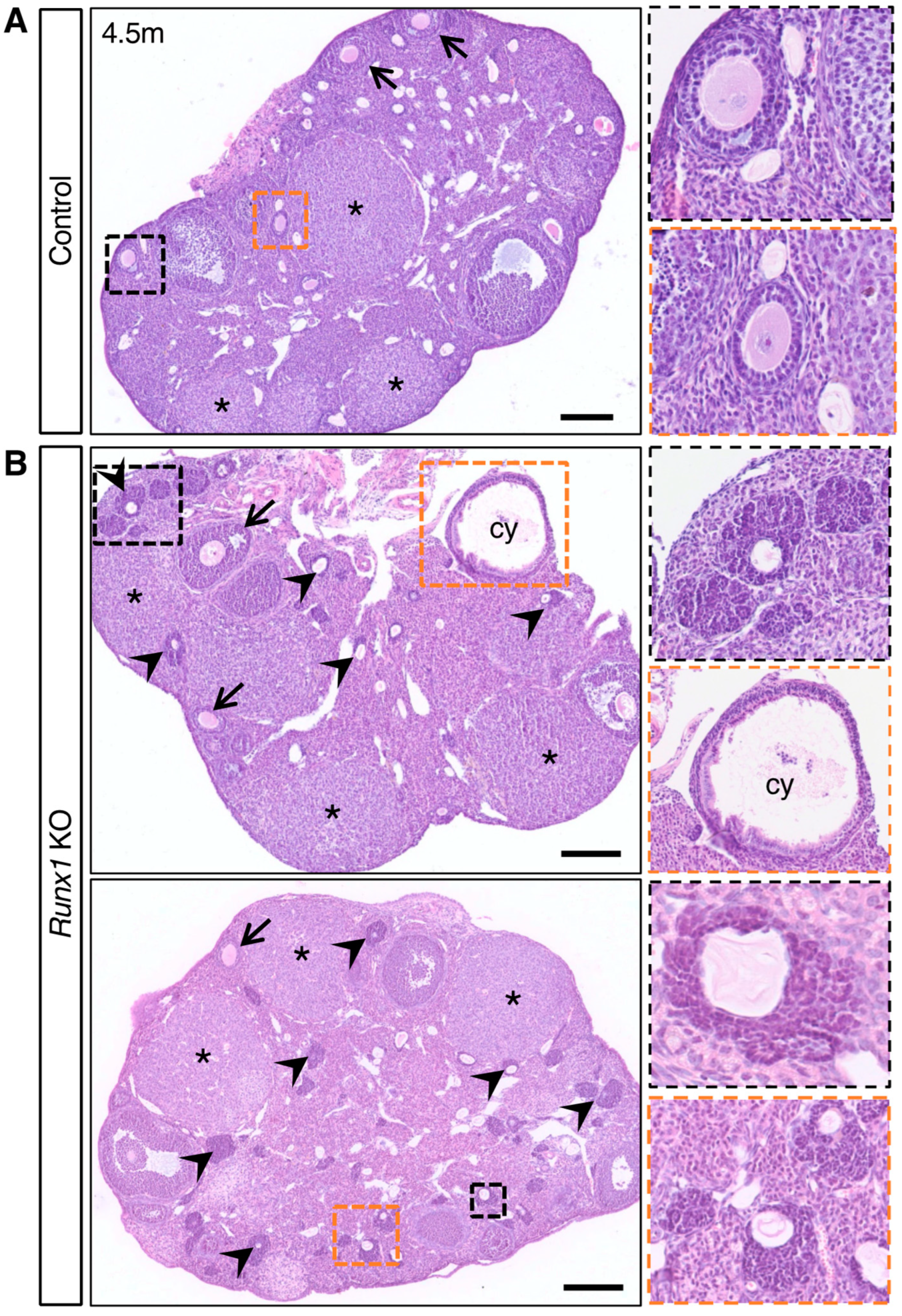
2.3. Young Runx1 KO Mice Present Ovarian Defects but No Apparent Tumors
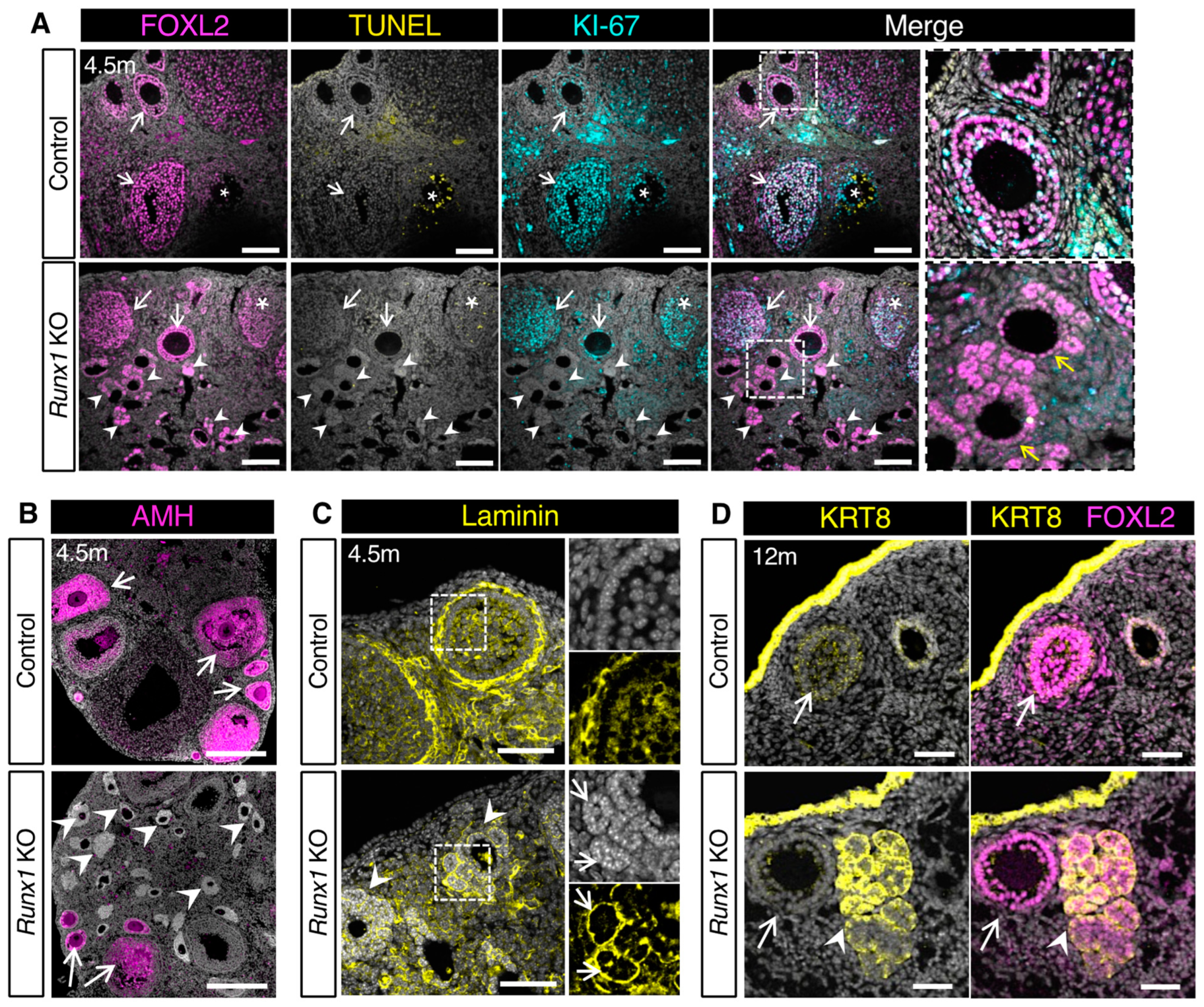
2.4. The Follicle-like Lesions in Runx1 KO Ovaries Are Composed of Quiescent Abnormal Granulosa Cells
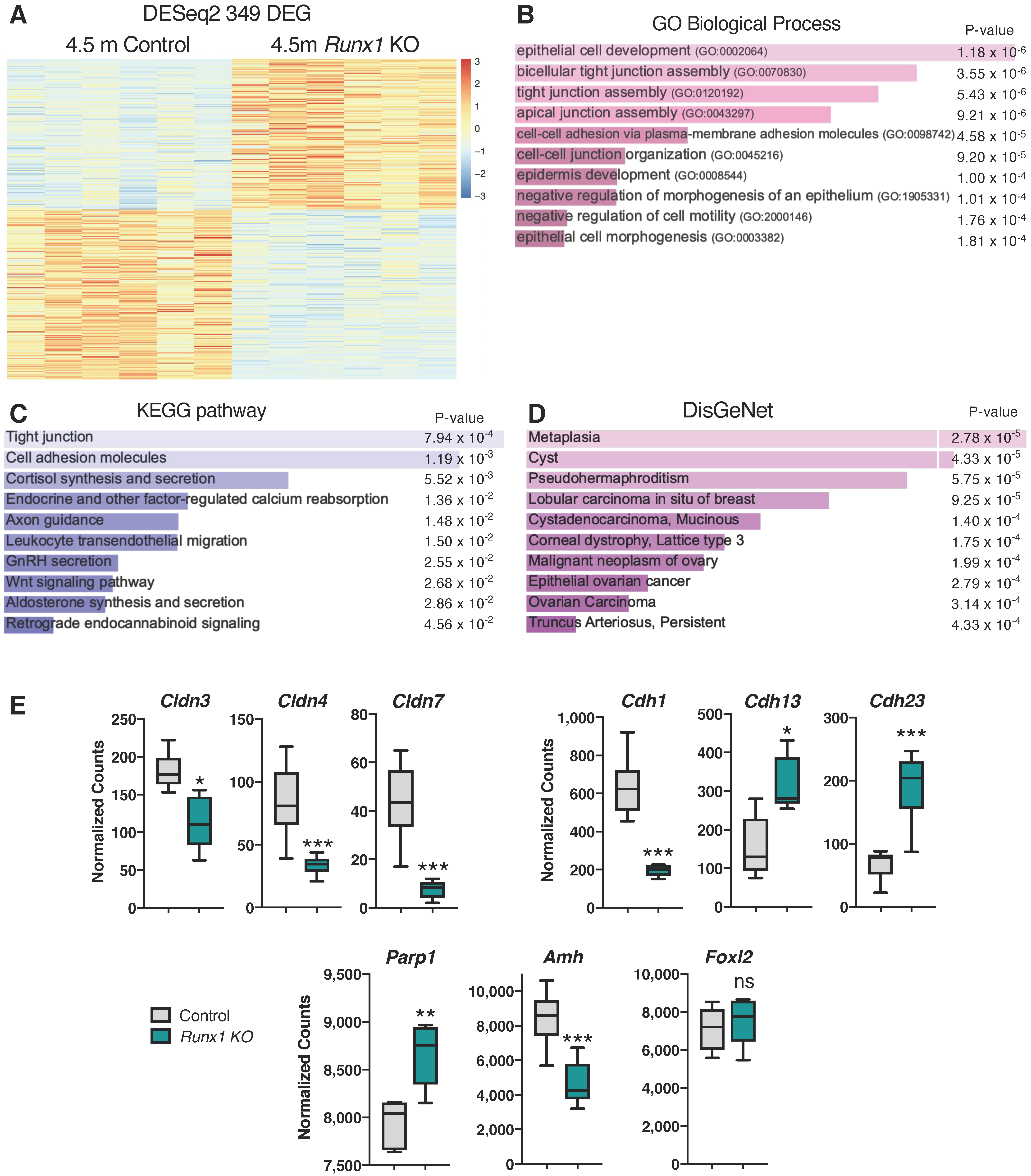
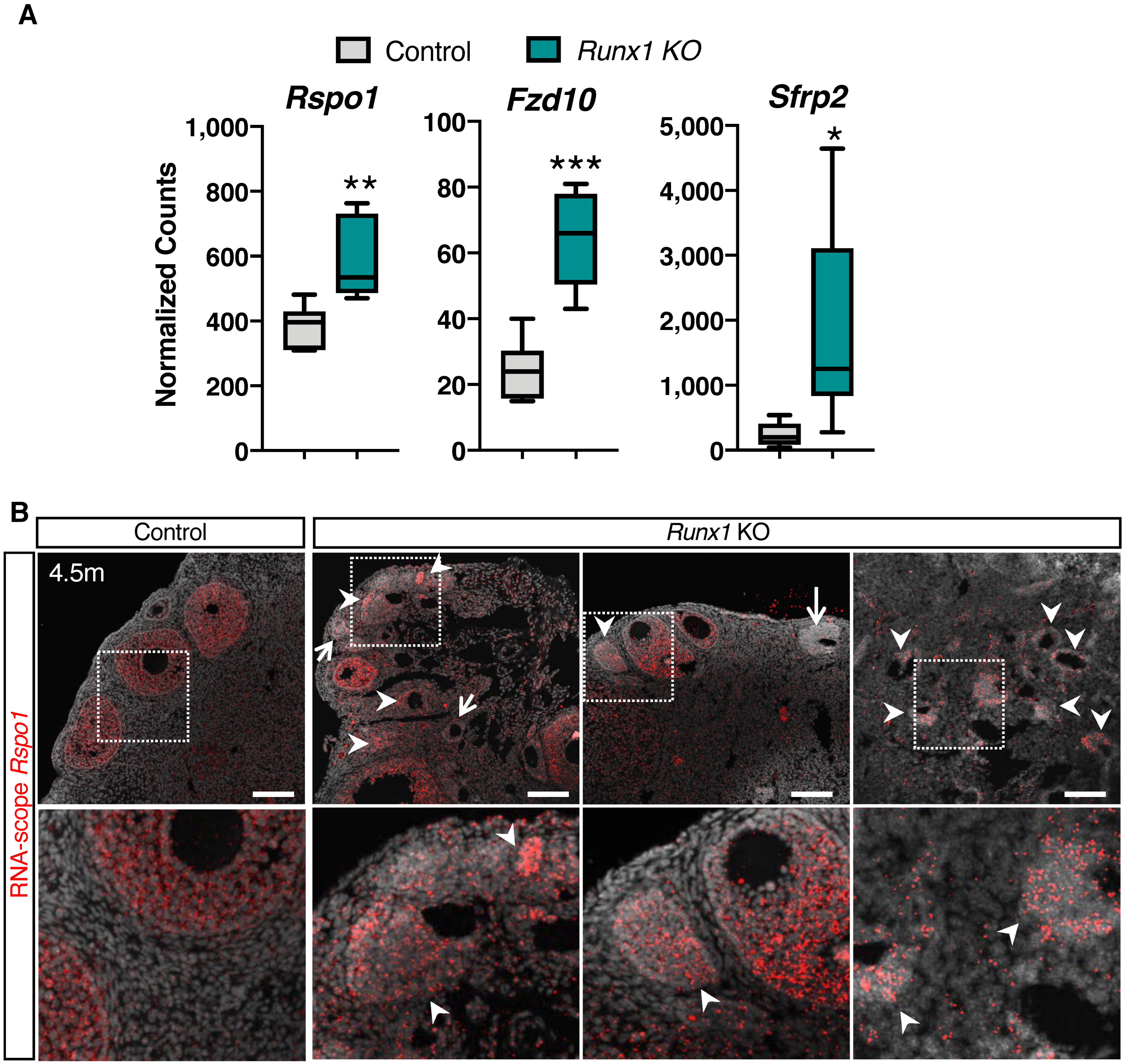
2.5. Differentially Expressed Genes in Runx1 KO Ovaries Are Associated with Epithelium Development, Tight Junctions, and Metaplasia
3. Discussion
4. Materials and Methods
4.1. Mouse Models
4.2. Immunofluorescence, Tunel Assay, and Histological Analyses
4.3. RNA Extraction and Real-time PCR Analysis
4.4. RNA Sequencing
4.5. RNAscope
4.6. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karnezis, A.N.; Cho, K.R.; Gilks, C.B.; Pearce, C.L.; Huntsman, D.G. The disparate origins of ovarian cancers: Pathogenesis and prevention strategies. Nat. Rev. Cancer 2017, 17, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Romero, I.; Bast, R.C., Jr. Minireview: Human ovarian cancer: Biology, current management, and paths to personalizing therapy. Endocrinology 2012, 153, 1593–1602. [Google Scholar] [CrossRef] [PubMed]
- Otalora-Otalora, B.A.; Henriquez, B.; Lopez-Kleine, L.; Rojas, A. RUNX family: Oncogenes or tumor suppressors (Review). Oncol. Rep. 2019, 42, 3–19. [Google Scholar] [CrossRef] [PubMed]
- De Braekeleer, E.; Ferec, C.; De Braekeleer, M. RUNX1 translocations in malignant hemopathies. Anticancer Res. 2009, 29, 1031–1037. [Google Scholar] [PubMed]
- Dowdy, C.R.; Xie, R.; Frederick, D.; Hussain, S.; Zaidi, S.K.; Vradii, D.; Javed, A.; Li, X.; Jones, S.N.; Lian, J.B.; et al. Definitive hematopoiesis requires Runx1 C-terminal-mediated subnuclear targeting and transactivation. Hum. Mol. Genet. 2010, 19, 1048–1057. [Google Scholar] [CrossRef]
- Sood, R.; Kamikubo, Y.; Liu, P. Role of RUNX1 in hematological malignancies. Blood 2017, 129, 2070–2082. [Google Scholar] [CrossRef]
- Lin, T.C. RUNX1 and cancer. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188715. [Google Scholar] [CrossRef]
- Riggio, A.I.; Blyth, K. The enigmatic role of RUNX1 in female-related cancers—Current knowledge & future perspectives. FEBS J. 2017, 284, 2345–2362. [Google Scholar] [CrossRef]
- Keita, M.; Bachvarova, M.; Morin, C.; Plante, M.; Gregoire, J.; Renaud, M.C.; Sebastianelli, A.; Trinh, X.B.; Bachvarov, D. The RUNX1 transcription factor is expressed in serous epithelial ovarian carcinoma and contributes to cell proliferation, migration and invasion. Cell Cycle 2013, 12, 972–986. [Google Scholar] [CrossRef]
- Nicol, B.; Grimm, S.A.; Chalmel, F.; Lecluze, E.; Pannetier, M.; Pailhoux, E.; Dupin-De-Beyssat, E.; Guiguen, Y.; Capel, B.; Yao, H.H. RUNX1 maintains the identity of the fetal ovary through an interplay with FOXL2. Nat. Commun. 2019, 10, 5116. [Google Scholar] [CrossRef]
- Gong, S.; Zheng, C.; Doughty, M.L.; Losos, K.; Didkovsky, N.; Schambra, U.B.; Nowak, N.J.; Joyner, A.; Leblanc, G.; Hatten, M.E.; et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 2003, 425, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Loffler, K.A.; Zarkower, D.; Koopman, P. Etiology of Ovarian Failure in Blepharophimosis Ptosis Epicanthus Inversus Syndrome: FOXL2 Is a Conserved, Early-Acting Gene in Vertebrate Ovarian Development. Endocrinology 2003, 144, 3237–3243. [Google Scholar] [CrossRef] [PubMed]
- Bingham, N.C.; Verma-Kurvari, S.; Parada, L.F.; Parker, K.L. Development of a steroidogenic factor 1/Cre transgenic mouse line. Genesis 2006, 44, 419–424. [Google Scholar] [CrossRef]
- Liu, Z.; Ren, Y.A.; Pangas, S.A.; Adams, J.; Zhou, W.; Castrillon, D.H.; Wilhelm, D.; Richards, J.S. FOXO1/3 and PTEN Depletion in Granulosa Cells Promotes Ovarian Granulosa Cell Tumor Development. Mol. Endocrinol. 2015, 29, 1006–1024. [Google Scholar] [CrossRef] [PubMed]
- De Cian, M.C.; Pauper, E.; Bandiera, R.; Vidal, V.P.; Sacco, S.; Gregoire, E.P.; Chassot, A.A.; Panzolini, C.; Wilhelm, D.; Pailhoux, E.; et al. Amplification of R-spondin1 signaling induces granulosa cell fate defects and cancers in mouse adult ovary. Oncogene 2017, 36, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Crosley, P.; Azad, A.K.; Gupta, N.; Gokul, N.; Xu, Z.; Weinfeld, M.; Postovit, L.M.; Pangas, S.A.; Hitt, M.M.; et al. RUNX3 Promotes the Tumorigenic Phenotype in KGN, a Human Granulosa Cell Tumor-Derived Cell Line. Int. J. Mol. Sci. 2019, 20, 3471. [Google Scholar] [CrossRef]
- Bhat, A.A.; Uppada, S.; Achkar, I.W.; Hashem, S.; Yadav, S.K.; Shanmugakonar, M.; Al-Naemi, H.A.; Haris, M.; Uddin, S. Tight Junction Proteins and Signaling Pathways in Cancer and Inflammation: A Functional Crosstalk. Front. Physiol. 2018, 9, 1942. [Google Scholar] [CrossRef]
- Niu, W.; Spradling, A.C. Two distinct pathways of pregranulosa cell differentiation support follicle formation in the mouse ovary. Proc. Natl. Acad. Sci. USA 2020, 117, 20015–20026. [Google Scholar] [CrossRef]
- Rastetter, R.H.; Bernard, P.; Palmer, J.S.; Chassot, A.A.; Chen, H.; Western, P.S.; Ramsay, R.G.; Chaboissier, M.C.; Wilhelm, D. Marker genes identify three somatic cell types in the fetal mouse ovary. Dev. Biol. 2014, 394, 242–252. [Google Scholar] [CrossRef]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016, 45, D833–D839. [Google Scholar] [CrossRef]
- Yuecheng, Y.; Hongmei, L.; Xiaoyan, X. Clinical evaluation of E-cadherin expression and its regulation mechanism in epithelial ovarian cancer. Clin. Exp. Metastasis 2006, 23, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Berx, G.; Becker, K.F.; Höfler, H.; van Roy, F. Mutations of the human E-cadherin (CDH1) gene. Hum. Mutat. 1998, 12, 226–237. [Google Scholar] [CrossRef]
- Ossovskaya, V.; Koo, I.C.; Kaldjian, E.P.; Alvares, C.; Sherman, B.M. Upregulation of Poly (ADP-Ribose) Polymerase-1 (PARP1) in Triple-Negative Breast Cancer and Other Primary Human Tumor Types. Genes Cancer 2010, 1, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Boerboom, D.; Paquet, M.; Hsieh, M.; Liu, J.; Jamin, S.P.; Behringer, R.R.; Sirois, J.; Taketo, M.M.; Richards, J.S. Misregulated Wnt/beta-catenin signaling leads to ovarian granulosa cell tumor development. Cancer Res. 2005, 65, 9206–9215. [Google Scholar] [CrossRef] [PubMed]
- Owens, G.E.; Keri, R.A.; Nilson, J.H. Ovulatory surges of human CG prevent hormone-induced granulosa cell tumor formation leading to the identification of tumor-associated changes in the transcriptome. Mol. Endocrinol. 2002, 16, 1230–1242. [Google Scholar] [CrossRef] [PubMed]
- Terasaki, H.; Saitoh, T.; Shiokawa, K.; Katoh, M. Frizzled-10, up-regulated in primary colorectal cancer, is a positive regulator of the WNT-beta-catenin-TCF signaling pathway. Int. J. Mol. Med. 2002, 9, 107–112. [Google Scholar]
- Vincent, K.M.; Postovit, L.M. A pan-cancer analysis of secreted Frizzled-related proteins: Re-examining their proposed tumour suppressive function. Sci. Rep. 2017, 7, 42719. [Google Scholar] [CrossRef]
- Chassot, A.A.; Ranc, F.; Gregoire, E.P.; Roepers-Gajadien, H.L.; Taketo, M.M.; Camerino, G.; de Rooij, D.G.; Schedl, A.; Chaboissier, M.C. Activation of beta-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum. Mol. Genet. 2008, 17, 1264–1277. [Google Scholar] [CrossRef]
- Ni, N.; Fang, X.; Mullens, D.A.; Cai, J.J.; Ivanov, I.; Bartholin, L.; Li, Q. Transcriptomic Profiling of Gene Expression Associated with Granulosa Cell Tumor Development in a Mouse Model. Cancers 2022, 14, 2184. [Google Scholar] [CrossRef]
- Irving-Rodgers, H.F.; Rodgers, R.J. Granulosa cell expression of basal lamina matrices: Call-Exner bodies and focimatrix. Ital. J. Anat. Embryol. 2005, 110, 225–230. [Google Scholar]
- Fan, H.Y.; Shimada, M.; Liu, Z.; Cahill, N.; Noma, N.; Wu, Y.; Gossen, J.; Richards, J.S. Selective expression of KrasG12D in granulosa cells of the mouse ovary causes defects in follicle development and ovulation. Development 2008, 135, 2127–2137. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.S.; Fan, H.Y.; Liu, Z.; Tsoi, M.; Lague, M.N.; Boyer, A.; Boerboom, D. Either Kras activation or Pten loss similarly enhance the dominant-stable CTNNB1-induced genetic program to promote granulosa cell tumor development in the ovary and testis. Oncogene 2012, 31, 1504–1520. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chimge, N.-O.; Little, G.H.; Baniwal, S.K.; Adisetiyo, H.; Xie, Y.; Zhang, T.; O’Laughlin, A.; Liu, Z.Y.; Ulrich, P.; Martin, A.; et al. RUNX1 prevents oestrogen-mediated AXIN1 suppression and β-catenin activation in ER-positive breast cancer. Nat. Commun. 2016, 7, 10751. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.P.; Köbel, M.; Senz, J.; Morin, R.D.; Clarke, B.A.; Wiegand, K.C.; Leung, G.; Zayed, A.; Mehl, E.; Kalloger, S.E.; et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N. Engl. J. Med. 2009, 360, 2719–2729. [Google Scholar] [CrossRef] [PubMed]
- Köbel, M.; Gilks, C.B.; Huntsman, D.G. Adult-Type Granulosa Cell Tumors and FOXL2 Mutation. Cancer Res. 2009, 69, 9160–9162. [Google Scholar] [CrossRef]
- Carles, A.; Trigo-Gonzalez, G.; Cao, Q.; Cheng, S.G.; Moksa, M.; Bilenky, M.; Huntsman, D.G.; Morin, G.B.; Hirst, M. The Pathognomonic FOXL2 C134W Mutation Alters DNA-Binding Specificity. Cancer Res. 2020, 80, 3480–3491. [Google Scholar] [CrossRef]
- Weis-Banke, S.E.; Lerdrup, M.; Kleine-Kohlbrecher, D.; Mohammad, F.; Sidoli, S.; Jensen, O.N.; Yanase, T.; Nakamura, T.; Iwase, A.; Stylianou, A.; et al. Mutant FOXL2(C134W) Hijacks SMAD4 and SMAD2/3 to Drive Adult Granulosa Cell Tumors. Cancer Res. 2020, 80, 3466–3479. [Google Scholar] [CrossRef]
- Caburet, S.; Anttonen, M.; Todeschini, A.L.; Unkila-Kallio, L.; Mestivier, D.; Butzow, R.; Veitia, R.A. Combined comparative genomic hybridization and transcriptomic analyses of ovarian granulosa cell tumors point to novel candidate driver genes. BMC Cancer 2015, 15, 251. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, J.; Gan, X.; Shen, F.; Yang, X.; Du, N.; Xia, D.; Liu, L.; Qiao, L.; Pan, J.; et al. LGR5 promotes epithelial ovarian cancer proliferation, metastasis, and epithelial-mesenchymal transition through the Notch1 signaling pathway. Cancer Med. 2018, 7, 3132–3142. [Google Scholar] [CrossRef]
- Ng, A.; Tan, S.; Singh, G.; Rizk, P.; Swathi, Y.; Tan, T.Z.; Huang, R.Y.; Leushacke, M.; Barker, N. Lgr5 marks stem/progenitor cells in ovary and tubal epithelia. Nat. Cell Biol. 2014, 16, 745–757. [Google Scholar] [CrossRef]
- Zhang, S.; Dolgalev, I.; Zhang, T.; Ran, H.; Levine, D.A.; Neel, B.G. Both fallopian tube and ovarian surface epithelium are cells-of-origin for high-grade serous ovarian carcinoma. Nat. Commun. 2019, 10, 5367. [Google Scholar] [CrossRef] [PubMed]
- Labidi-Galy, S.I.; Papp, E.; Hallberg, D.; Niknafs, N.; Adleff, V.; Noe, M.; Bhattacharya, R.; Novak, M.; Jones, S.; Phallen, J.; et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat. Commun. 2017, 8, 1093. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Stacy, T.; Binder, M.; Marin-Padilla, M.; Sharpe, A.H.; Speck, N.A. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA 1996, 93, 3444–3449. [Google Scholar] [CrossRef] [PubMed]
- Taniuchi, I.; Osato, M.; Egawa, T.; Sunshine, M.J.; Bae, S.C.; Komori, T.; Ito, Y.; Littman, D.R. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 2002, 111, 621–633. [Google Scholar] [CrossRef]
- Foley, K.G.; Pritchard, M.T.; Duncan, F.E. Macrophage-derived multinucleated giant cells: Hallmarks of the aging ovary. Reproduction 2021, 161, V5–V9. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bridges, K.; Yao, H.H.-C.; Nicol, B. Loss of Runx1 Induces Granulosa Cell Defects and Development of Ovarian Tumors in the Mouse. Int. J. Mol. Sci. 2022, 23, 14442. https://doi.org/10.3390/ijms232214442
Bridges K, Yao HH-C, Nicol B. Loss of Runx1 Induces Granulosa Cell Defects and Development of Ovarian Tumors in the Mouse. International Journal of Molecular Sciences. 2022; 23(22):14442. https://doi.org/10.3390/ijms232214442
Chicago/Turabian StyleBridges, Kamiya, Humphrey H.-C. Yao, and Barbara Nicol. 2022. "Loss of Runx1 Induces Granulosa Cell Defects and Development of Ovarian Tumors in the Mouse" International Journal of Molecular Sciences 23, no. 22: 14442. https://doi.org/10.3390/ijms232214442
APA StyleBridges, K., Yao, H. H.-C., & Nicol, B. (2022). Loss of Runx1 Induces Granulosa Cell Defects and Development of Ovarian Tumors in the Mouse. International Journal of Molecular Sciences, 23(22), 14442. https://doi.org/10.3390/ijms232214442



