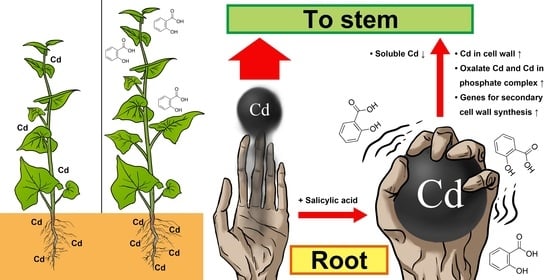
Salicylic Acid Enhances Cadmium Tolerance and Reduces Its Shoot Accumulation in Fagopyrum tataricum Seedlings by Promoting Root Cadmium Retention and Mitigating Oxidative Stress
Abstract
1. Introduction
2. Results
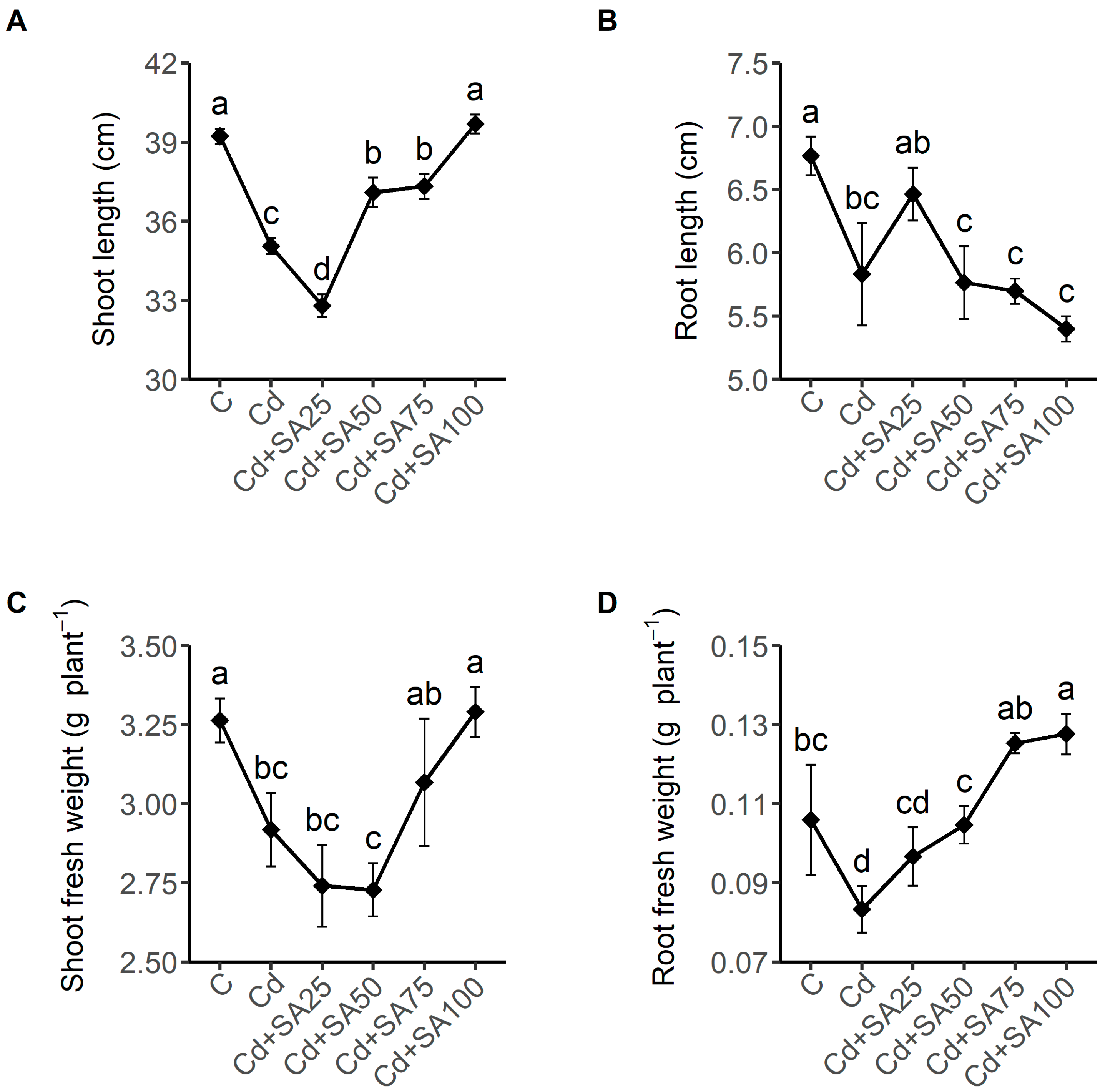
2.1. SA Promotes Growth and Mitigates Oxidative Stress of Tartary Buckwheat under Cadmium Stress
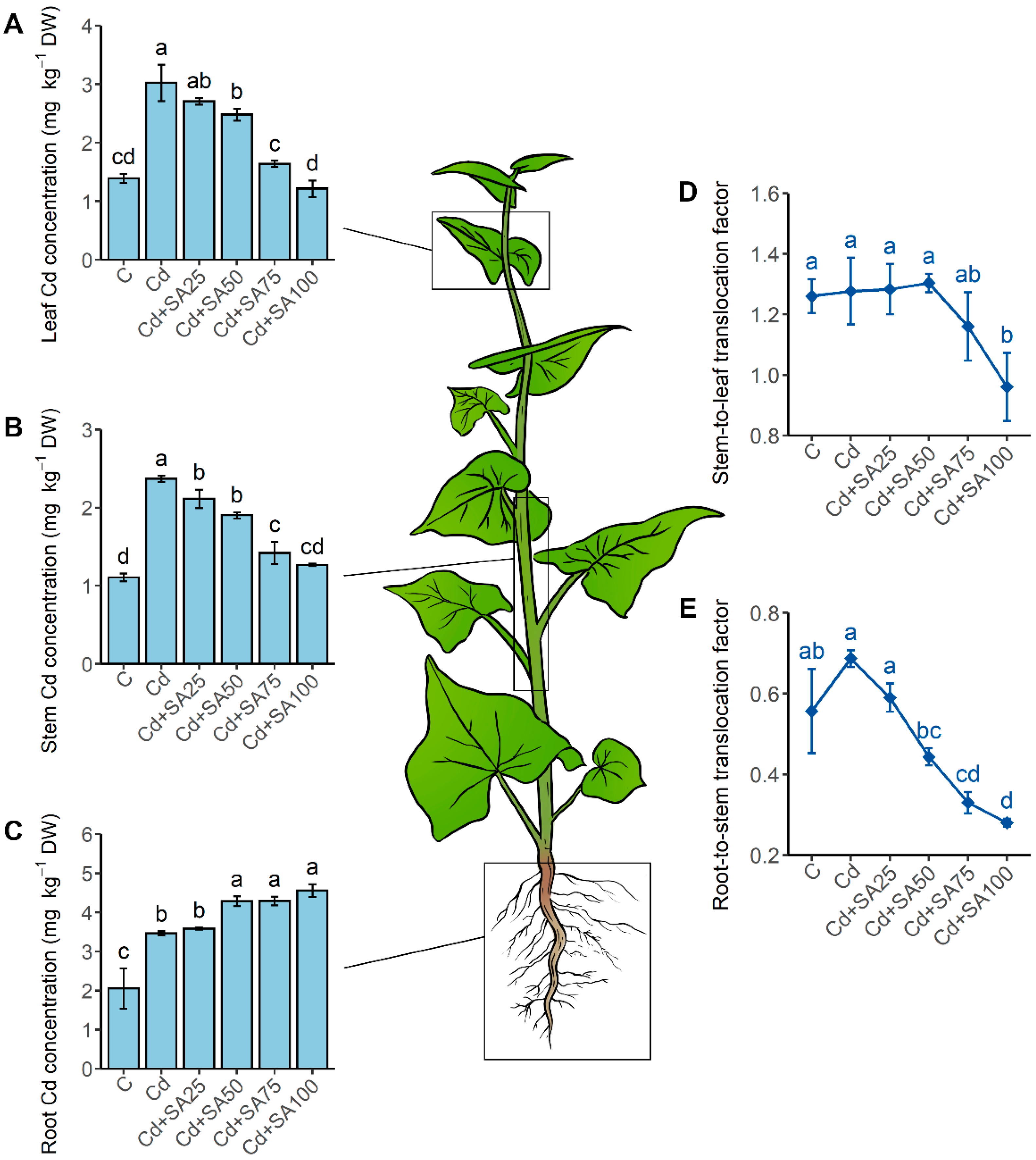
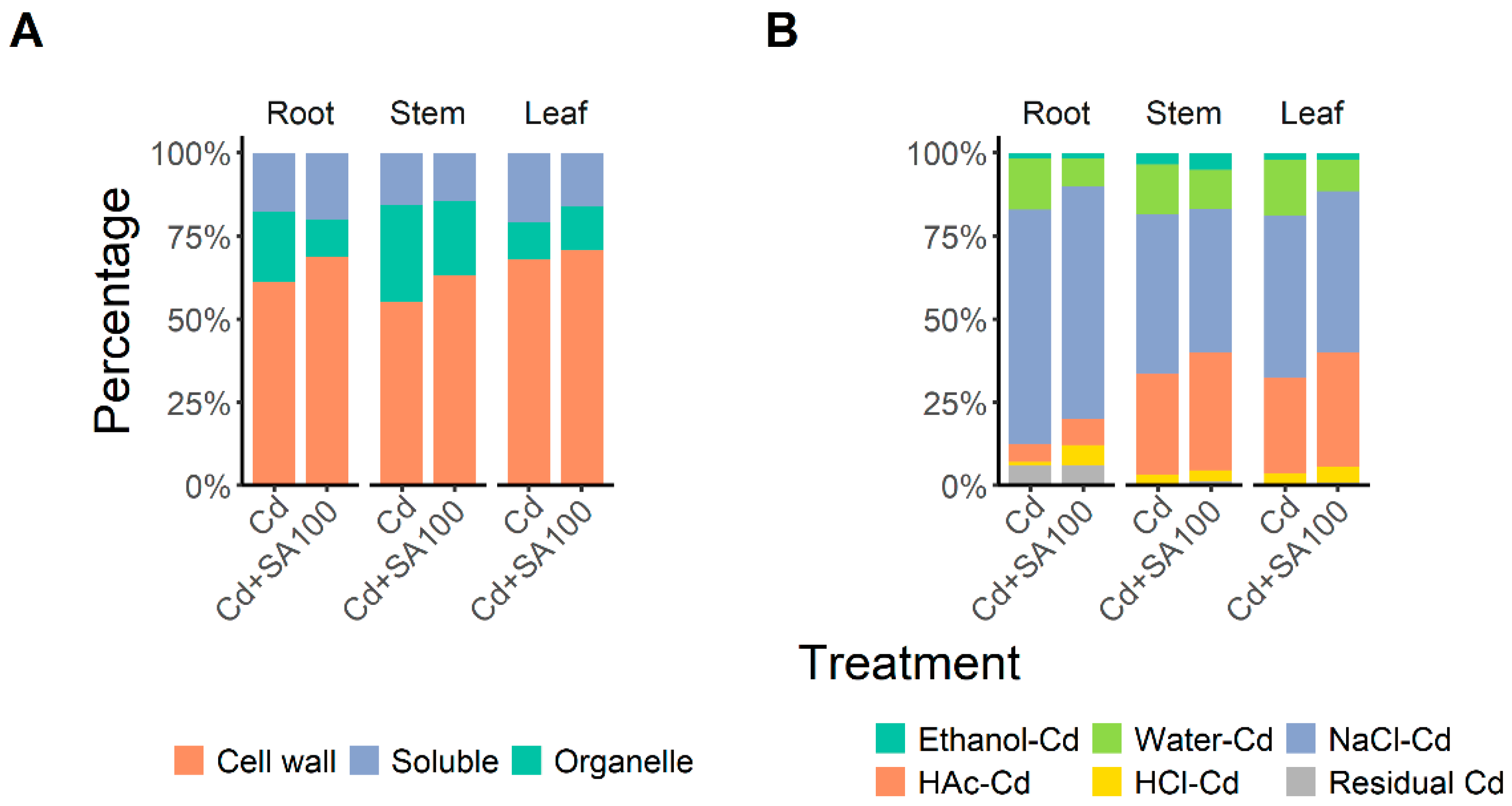
2.2. SA Promotes Root Cd Retention and Enhances Cd Partitioning in the Cell Wall and Insoluble Cd Chemical Forms
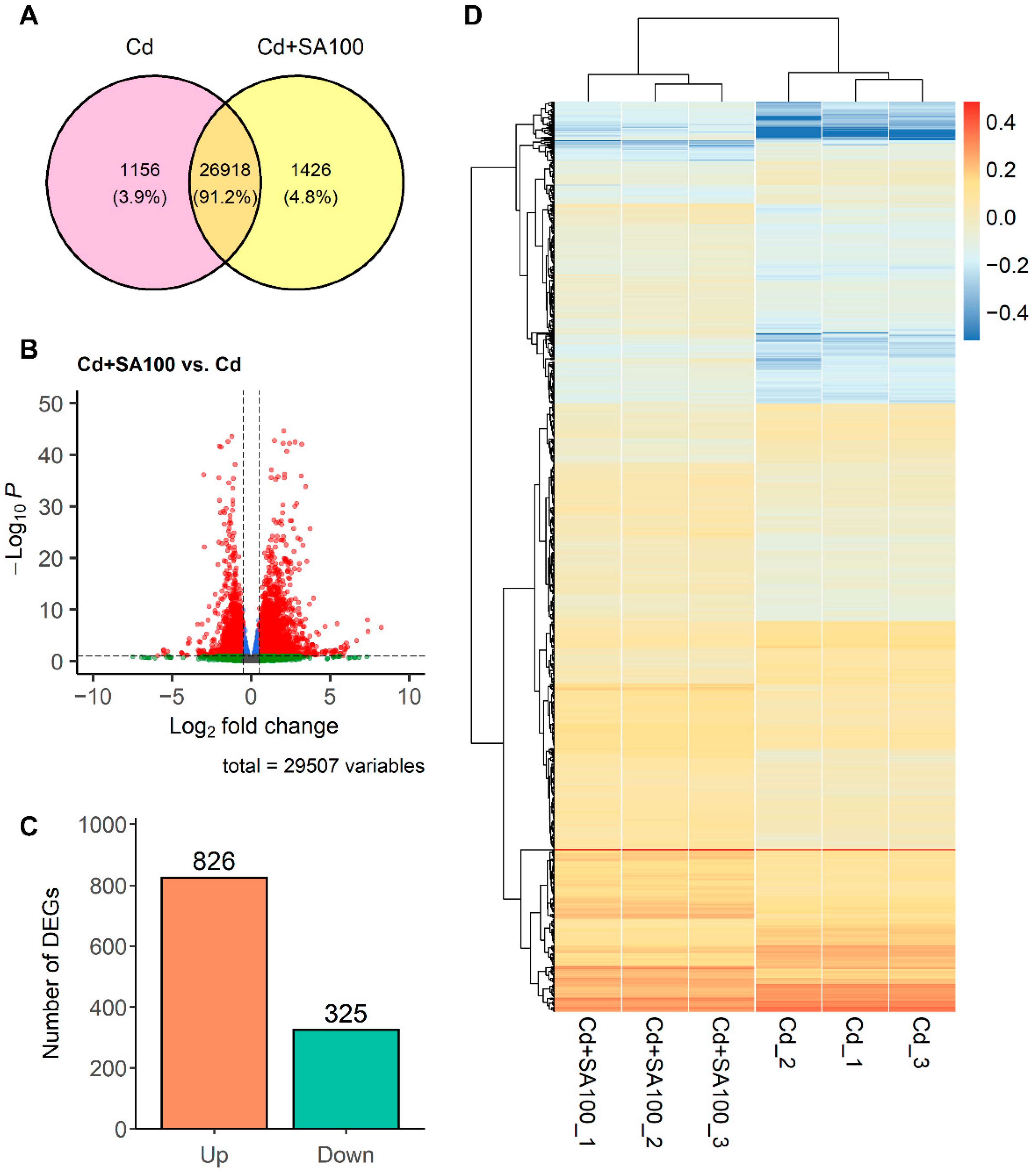
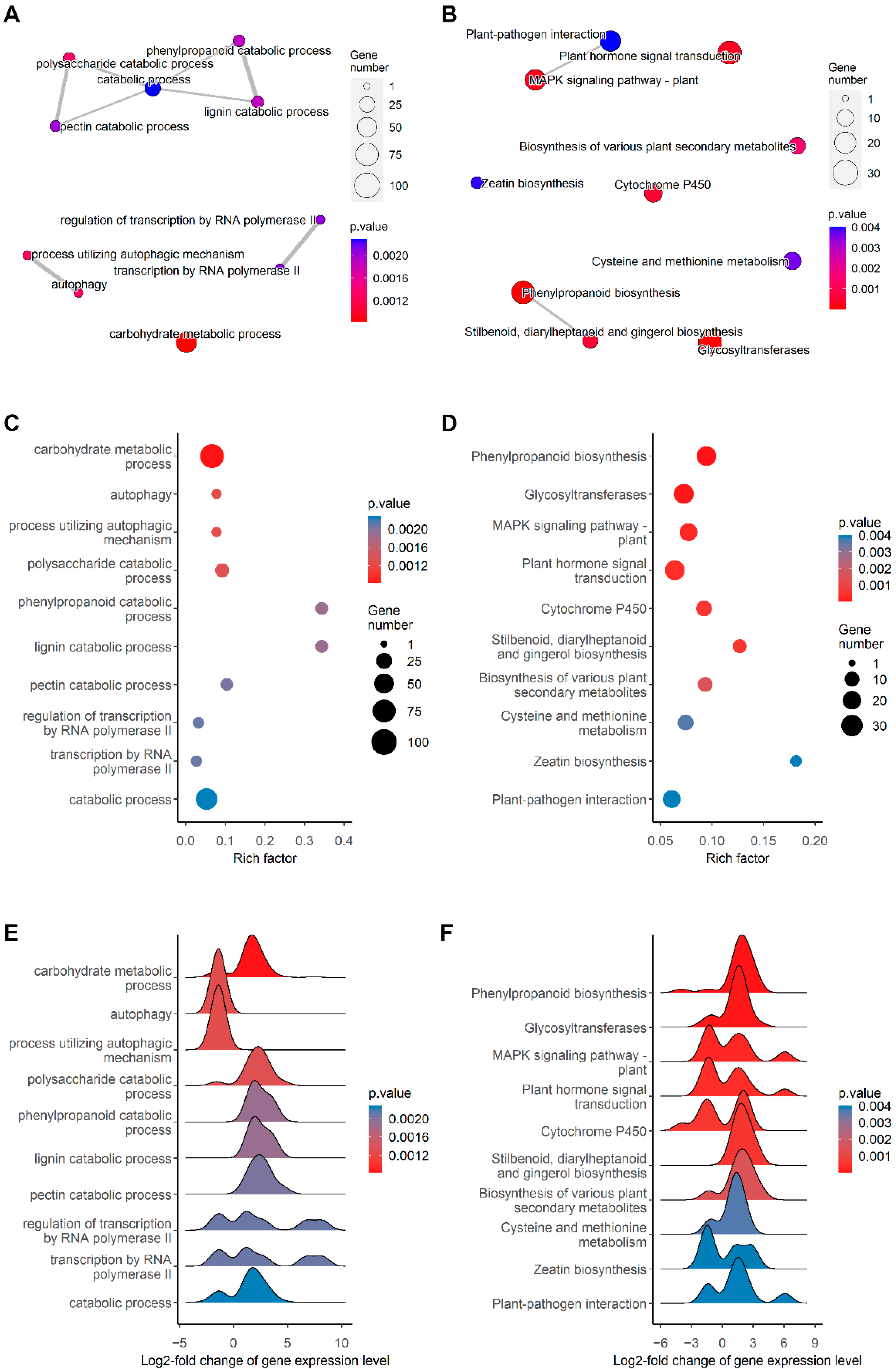
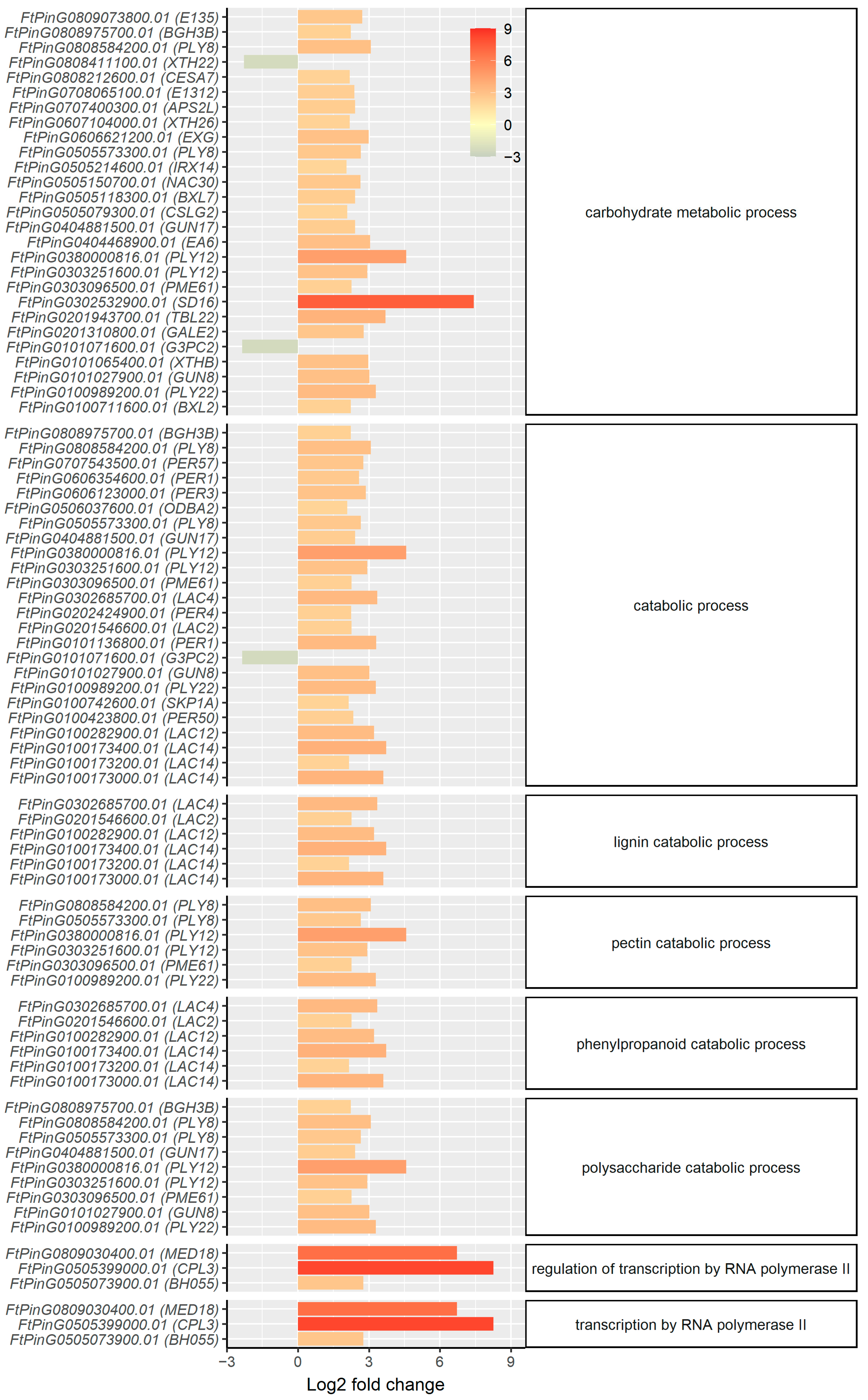
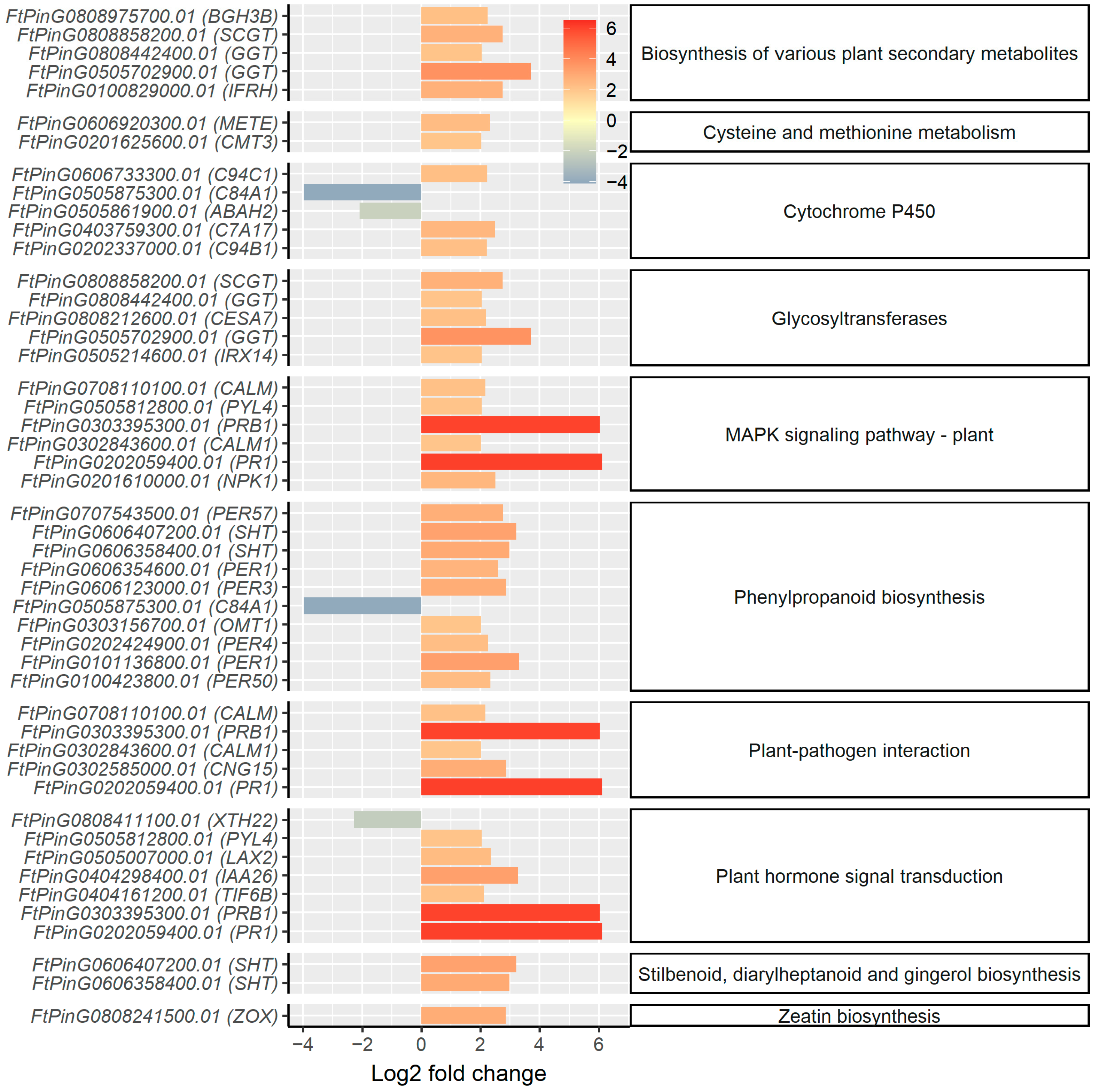
2.3. mRNA Profiling Reveals Upregulation of Genes Involved in Secondary Cell Wall Synthesis and Oxidative Stress Response in the Root
3. Discussion
4. Materials and Methods
4.1. Soil Preparation
4.2. Pot Experiment of Tartary Buckwheat
4.3. Measurement of Cd in Organ Level and Subcellular Fractions
4.4. Measurement of Cd Chemical Form Distribution
4.5. Measurement of H2O2, MDA, and Proline Content
4.6. SOD, POD, and CAT Enzyme Activity Assay
4.7. Transcriptome Analysis of Root
4.8. Statistical Analysis and Data Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shahid, M.; Dumat, C.; Khalid, S.; Niazi, N.K.; Antunes, P.M.C. Cadmium Bioavailability, Uptake, Toxicity and Detoxification in Soil-Plant System. In Reviews of Environmental Contamination and Toxicology; de Voogt, P., Ed.; Springer International Publishing: Cham, Switzerland, 2016; Volume 241, pp. 73–137. ISBN 978-3-319-46944-7. [Google Scholar]
- Li, X.; Zhou, D. A Meta-Analysis on Phenotypic Variation in Cadmium Accumulation of Rice and Wheat: Implications for Food Cadmium Risk Control. Pedosphere 2019, 29, 545–553. [Google Scholar] [CrossRef]
- Arif, N.; Sharma, N.C.; Yadav, V.; Ramawat, N.; Dubey, N.K.; Tripathi, D.K.; Chauhan, D.K.; Sahi, S. Understanding Heavy Metal Stress in a Rice Crop: Toxicity, Tolerance Mechanisms, and Amelioration Strategies. J. Plant Biol. 2019, 62, 239–253. [Google Scholar] [CrossRef]
- El Rasafi, T.; Oukarroum, A.; Haddioui, A.; Song, H.; Kwon, E.E.; Bolan, N.; Tack, F.M.G.; Sebastian, A.; Prasad, M.N.V.; Rinklebe, J. Cadmium Stress in Plants: A Critical Review of the Effects, Mechanisms, and Tolerance Strategies. Crit. Rev. Environ. Sci. Technol. 2022, 52, 675–726. [Google Scholar] [CrossRef]
- Lu, Y.; Song, S.; Wang, R.; Liu, Z.; Meng, J.; Sweetman, A.J.; Jenkins, A.; Ferrier, R.C.; Li, H.; Luo, W.; et al. Impacts of Soil and Water Pollution on Food Safety and Health Risks in China. Environ. Int. 2015, 77, 5–15. [Google Scholar] [CrossRef]
- Wang, J.; Su, J.; Li, Z.; Liu, B.; Cheng, G.; Jiang, Y.; Li, Y.; Zhou, S.; Yuan, W. Source Apportionment of Heavy Metal and Their Health Risks in Soil-Dustfall-Plant System Nearby a Typical Non-Ferrous Metal Mining Area of Tongling, Eastern China. Environ. Pollut. 2019, 254, 113089. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, P.; Zhao, F.-J. Dietary Cadmium Exposure, Risks to Human Health and Mitigation Strategies. Crit. Rev. Environ. Sci. Technol. 2022, 1–25. [Google Scholar] [CrossRef]
- Saini, S.; Kaur, N.; Pati, P.K. Phytohormones: Key Players in the Modulation of Heavy Metal Stress Tolerance in Plants. Ecotoxicol. Environ. Saf. 2021, 223, 112578. [Google Scholar] [CrossRef]
- Dai, Z.-H.; Guan, D.-X.; Bundschuh, J.; Ma, L.Q. Roles of Phytohormones in Mitigating Abiotic Stress in Plants Induced by Metal(Loid)s As, Cd, Cr, Hg, and Pb. Crit. Rev. Environ. Sci. Technol. 2022, 1–21. [Google Scholar] [CrossRef]
- Sytar, O.; Kumari, P.; Yadav, S.; Brestic, M.; Rastogi, A. Phytohormone Priming: Regulator for Heavy Metal Stress in Plants. J. Plant Growth Regul. 2019, 38, 739–752. [Google Scholar] [CrossRef]
- Li, Q.; Wang, G.; Wang, Y.; Yang, D.; Guan, C.; Ji, J. Foliar Application of Salicylic Acid Alleviate the Cadmium Toxicity by Modulation the Reactive Oxygen Species in Potato. Ecotoxicol. Environ. Saf. 2019, 172, 317–325. [Google Scholar] [CrossRef]
- Jini, D.; Joseph, B. Physiological Mechanism of Salicylic Acid for Alleviation of Salt Stress in Rice. Rice Sci. 2017, 24, 97–108. [Google Scholar] [CrossRef]
- Faried, H.N.; Ayyub, C.M.; Amjad, M.; Ahmed, R.; Wattoo, F.M.; Butt, M.; Bashir, M.; Shaheen, M.R.; Waqas, M.A. Salicylic Acid Confers Salt Tolerance in Potato Plants by Improving Water Relations, Gaseous Exchange, Antioxidant Activities and Osmoregulation: Salt Stress Alleviation in Potato by Foliar Application of Salicylic Acid. J. Sci. Food Agric. 2017, 97, 1868–1875. [Google Scholar] [CrossRef]
- Gharib, F.; Hegazi, A. Salicylic Acid Ameliorates Germination, Seedling Growth, Phytohormone and Enzymes Activity in Bean (Phaseolus vulgaris L.) under Cold Stress. J. Am. Sci. 2010, 6, 675–683. [Google Scholar]
- Shakirova, F.M.; Allagulova, C.R.; Maslennikova, D.R.; Klyuchnikova, E.O.; Avalbaev, A.M.; Bezrukova, M.V. Salicylic Acid-Induced Protection against Cadmium Toxicity in Wheat Plants. Environ. Exp. Bot. 2016, 122, 19–28. [Google Scholar] [CrossRef]
- Biareh, V.; Shekari, F.; Sayfzadeh, S.; Zakerin, H.; Hadidi, E.; Beltrão, J.G.T.; Mastinu, A. Physiological and Qualitative Response of Cucurbita pepo L. to Salicylic Acid under Controlled Water Stress Conditions. Horticulturae 2022, 8, 79. [Google Scholar] [CrossRef]
- Ahmad, A.; Aslam, Z.; Naz, M.; Hussain, S.; Javed, T.; Aslam, S.; Raza, A.; Ali, H.M.; Siddiqui, M.H.; Salem, M.Z.M.; et al. Exogenous Salicylic Acid-Induced Drought Stress Tolerance in Wheat (Triticum aestivum L.) Grown under Hydroponic Culture. PLoS ONE 2021, 16, e0260556. [Google Scholar] [CrossRef]
- Zhong, Q.; Hu, H.; Fan, B.; Zhu, C.; Chen, Z. Biosynthesis and Roles of Salicylic Acid in Balancing Stress Response and Growth in Plants. Int. J. Mol. Sci. 2021, 22, 11672. [Google Scholar] [CrossRef]
- Khalvandi, M.; Siosemardeh, A.; Roohi, E.; Keramati, S. Salicylic Acid Alleviated the Effect of Drought Stress on Photosynthetic Characteristics and Leaf Protein Pattern in Winter Wheat. Heliyon 2021, 7, e05908. [Google Scholar] [CrossRef]
- Hongna, C.; Leyuan, T.; Junmei, S.; Xiaori, H.; Xianguo, C. Exogenous Salicylic Acid Signal Reveals an Osmotic Regulatory Role in Priming the Seed Germination of Leymus Chinensis under Salt-Alkali Stress. Environ. Exp. Bot. 2021, 188, 104498. [Google Scholar] [CrossRef]
- Liu, Z.; Ding, Y.; Wang, F.; Ye, Y.; Zhu, C. Role of Salicylic Acid in Resistance to Cadmium Stress in Plants. Plant Cell Rep. 2016, 35, 719–731. [Google Scholar] [CrossRef]
- Pan, J.; Guan, M.; Xu, P.; Chen, M.; Cao, Z. Salicylic Acid Reduces Cadmium (Cd) Accumulation in Rice (Oryza sativa L.) by Regulating Root Cell Wall Composition via Nitric Oxide Signaling. Sci. Total Environ. 2021, 797, 149202. [Google Scholar] [CrossRef]
- Ali, E.; Maodzeka, A.; Hussain, N.; Shamsi, I.H.; Jiang, L. The Alleviation of Cadmium Toxicity in Oilseed Rape (Brassica Napus) by the Application of Salicylic Acid. Plant Growth Regul. 2015, 75, 641–655. [Google Scholar] [CrossRef]
- Guo, B.; Liu, C.; Li, H.; Yi, K.; Ding, N.; Li, N.; Lin, Y.; Fu, Q. Endogenous Salicylic Acid Is Required for Promoting Cadmium Tolerance of Arabidopsis by Modulating Glutathione Metabolisms. J. Hazard. Mater. 2016, 316, 77–86. [Google Scholar] [CrossRef]
- Lu, Q.; Zhang, T.; Zhang, W.; Su, C.; Yang, Y.; Hu, D.; Xu, Q. Alleviation of Cadmium Toxicity in Lemna Minor by Exogenous Salicylic Acid. Ecotoxicol. Environ. Saf. 2018, 147, 500–508. [Google Scholar] [CrossRef]
- Hediji, H.; Kharbech, O.; Massoud, M.B.; Boukari, N.; Debez, A.; Chaibi, W.; Chaoui, A.; Djebali, W. Salicylic Acid Mitigates Cadmium Toxicity in Bean (Phaseolus vulgaris L.) Seedlings by Modulating Cellular Redox Status. Environ. Exp. Bot. 2021, 186, 104432. [Google Scholar] [CrossRef]
- Gondor, O.K.; Janda, T.; Soós, V.; Pál, M.; Majláth, I.; Adak, M.K.; Balázs, E.; Szalai, G. Salicylic Acid Induction of Flavonoid Biosynthesis Pathways in Wheat Varies by Treatment. Front. Plant Sci. 2016, 7, 1447. [Google Scholar] [CrossRef]
- Jia, H.; Wang, X.; Wei, T.; Wang, M.; Liu, X.; Hua, L.; Ren, X.; Guo, J.; Li, J. Exogenous Salicylic Acid Regulates Cell Wall Polysaccharides Synthesis and Pectin Methylation to Reduce Cd Accumulation of Tomato. Ecotoxicol. Environ. Saf. 2021, 207, 111550. [Google Scholar] [CrossRef]
- Riaz, M.; Kamran, M.; Rizwan, M.; Ali, S.; Parveen, A.; Malik, Z.; Wang, X. Cadmium Uptake and Translocation: Selenium and Silicon Roles in Cd Detoxification for the Production of Low Cd Crops: A Critical Review. Chemosphere 2021, 273, 129690. [Google Scholar] [CrossRef]
- Kim, S.-J.; Zaidul, I.S.M.; Suzuki, T.; Mukasa, Y.; Hashimoto, N.; Takigawa, S.; Noda, T.; Matsuura-Endo, C.; Yamauchi, H. Comparison of Phenolic Compositions between Common and Tartary Buckwheat (Fagopyrum) Sprouts. Food Chem. 2008, 110, 814–820. [Google Scholar] [CrossRef]
- Bonafaccia, G.; Marocchini, M.; Kreft, I. Composition and Technological Properties of the Flour and Bran from Common and Tartary Buckwheat. Food Chem. 2003, 80, 9–15. [Google Scholar] [CrossRef]
- Vombergar, B.; Tašner, L.; Horvat, M.; Vorih, S.; Pem, N.; Golob, S.; Kovač, T. Buckwheat—Challenges in Nutrition and Technology/Ajda—Izzivi v Tehnologiji in Prehrani. Fagopyrum 2022, 39, 33–42. [Google Scholar] [CrossRef]
- Pirzadah, T.B.; Malik, B.; Tahir, I.; Irfan, Q.M.; Rehman, R.U. Characterization of Mercury-Induced Stress Biomarkers in Fagopyrum Tataricum Plants. Int. J. Phytoremediat. 2018, 20, 225–236. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, Q.; Li, J.; Xiong, J.; Zhou, L.; He, S.; Zhang, J.; Chen, Z.; He, S.; Liu, H. Effects of Exogenous Sulfur on Alleviating Cadmium Stress in Tartary Buckwheat. Sci. Rep. 2019, 9, 7397. [Google Scholar] [CrossRef]
- Pirzadah, T.B.; Malik, B.; Tahir, I.; Hakeem, K.R.; Alharby, H.F.; Rehman, R.U. Lead Toxicity Alters the Antioxidant Defense Machinery and Modulate the Biomarkers in Tartary Buckwheat Plants. Int. Biodeterior. Biodegrad. 2020, 151, 104992. [Google Scholar] [CrossRef]
- Wang, L.; Zheng, B.; Yuan, Y.; Xu, Q.; Chen, P. Transcriptome Profiling of Fagopyrum Tataricum Leaves in Response to Lead Stress. BMC Plant Biol. 2020, 20, 54. [Google Scholar] [CrossRef]
- Li, Z.; Li, Z.; Huang, Y.; Jiang, Y.; Liu, Y.; Wen, W.; Li, H.; Shao, J.; Wang, C.; Zhu, X. Antioxidant Capacity, Metal Contents, and Their Health Risk Assessment of Tartary Buckwheat Teas. ACS Omega 2020, 5, 9724–9732. [Google Scholar] [CrossRef]
- Emamverdian, A.; Ding, Y.; Mokhberdoran, F. The Role of Salicylic Acid and Gibberellin Signaling in Plant Responses to Abiotic Stress with an Emphasis on Heavy Metals. Plant Signal. Behav. 2020, 15, 1777372. [Google Scholar] [CrossRef]
- Sharma, A.; Sidhu, G.P.S.; Araniti, F.; Bali, A.S.; Shahzad, B.; Tripathi, D.K.; Brestic, M.; Skalicky, M.; Landi, M. The Role of Salicylic Acid in Plants Exposed to Heavy Metals. Molecules 2020, 25, 540. [Google Scholar] [CrossRef]
- He, J.; Ren, Y.; Pan, X.; Yan, Y.; Zhu, C.; Jiang, D. Salicylic Acid Alleviates the Toxicity Effect of Cadmium on Germination, Seedling Growth, and Amylase Activity of Rice. J. Plant Nutr. Soil Sci. 2010, 173, 300–305. [Google Scholar] [CrossRef]
- Khan, I.; Seleiman, M.F.; Chattha, M.U.; Jalal, R.S.; Mahmood, F.; Hassan, F.A.S.; Izzet, W.; Alhammad, B.A.; Ali, E.F.; Roy, R.; et al. Enhancing Antioxidant Defense System of Mung Bean with a Salicylic Acid Exogenous Application to Mitigate Cadmium Toxicity. Not. Bot. Horti Agrobot. Cluj-Napoca 2021, 49, 12303. [Google Scholar] [CrossRef]
- Moussa, H.R.; El-Gamal, S.M. Effect of Salicylic Acid Pretreatment on Cadmium Toxicity in Wheat. Biol. Plant. 2010, 54, 315–320. [Google Scholar] [CrossRef]
- Faraz, A.; Faizan, M.; Sami, F.; Siddiqui, H.; Hayat, S. Supplementation of Salicylic Acid and Citric Acid for Alleviation of Cadmium Toxicity to Brassica Juncea. J. Plant Growth Regul. 2020, 39, 641–655. [Google Scholar] [CrossRef]
- Gul, F.; Arfan, M.; Shahbaz, M.; Basra, S.M.A. Salicylic Acid Seed Priming Modulates Morphology, Nutrient Relations and Photosynthetic Attributes of Wheat Grown under Cadmium Stress. Int. J. Agric. Biol. 2020, 23, 9. [Google Scholar]
- Zhu, C.Q.; Hu, W.J.; Cao, X.C.; Zhu, L.F.; Bai, Z.G.; Huang, J.; Liang, Q.D.; Jin, Q.Y.; Zhang, J.H. Role of Salicylic Acid in Alleviating the Inhibition of Root Elongation by Suppressing Ethylene Emission in Rice under Al Toxicity Conditions. Plant Growth Regul. 2020, 90, 475–487. [Google Scholar] [CrossRef]
- Bagautdinova, Z.Z.; Omelyanchuk, N.; Tyapkin, A.V.; Kovrizhnykh, V.V.; Lavrekha, V.V.; Zemlyanskaya, E.V. Salicylic Acid in Root Growth and Development. Int. J. Mol. Sci. 2022, 23, 2228. [Google Scholar] [CrossRef]
- Li, C.; Liu, Z.; Zhang, Q.; Wang, R.; Xiao, L.; Ma, H.; Chong, K.; Xu, Y. SKP1 Is Involved in Abscisic Acid Signalling to Regulate Seed Germination, Stomatal Opening and Root Growth in Arabidopsis Thaliana: SKP1 Is Involved in ABA Signalling. Plant Cell Environ. 2012, 35, 952–965. [Google Scholar] [CrossRef]
- Yang, X.; Kim, M.Y.; Ha, J.; Lee, S.-H. Overexpression of the Soybean NAC Gene GmNAC109 Increases Lateral Root Formation and Abiotic Stress Tolerance in Transgenic Arabidopsis Plants. Front. Plant Sci. 2019, 10, 1036. [Google Scholar] [CrossRef]
- Sytar, O.; Kumar, A.; Latowski, D.; Kuczynska, P.; Strzałka, K.; Prasad, M.N.V. Heavy Metal-Induced Oxidative Damage, Defense Reactions, and Detoxification Mechanisms in Plants. Acta Physiol. Plant. 2013, 35, 985–999. [Google Scholar] [CrossRef]
- Wani, A.B.; Chadar, H.; Wani, A.H.; Singh, S.; Upadhyay, N. Salicylic Acid to Decrease Plant Stress. Environ. Chem. Lett. 2017, 15, 101–123. [Google Scholar] [CrossRef]
- Tognolli, M.; Penel, C.; Greppin, H.; Simon, P. Analysis and Expression of the Class III Peroxidase Large Gene Family in Arabidopsis Thaliana. Gene 2002, 288, 129–138. [Google Scholar] [CrossRef]
- Singh, S.; Singh, V.P.; Prasad, S.M.; Sharma, S.; Ramawat, N.; Dubey, N.K.; Tripathi, D.K.; Chauhan, D.K. Interactive Effect of Silicon (Si) and Salicylic Acid (SA) in Maize Seedlings and Their Mechanisms of Cadmium (Cd) Toxicity Alleviation. J. Plant Growth Regul. 2019, 38, 1587–1597. [Google Scholar] [CrossRef]
- Popova, L.P.; Maslenkova, L.T.; Yordanova, R.Y.; Ivanova, A.P.; Krantev, A.P.; Szalai, G.; Janda, T. Exogenous Treatment with Salicylic Acid Attenuates Cadmium Toxicity in Pea Seedlings. Plant Physiol. Biochem. 2009, 47, 224–231. [Google Scholar] [CrossRef]
- Lu, H.; Li, Z.; Wu, J.; Shen, Y.; Li, Y.; Zou, B.; Tang, Y.; Zhuang, P. Influences of Calcium Silicate on Chemical Forms and Subcellular Distribution of Cadmium in Amaranthus hypochondriacus L. Sci. Rep. 2017, 7, 40583. [Google Scholar] [CrossRef]
- Bai, X.; Dong, Y.; Kong, J.; Xu, L.; Liu, S. Effects of Application of Salicylic Acid Alleviates Cadmium Toxicity in Perennial Ryegrass. Plant Growth Regul. 2015, 75, 695–706. [Google Scholar] [CrossRef]
- Wang, Q.; Liang, X.; Dong, Y.; Xu, L.; Zhang, X.; Kong, J.; Liu, S. Effects of Exogenous Salicylic Acid and Nitric Oxide on Physiological Characteristics of Perennial Ryegrass Under Cadmium Stress. J. Plant Growth Regul. 2013, 32, 721–731. [Google Scholar] [CrossRef]
- DalCorso, G.; Farinati, S.; Maistri, S.; Furini, A. How Plants Cope with Cadmium: Staking All on Metabolism and Gene Expression. J. Integr. Plant Biol. 2008, 50, 1268–1280. [Google Scholar] [CrossRef]
- Angulo-Bejarano, P.I.; Puente-Rivera, J.; Cruz-Ortega, R. Metal and Metalloid Toxicity in Plants: An Overview on Molecular Aspects. Plants 2021, 10, 635. [Google Scholar] [CrossRef]
- Bernal, M.; Krämer, U. Involvement of Arabidopsis Multi-Copper Oxidase-Encoding LACCASE12 in Root-to-Shoot Iron Partitioning: A Novel Example of Copper-Iron Crosstalk. Front. Plant Sci. 2021, 12, 688318. [Google Scholar] [CrossRef]
- Jarosz-Wilkołazka, A.; Grąz, M.; Braha, B.; Menge, S.; Schlosser, D.; Krauss, G.-J. Species-Specific Cd-Stress Response in the White Rot Basidiomycetes Abortiporus Biennis and Cerrena Unicolor. Biometals 2006, 19, 39–49. [Google Scholar] [CrossRef]
- Kobyletska, M.; Kavulych, Y.; Romanyuk, N.; Korchynska, O.; Terek, O. Exogenous Salicylic Acid Modifies Cell Wall Lignification, Total Phenolic Content, PAL-Activity in Wheat (Triticum aestivum L.) and Buckwheat (Fagopyrum Esculentum Moench) Plants under Cadmium Chloride Impact. Biointerface Res. Appl. Chem. 2022, 13, 117. [Google Scholar] [CrossRef]
- Lüthje, S.; Martinez-Cortes, T. Membrane-Bound Class III Peroxidases: Unexpected Enzymes with Exciting Functions. Int. J. Mol. Sci. 2018, 19, 2876. [Google Scholar] [CrossRef]
- Ahmed, W.; Imran, M.; Yaseen, M.; Haq, T.U.; Jamshaid, M.U.; Rukh, S.; Ikram, R.M.; Ali, M.; Ali, A.; Maqbool, M.; et al. Role of Salicylic Acid in Regulating Ethylene and Physiological Characteristics for Alleviating Salinity Stress on Germination, Growth and Yield of Sweet Pepper. PeerJ 2020, 8, e8475. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Ye, Z.-H. Global Analysis of Direct Targets of Secondary Wall NAC Master Switches in Arabidopsis. Mol. Plant 2010, 3, 1087–1103. [Google Scholar] [CrossRef]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic Acid, a Multifaceted Hormone to Combat Disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef]
- Aires, E.S.; Ferraz, A.K.L.; Carvalho, B.L.; Teixeira, F.P.; Putti, F.F.; de Souza, E.P.; Rodrigues, J.D.; Ono, E.O. Foliar Application of Salicylic Acid to Mitigate Water Stress in Tomato. Plants 2022, 11, 1775. [Google Scholar] [CrossRef]
- Barros, T.C.; de Mello Prado, R.; Roque, C.G.; Arf, M.V.; Vilela, R.G. Silicon and Salicylic Acid in the Physiology and Yield of Cotton. J. Plant Nutr. 2019, 42, 458–465. [Google Scholar] [CrossRef]
- El–Bially, M.E.; Saudy, H.S.; Hashem, F.A.; El–Gabry, Y.A.; Shahin, M.G. Salicylic Acid as a Tolerance Inducer of Drought Stress on Sunflower Grown in Sandy Soil. Gesunde Pflanz. 2022, 74, 603–613. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Matin, M.A.; Fardus, J.; Hasanuzzaman, M.; Hossain, M.S.; Parvin, K. Foliar Application of Salicylic Acid Improves Growth and Yield Attributes by Upregulating the Antioxidant Defense System in Brassica Campestris Plants Grown in Lead-Amended Soils. Acta Agrobot. 2019, 72, 2. [Google Scholar] [CrossRef]
- Majumdar, S.; Sachdev, S.; Kundu, R. Salicylic Acid Mediated Reduction in Grain Cadmium Accumulation and Amelioration of Toxicity in Oryza sativa L. Cv Bandana. Ecotoxicol. Environ. Saf. 2020, 205, 111167. [Google Scholar] [CrossRef]
- Naeem, M.; Basit, A.; Ahmad, I.; Mohamed, H.I.; Wasila, H. Effect of Salicylic Acid and Salinity Stress on the Performance of Tomato Plants. Gesunde Pflanz. 2020, 72, 393–402. [Google Scholar] [CrossRef]
- Safar-Noori, M.; Assaha, D.V.M.; Saneoka, H. Effect of Salicylic Acid and Potassium Application on Yield and Grain Nutritional Quality of Wheat under Drought Stress Condition. Cereal Res. Commun. 2018, 46, 558–568. [Google Scholar] [CrossRef]
- Tahjib-Ul-Arif, M.; Siddiqui, M.N.; Sohag, A.A.M.; Sakil, M.A.; Rahman, M.M.; Polash, M.A.S.; Mostofa, M.G.; Tran, L.-S.P. Salicylic Acid-Mediated Enhancement of Photosynthesis Attributes and Antioxidant Capacity Contributes to Yield Improvement of Maize Plants Under Salt Stress. J. Plant Growth Regul. 2018, 37, 1318–1330. [Google Scholar] [CrossRef]
- Khan, S.; Naushad, M.; Lima, E.C.; Zhang, S.; Shaheen, S.M.; Rinklebe, J. Global Soil Pollution by Toxic Elements: Current Status and Future Perspectives on the Risk Assessment and Remediation Strategies—A Review. J. Hazard. Mater. 2021, 417, 126039. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, C.W.A.D.; Biondi, C.M.; da Silva, F.B.V.; Lima, L.H.V. Using Plants to Remediate or Manage Metal-Polluted Soils: An Overview on the Current State of Phytotechnologies. Acta Sci. Agron. 2021, 43, e58283. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Q.; Liao, X.; Li, X.; Zheng, S.; Zhao, F. Phytoexclusion of Heavy Metals Using Low Heavy Metal Accumulating Cultivars: A Green Technology. J. Hazard. Mater. 2021, 413, 125427. [Google Scholar] [CrossRef]
- World Reference Base for Soil Resources 2014: International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; FAO: Rome, Italy, 2014; ISBN 978-92-5-108369-7.
- National Environmental Protection Bureau, State Bureau of Technical Supervision. Soil Environmental Quality Risk Control Standard for Soil Contamination of Agricultural Land; People’s Republic of China: Ministry of Ecology and Environment: Beijing, China, 2018.
- Weigel, H.J.; Jäger, H.J. Subcellular Distribution and Chemical Form of Cadmium in Bean Plants. Plant Physiol. 1980, 65, 480–482. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Hasanuzzaman, M.; Fujita, M. Up-Regulation of Antioxidant and Glyoxalase Systems by Exogenous Glycinebetaine and Proline in Mung Bean Confer Tolerance to Cadmium Stress. Physiol. Mol. Biol. Plants 2010, 16, 259–272. [Google Scholar] [CrossRef]
- Hossain, M.A.; Fujita, M. Evidence for a Role of Exogenous Glycinebetaine and Proline in Antioxidant Defense and Methylglyoxal Detoxification Systems in Mung Bean Seedlings under Salt Stress. Physiol. Mol. Biol. Plants 2010, 16, 19–29. [Google Scholar] [CrossRef]
- Trotel, P.; Bouchereau, A.; Niogret, M.F.; Larher, F. The Fate of Osmo-Accumulated Proline in Leaf Discs of Rape (Brassica napus L.) Incubated in a Medium of Low Osmolarity. Plant Sci. 1996, 118, 31–45. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, X.; Ma, B.; Gao, Q.; Du, H.; Han, Y.; Li, Y.; Cao, Y.; Qi, M.; Zhu, Y.; et al. The Tartary Buckwheat Genome Provides Insights into Rutin Biosynthesis and Abiotic Stress Tolerance. Mol. Plant 2017, 10, 1224–1237. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of MRNA Abundance Using RNA-Seq Data: RPKM Measure Is Inconsistent among Samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Madden, T. The BLAST Sequence Analysis Tool. In the NCBI Handbook [Internet]; National Center for Biotechnology Information: Bethesda, MD, USA, 2002; p. 229. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Schurch, N.J.; Schofield, P.; Gierliński, M.; Cole, C.; Sherstnev, A.; Singh, V.; Wrobel, N.; Gharbi, K.; Simpson, G.G.; Owen-Hughes, T.; et al. How Many Biological Replicates Are Needed in an RNA-Seq Experiment and Which Differential Expression Tool Should You Use? RNA 2016, 22, 839–851. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Yan, L. Ggvenn: Draw Venn Diagram by “Ggplot2”. 2021. Available online: https://CRAN.R-project.org/package=ggvenn (accessed on 8 October 2022).
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. 2022. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 8 October 2022).
- Yu, G. Enrichplot: Visualization of Functional Enrichment Result. 2022. Available online: https://yulab-smu.top/biomedical-knowledge-mining-book/ (accessed on 8 October 2022).
- Wilke, C.O. Ggridges: Ridgeline Plots in “Ggplot2”. 2022. Available online: https://CRAN.R-project.org/package=ggridges (accessed on 8 October 2022).
- Kolde, R. Pheatmap: Pretty Heatmaps. 2019. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 8 October 2022).
| Treatment | H2O2 Content (µmol g−1) | MDA Content (nmol g−1) | Pro Content (µg g−1) | POD Activity (U g−1) | SOD Activity (U g−1) | CAT Activity (U g−1) |
|---|---|---|---|---|---|---|
| C | 0.56 ± 0.01 c | 2.5 ± 0.3 c | 80 ± 1 ab | 768 ± 106 a | 631 ± 25 b | 393 ± 24 ab |
| Cd | 0.73 ± 0.03 b | 5.7 ± 0.1 a | 51 ± 2 c | 399 ± 22 cd | 509 ± 17 c | 181 ± 81 c |
| Cd + SA25 | 0.88 ± 0.04 a | 5.8 ± 0.5 a | 67 ± 1 bc | 531 ± 10 bc | 629 ± 18 b | 329 ± 39 abc |
| Cd + SA50 | 0.73 ± 0.03 b | 5.1 ± 0.2 a | 81 ± 6 ab | 385 ± 23 d | 692 ± 43 ab | 270 ± 138 bc |
| Cd + SA75 | 0.5 ± 0.05 c | 3.6 ± 0.2 b | 81 ± 6 ab | 492 ± 40 bcd | 727 ± 5 ab | 373 ± 50 abc |
| Cd + SA100 | 0.45 ± 0.03 c | 3.2 ± 0.2 bc | 98 ± 10 a | 601 ± 23 b | 797 ± 83 a | 526 ± 47 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, S.; Wang, K.; Li, Z.; Li, H.; Shao, J.; Zhu, X. Salicylic Acid Enhances Cadmium Tolerance and Reduces Its Shoot Accumulation in Fagopyrum tataricum Seedlings by Promoting Root Cadmium Retention and Mitigating Oxidative Stress. Int. J. Mol. Sci. 2022, 23, 14746. https://doi.org/10.3390/ijms232314746
Luo S, Wang K, Li Z, Li H, Shao J, Zhu X. Salicylic Acid Enhances Cadmium Tolerance and Reduces Its Shoot Accumulation in Fagopyrum tataricum Seedlings by Promoting Root Cadmium Retention and Mitigating Oxidative Stress. International Journal of Molecular Sciences. 2022; 23(23):14746. https://doi.org/10.3390/ijms232314746
Chicago/Turabian StyleLuo, Siwei, Kaiyi Wang, Zhiqiang Li, Hanhan Li, Jirong Shao, and Xuemei Zhu. 2022. "Salicylic Acid Enhances Cadmium Tolerance and Reduces Its Shoot Accumulation in Fagopyrum tataricum Seedlings by Promoting Root Cadmium Retention and Mitigating Oxidative Stress" International Journal of Molecular Sciences 23, no. 23: 14746. https://doi.org/10.3390/ijms232314746
APA StyleLuo, S., Wang, K., Li, Z., Li, H., Shao, J., & Zhu, X. (2022). Salicylic Acid Enhances Cadmium Tolerance and Reduces Its Shoot Accumulation in Fagopyrum tataricum Seedlings by Promoting Root Cadmium Retention and Mitigating Oxidative Stress. International Journal of Molecular Sciences, 23(23), 14746. https://doi.org/10.3390/ijms232314746