EGFR-Targeted Cellular Delivery of Therapeutic Nucleic Acids Mediated by Boron Clusters
Abstract
:1. Introduction
2. Results
2.1. Chemistry
2.2. Pull-Down Assay to Identify the Pool of Proteins Interacting with the Boron Cluster Bait and Proteomic Analysis by Liquid Chromatography-Mass Spectrometry LC-MS
2.3. Cellular Accumulation of Boron Is Modulated by EGFR Overexpressed in Cancer Cells
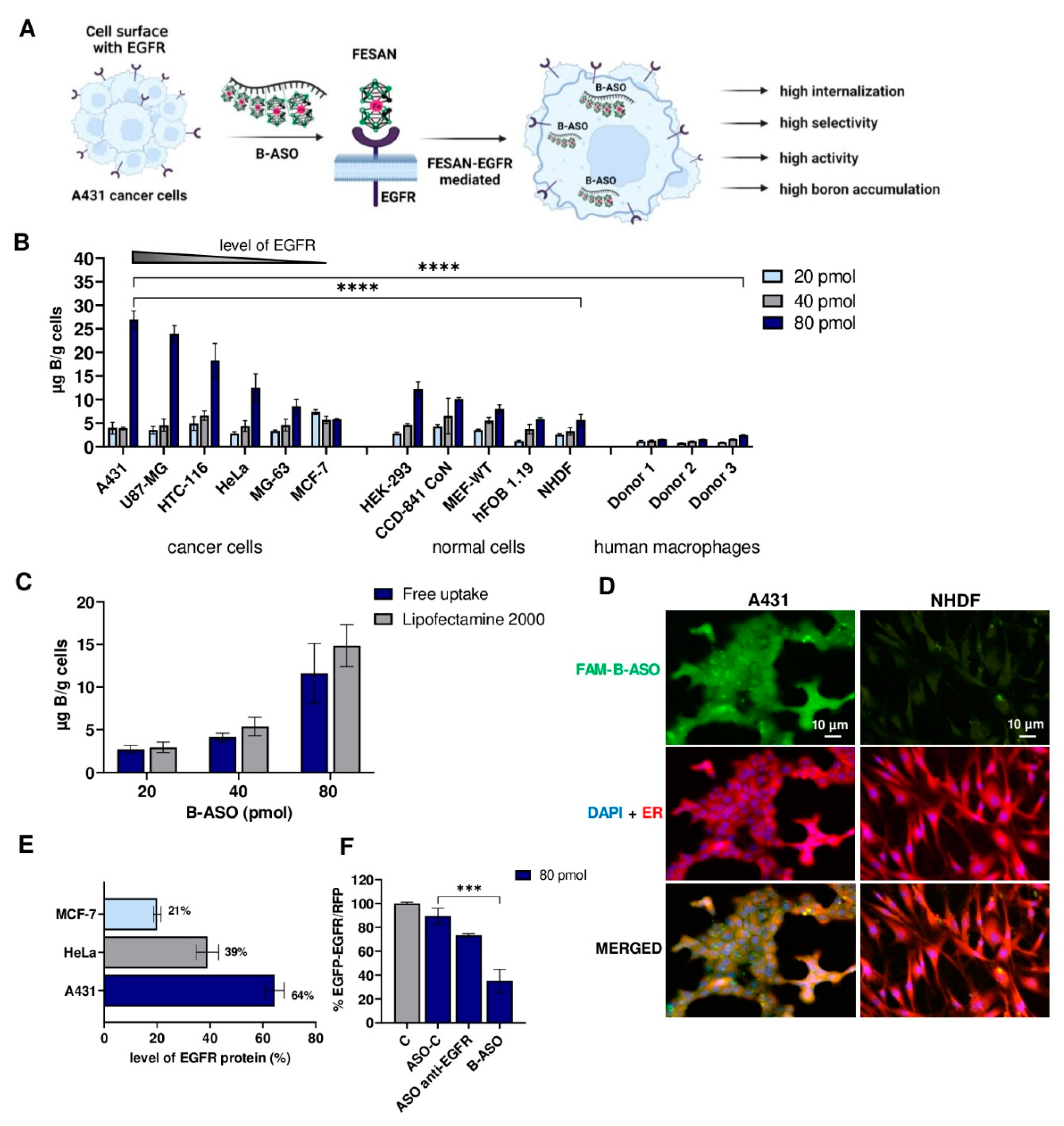
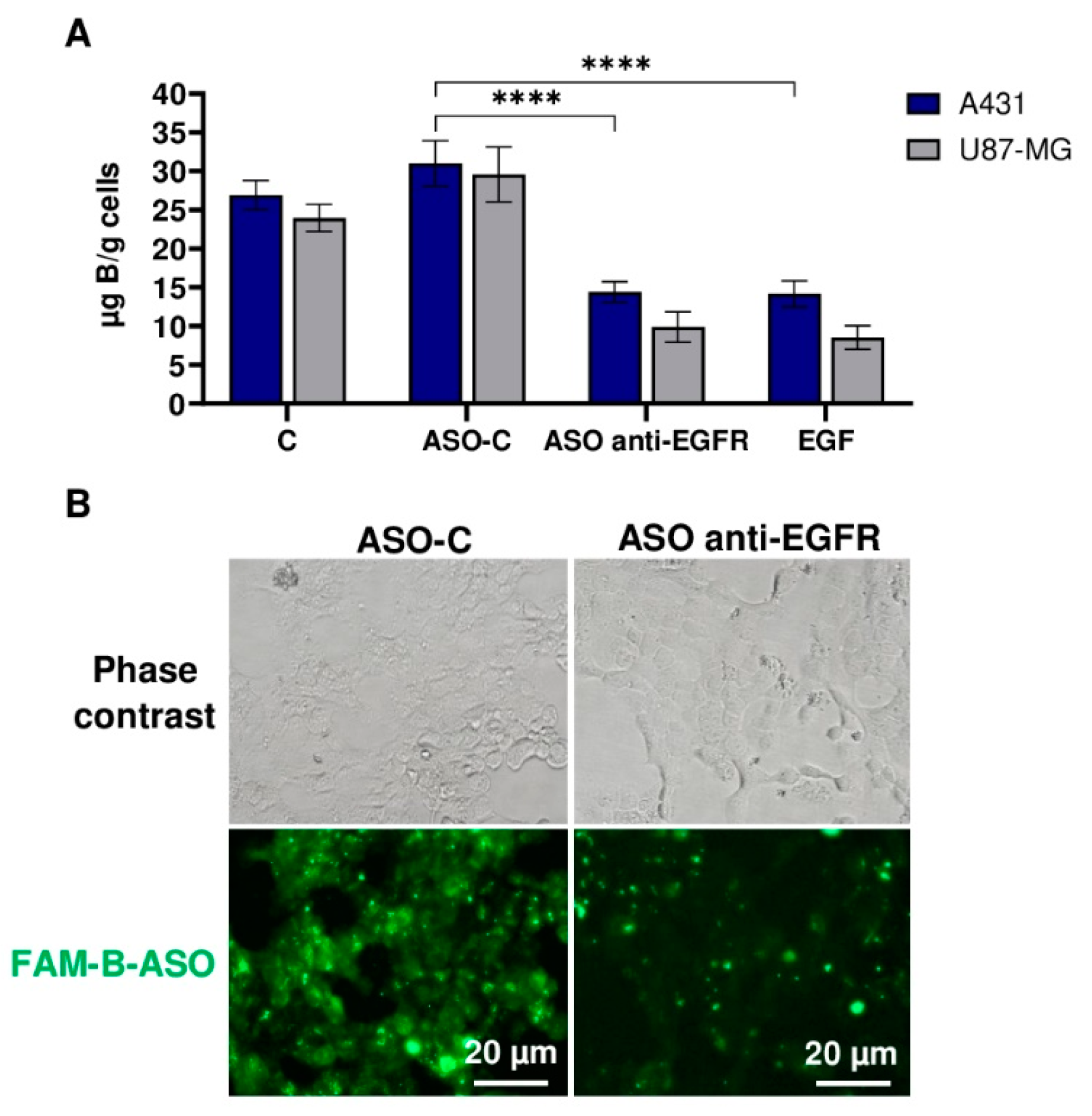
2.4. Reduction of EGFR Level in Cancer Cells Decreases the Free Uptake of B-ASO
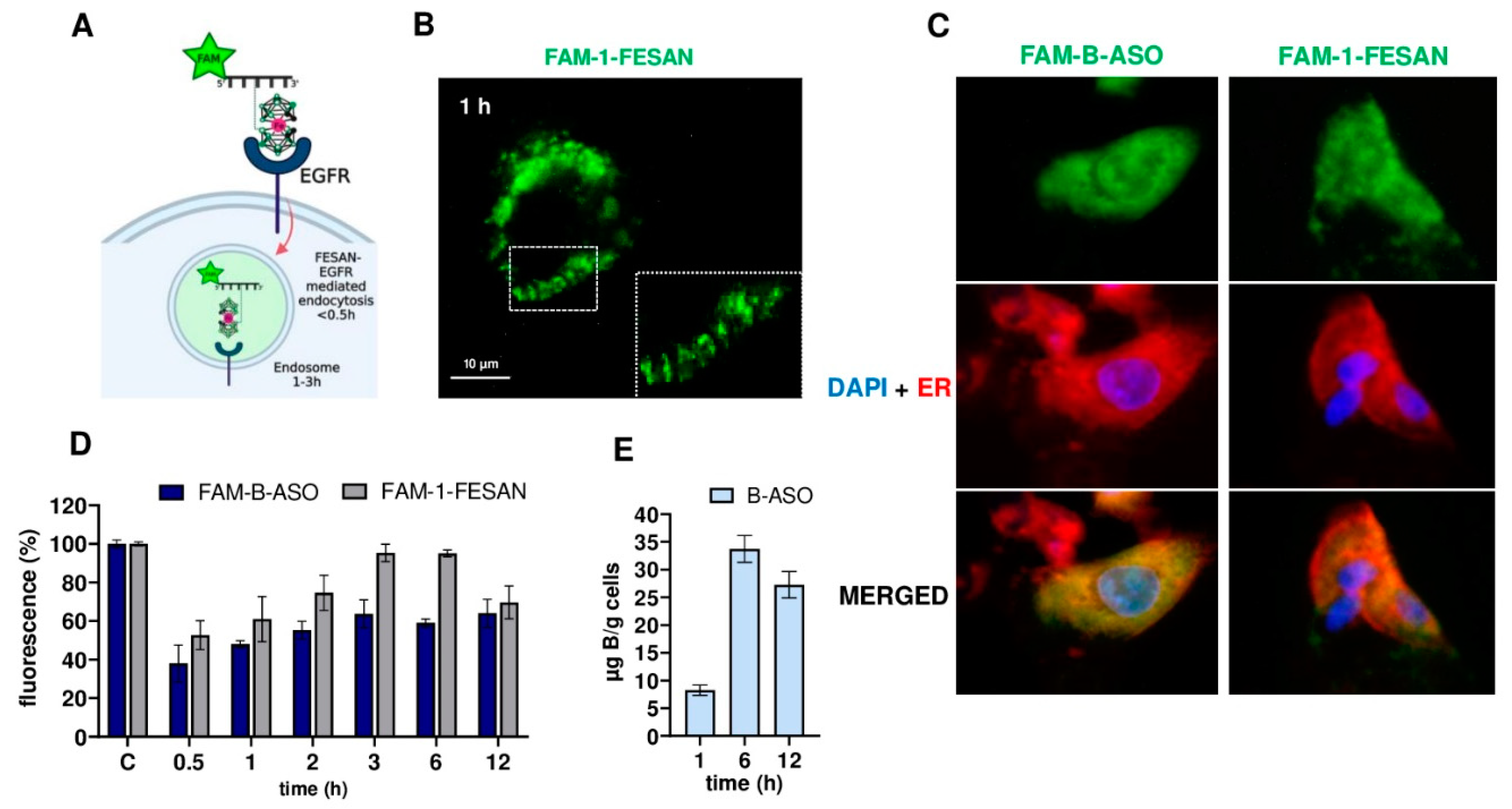
2.5. Time Course of the Free Uptake of Boron Cluster Decorated FAM-B-ASO or FAM-1-FESAN
3. Discussion
4. Materials and Methods
4.1. Synthesis of Oligonucleotides
4.2. Synthesis and Purification of Oligonucleotides Modified with Boron Cluster
4.3. RP-HPLC Analysis
4.4. Pull-Down Assay Using Biotinylated BIOT-1 Probe Conjugated with Boron Clusters (FESAN or 1,2-DCDDB)
4.5. Cell Lines and Culture Conditions
4.6. Determination of the Boron Content in Cancer and Normal Cells by ICP MS Measurements
4.7. Human Primary Monocyte-Derived Macrophages and Measurement of Free Uptake B-ASO by Using ICP MS Analysis
4.8. Dual Fluorescence Assay (DFA) of Silencing Activity of B-ASOs
4.9. Microscopic Analysis of the FAM-B-ASO and FAM-1-FESAN Uptake by Cancer and Normal Cells
4.10. Reduction and Blocking of EGFR in Cancer Cells
- (I)
- After 24 h of incubation, the cells should be 80% confluent. Directly before the transfection, the cell media was replaced with antibiotic-free medium (100 µL per well). Cells were transfected using the Lipofectamine 2000 transfection reagent at a 2:1 ratio (2 μL of Lipofectamine 2000 per 1 μg of nucleic acid ASO anti-EGFR or ASO-C at 200nM) according to the manufacturer’s protocol. After 5 h of incubation, the transfection mixture was replaced with 200 µL of fresh full medium. After 36 h of incubation, the media was replaced with 100 µL basic media (without FBS) containing aqueous solutions of B-ASO (80 pmol/well). Then, the cells were incubated for 12 h at 37 °C and 5% CO2. After 12 h of incubation, the media was removed. The cells were treated with 20 µL of trypsin per well for 1.5 h. Then, the cell was diluted with ultrapure water up to 1.5 mL volume for ICP MS analysis which was described above.
- (II)
- After 24 h, the full medium was mixed with EGF (final concentration 10 ng/mL) and incubated for 1 h at 37 °C and 5% CO2. Then, the mixture was replaced with 100 µL basic media (without FBS) containing B-ASO (80 pmol/well), and the cells were incubated for 12 h at 37 °C and 5% CO2.
4.11. Time Course Analysis of Cellular Uptake of FAM-B-ASO and FAM-1-FESAN
4.12. Western Blot Analysis of EGFR Protein Level in A431, HeLa and MCF-7 Cells
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schlessinger, J. Receptor tyrosine kinases: Legacy of the first two decades. Cold Spring Harb. Perspect. Biol. 2014, 6, a008912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purba, E.R.; Saita, E.I.; Maruyama, I.N. Activation of the EGF receptor by ligand binding and oncogenic mutations: The “rotation model”. Cells 2017, 6, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Y.; Mohammad, I.S.; Liu, Z. Overview of the STAT-3 signaling pathway in cancer and the development of specific inhibitors (Review). Oncol. Lett. 2020, 19, 2585–2594. [Google Scholar] [CrossRef]
- Adrain, C.; Freeman, M. Regulation of receptor tyrosine kinase ligand processing. Cold Spring Harb. Perspect. Biol. 2014, 6, a008995. [Google Scholar] [CrossRef] [Green Version]
- Shao, Q.; Zhu, W. Ligand binding effects on the activation of the EGFR extracellular domain. Phys. Chem. Chem. Phys. 2019, 21, 8141–8151. [Google Scholar] [CrossRef]
- Freed, D.M.; Bessman, N.J.; Kiyatkin, A.; Salazar-Cavazos, E.; Byrne, P.O.; Moore, J.O.; Valley, C.C.; Ferguson, K.M.; Leahy, D.J.; Lidke, D.S.; et al. EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics. Cell 2017, 171, 683–695. [Google Scholar] [CrossRef] [Green Version]
- Sigismund, S.; Avanzato, D.; Lanzetti, L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018, 12, 3–20. [Google Scholar] [CrossRef] [Green Version]
- Chandra, P.; Ticku, S.; Pallavi, B.K.; Rudresh, K.B.; Mansabdar, P.; Rajaram, P. Epidermal growth factor receptor: Role in human cancer. Indian J. Dent. Res. 2017, 28, 687–694. [Google Scholar] [CrossRef]
- Lee, H.J.; Seo, A.N.; Kim, E.J.; Jang, M.H.; Kim, Y.J.; Kim, J.H.; Kim, S.W.; Ryu, H.S.; Park, I.A.; Im, S.A.; et al. Prognostic and predictive values of EGFR overexpression and EGFR copy number alteration in HER2-positive breast cancer. Br. J. Cancer 2015, 112, 103–111. [Google Scholar] [CrossRef] [Green Version]
- Saade, F.S.; Mahfouz, R.; Assi, H.I. EGFR as a clinical marker in glioblastomas and other gliomas. Int. J. Biol. Markers 2018, 33, 22–32. [Google Scholar] [CrossRef]
- Hashmi, A.A.; Hussain, Z.F.; Irfan, M.; Khan, E.Y.; Faridi, N.; Naqvi, H.; Khan, A.; Edhi, M.M. Prognostic significance of epidermal growth factor receptor (EGFR) over expression in urothelial carcinoma of urinary bladder. BMC Urol. 2018, 18, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegelin, M.D.; Borczuk, A.C. Epidermal growth factor receptor mutations in lung adenocarcinoma. Lab. Investig. 2014, 94, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Zeng, F.; Harris, R.C. Epidermal growth factor, from gene organization to bedside. Semin. Cell Dev. Biol. 2014, 28, 2–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, H.K.; Cvrljevic, A.N.; Johns, T.G. The epidermal growth factor receptor variant III (EGFRvIII): Where wild things are altered. FEBS J. 2013, 280, 5350–5370. [Google Scholar] [CrossRef] [PubMed]
- Moreira, D.; Sampath, S.; Won, H.; White, S.V.; Su, Y.L.; Alcantara, M.; Wang, C.; Lee, P.; Maghami, E.; Massarelli, E.; et al. Myeloid cell-targeted STAT3 inhibition sensitizes head and neck cancers to radiotherapy and T cell-mediated immunity. J. Clin. Investig. 2021, 131, e137001. [Google Scholar] [CrossRef] [PubMed]
- Adamus, T.; Hung, C.Y.; Yu, C.; Kang, E.; Hammad, M.; Flores, L.; Nechaev, S.; Zhang, Q.; Gonzaga, J.M.; Muthaiyah, K.; et al. Glioma-targeted delivery of exosome-encapsulated antisense oligonucleotides using neural stem cells. Mol. Ther.- Nucleic Acids 2022, 27, 611–620. [Google Scholar] [CrossRef]
- Fan, Q.W.; Cheng, C.K.; Gustafson, W.C.; Charron, E.; Zipper, P.; Wong, R.A.; Chen, J.; Lau, J.; Knobbe-Thomsen, C.; Weller, M.; et al. EGFR Phosphorylates Tumor-Derived EGFRvIII Driving STAT3/5 and Progression in Glioblastoma. Cancer Cell 2013, 24, 438–449. [Google Scholar] [CrossRef] [Green Version]
- Maliszewska-Olejniczak, K.; Kaniowski, D.; Araszkiewicz, M.; Tymińska, K.; Korgul, A. Molecular Mechanisms of Specific Cellular DNA Damage Response and Repair Induced by the Mixed Radiation Field During Boron Neutron Capture Therapy. Front. Oncol. 2021, 11, 676575. [Google Scholar] [CrossRef]
- Xu, M.J.; Johnson, D.E.; Grandis, J.R. EGFR-targeted therapies in the post-genomic era. Cancer Metastasis Rev. 2017, 36, 463–473. [Google Scholar] [CrossRef]
- Guardiola, S.; Varese, M.; Sánchez-Navarro, M.; Giralt, E.A. Third Shot at EGFR: New Opportunities in Cancer Therapy. Trends Pharmacol. Sci. 2019, 40, 941–955. [Google Scholar] [CrossRef]
- García-Foncillas, J.; Sunakawa, Y.; Aderka, D.; Wainberg, Z.; Ronga, P.; Witzler, P.; Stintzing, S. Distinguishing Features of Cetuximab and Panitumumab in Colorectal Cancer and Other Solid Tumors. Front. Oncol. 2019, 9, 849. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, T.; Ohba, M.; Ohmori, T. Molecular-targeted therapies for epidermal growth factor receptor and its resistance mechanismse. Int. J. Mol. Sci. 2017, 18, 2420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebenryter-Olbińska, K.; Kaniowski, D.; Sobczak, M.; Wojtczak, B.A.; Janczak, S.; Wielgus, E.; Nawrot, B.; Leśnikowski, Z.J. Versatile Method for the Site-Specific Modification of DNA with Boron Clusters: Anti-Epidermal Growth Factor Receptor (EGFR) Antisense Oligonucleotide Case. Chem. A Eur. J. 2017, 23, 16535–16546. [Google Scholar] [CrossRef] [PubMed]
- Kaniowski, D.; Ebenryter-Olbińska, K.; Sobczak, M.; Wojtczak, B.; Janczak, S.; Leśnikowski, Z.J.; Nawrot, B. High boron-loaded DNA-oligomers as potential boron neutron capture therapy and antisense oligonucleotide dual-action anticancer agents. Molecules 2017, 22, 1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaniowski, D.; Kulik, K.; Ebenryter-Olbińska, K.; Wielgus, E.; Leśnikowski, Z.; Nawrot, B. Metallacarborane Complex Boosts the Rate of DNA Oligonucleotide Hydrolysis in the Reaction Catalyzed by Snake Venom Phosphodiesterase. Biomolecules 2020, 5, 718. [Google Scholar] [CrossRef]
- Kaniowski, D.; Ebenryter-Olbińska, K.; Kulik, K.; Janczak, S.; Maciaszek, A.; Bednarska-Szczepaniak, K.; Nawrot, B.; Leśnikowski, Z. Boron clusters as a platform for new materials: Composites of nucleic acids and oligofunctionalized carboranes (C2B10H12) and their assembly into functional nanoparticles. Nanoscale 2020, 12, 103–114. [Google Scholar] [CrossRef]
- Kaniowski, D.; Ebenryter-Olbińska, K.; Kulik, K.; Suwara, J.; Cypryk, W.; Jakóbik-Kolon, A.; Leśnikowski, Z.; Nawrot, B. Composites of nucleic acids and boron clusters (C2B10H12) as functional nanoparticles for downregulation of EGFR oncogene in cancer cells. Int. J. Mol. Sci. 2021, 22, 4863. [Google Scholar] [CrossRef]
- Gudanis, D.; Kaniowski, D.; Kulik, K.; Baranowski, D.; Gdaniec, Z.; Nawrot, B. Formation of an RNA Quadruplex-Duplex Hybrid in Living Cells between mRNA of the Epidermal Growth Factor Receptor (EGFR) and a G-Rich Antisense Oligoribonucleotide. Cells 2020, 11, 2375. [Google Scholar] [CrossRef]
- Satpathy, M.; Mezencev, R.; Wang, L.; McDonald, J.F. Targeted in vivo delivery of EGFR siRNA inhibits ovarian cancer growth and enhances drug sensitivity. Sci. Rep. 2016, 6, 36518. [Google Scholar] [CrossRef]
- Le, B.T.; Paul, S.; Jastrzebska, K.; Langer, H.; Caruthers, M.H.; Veedu, R.N. Thiomorpholino oligonucleotides as a robust class of next generation platforms for alternate mRNA splicing. Proc. Natl. Acad. Sci. USA 2022, 119, e2207956119. [Google Scholar] [CrossRef]
- Quemener, A.M.; Centomo, M.L.; Sax, S.L.; Panella, R. Small Drugs, Huge Impact: The Extraordinary Impact of Antisense Oligonucleotides in Research and Drug Development. Molecules 2022, 27, 536. [Google Scholar] [CrossRef] [PubMed]
- Byun, M.J.; Lim, J.; Kim, S.N.; Park, D.H.; Kim, T.H.; Park, W.; Park, C.G. Advances in Nanoparticles for Effective Delivery of RNA Therapeutics. Biochip J. 2022, 16, 128–145. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Feng, S.S.; Guo, Y. Antibody engineering promotes nanomedicine for cancer treatment. Nanomedicine 2010, 5, 1141–1145. [Google Scholar] [CrossRef] [Green Version]
- Dugal-Tessier, J.; Thirumalairajan, S.; Jain, N. Antibody-oligonucleotide conjugates: A twist to antibody-drug conjugates. J. Clin. Med. 2021, 10, 838. [Google Scholar] [CrossRef]
- Carpenter, G.; Cohen, S. Epidermal growth factor. J. Biol. Chem. 1990, 265, 7709–7712. [Google Scholar] [CrossRef]
- Haigler, H.; Ash, J.F.; Singer, S.J.; Cohen, S. Visualization by fluorescence of the binding and internalization of epidermal growth factor in human carcinoma cells A-431. Proc. Natl. Acad. Sci. USA 1978, 75, 3317–3321. [Google Scholar] [CrossRef] [Green Version]
- Ulfo, L.; Costantini, P.E.; Di Giosia, M.; Danielli, A.; Calvaresi, M. EGFR-Targeted Photodynamic Therapy. Pharmaceutics 2022, 2, 241. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, G.; Guo, L.; Wang, H.; Fu, Y.; Luo, Y. Enhancement of tumor uptake and therapeutic efficacy of EGFR-targeted antibody cetuximab and antibody-drug conjugates by cholesterol sequestration. Int. J. Cancer 2015, 136, 182–194. [Google Scholar] [CrossRef]
- de Paiva, I.M.; Vakili, M.R.; Soleimani, A.H.; Tabatabaei Dakhili, S.A.; Munira, S.; Paladino, M.; Martin, G.; Jirik, F.R.; Hall, D.G.; Weinfeld, M.; et al. Biodistribution and Activity of EGFR Targeted Polymeric Micelles Delivering a New Inhibitor of DNA Repair to Orthotopic Colorectal Cancer Xenografts with Metastasis. Mol. Pharm. 2021, 6, 1825–1838. [Google Scholar] [CrossRef]
- Kim, J.S.; Kim, M.W.; Kang, S.J.; Jeong, H.Y.; Park, S.I.; Lee, Y.K.; Kim, H.S.; Kim, K.S.; Park, Y.S. Tumor-specific delivery of therapeutic siRNAs by anti-EGFR immunonanoparticles. Int. J. Nanomed. 2018, 13, 4817–4830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matuszewski, M.; Kiliszek, A.; Rypniewski, W.; Lesnikowski, Z.J.; Olejniczak, A.B. Nucleoside bearing boron clusters and their phosphoramidites-building blocks for modified oligonucleotide synthesis. New J. Chem. 2015, 39, 1202–1221. [Google Scholar] [CrossRef]
- Wojtczak, B.A.; Olejniczak, A.B.; Lesnikowski, Z.J. Nucleoside modification with boron clusters and their metal complexes. Curr. Protoc. Nucleic Acid Chem. 2009, 4, 1–26. [Google Scholar] [CrossRef]
- Kaniowski, D.; Kulik, K.; Suwara, J.; Ebenryter-Olbińska, K.; Nawrot, B. Boron Clusters as Enhancers of RNase H Activity in the Smart Strategy of Gene Silencing by Antisense Oligonucleotides. Int. J. Mol. Sci. 2022, 23, 12190. [Google Scholar] [CrossRef] [PubMed]
- Meldal, M.; Tomøe, C.W. Cu-catalyzed azide—Alkyne cycloaddition. Chem. Rev. 2008, 108, 2952–3015. [Google Scholar] [CrossRef] [PubMed]
- Amblard, F.; Cho, J.H.; Schinazi, R.F. Cu(I)-Catalyzed Huisgen Azide−Alkyne 1,3-Dipolar Cycloaddition Reaction in Nucleoside, Nucleotide, and Oligonucleotide Chemistry. Chem. Rev. 2009, 109, 4207–4220. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Du, F.; Tang, L.; Xu, J.; Zhao, Y.; Wu, X.; Li, M.; Shen, J.; Wen, Q.; Cho, C.H.; et al. Carboranes as unique pharmacophores in antitumor medicinal chemistry. Mol. Ther. Oncolytics 2022, 24, 400–416. [Google Scholar] [CrossRef]
- Horiguchi, H.; Sato, T.; Kumada, H.; Yamamoto, T.; Sakae, T. Estimation of relative biological effectiveness for boron neutron capture therapy using the PHITS code coupled with a microdosimetric kinetic model. J. Radiat. Res. 2015, 56, 382–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerecetto, H.; Couto, M. Medicinal chemistry of boron-bearing compounds for BNCT- glioma treatment: Current challenges and perspectives. In Glioma—Contemporary Diagnostic and Therapeutic Approaches; Omerhodžić, I., Arnautović, K., Eds.; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef] [Green Version]
- Kraja, I.; Bing, R.; Hiwatashi, N.; Rousseau, B.; Nalband, D.; Kirshenbaum, K.; Branski, R.C. Preliminary study of a novel transfection modality for in vivo siRNA delivery to vocal fold fibroblasts. Laryngoscope 2017, 127, E231–E237. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, S.; Yin, L.; Yang, Y.; Guan, Y.; Wang, W.; Xu, H.; Tao, N. Quantification of Epidermal Growth Factor Receptor Expression Level and Binding Kinetics on Cell Surfaces by Surface Plasmon Resonance Imaging. Anal. Chem. 2015, 87, 9960–9965. [Google Scholar] [CrossRef] [Green Version]
- Thomas, R.; Weihua, Z. Rethink of EGFR in Cancer with Its Kinase Independent Function on Board. Front. Oncol. 2019, 9, 800. [Google Scholar] [CrossRef]
- Goszczyński, T.M.; Fink, K.; Kowalski, K.; Leśnikowski, Z.J.; Boratyński, J. Interactions of Boron Clusters and their Derivatives with Serum Albumin. Sci. Rep. 2017, 7, 9800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barba-Bon, A.; Salluce, G.; Lostalé-Seijo, I.; Assaf, K.I.; Hennig, A.; Montenegro, J.; Nau, W.M. Boron clusters as broadband membrane carriers. Nature 2022, 603, 637–642. [Google Scholar] [CrossRef]
- Couto, M.; Alamón, C.; Nievas, S.; Perona, M.; Dagrosa, M.A.; Teixidor, F.; Cabral, P.; Viñas, C.; Cerecetto, H. Bimodal Therapeutic Agents Against Glioblastoma, One of the Most Lethal Forms of Cancer. Chem. A Eur. J. 2020, 26, 14335–14340. [Google Scholar] [CrossRef]
- Couto, M.; Alamón, C.; Sánchez, C.; Dávila, B.; Fernández, M.; Lecot, N.; Cabral, P.; Teixidor, F.; Viñas, C.; Cerecetto, H. Carboranylanilinoquinazoline stage in the drug-development pipeline. Future Med. Chem. 2019, 11, 2273–2285. [Google Scholar] [CrossRef]
- Mamot, C.; Drummond, D.C.; Greiser, U.; Hong, K.; Kirpotin, D.B.; Marks, J.D.; Park, J.W. Epidermal Growth Factor Receptor (EGFR)-targeted Immunoliposomes Mediate Specific and Efficient Drug Delivery to EGFR- and EGFRvIII-overexpressing Tumor Cells. Cancer Res. 2003, 63, 3154–3161. [Google Scholar] [PubMed]
- Mi, P.; Yanagie, H.; Dewi, N.; Yen, H.C.; Liu, X.; Suzuki, M.; Sakurai, Y.; Ono, K.; Takahashi, H.; Cabral, H.; et al. Block copolymer-boron cluster conjugate for effective boron neutron capture therapy of solid tumors. J. Control. Release 2017, 254, 1–9. [Google Scholar] [CrossRef]
- Fuentes, I.; García-Mendiola, T.; Sato, S.; Pita, M.; Nakamura, H.; Lorenzo, E.; Teixidor, F.; Marques, F.; Viñas, C. Metallacarboranes on the Road to Anticancer Therapies: Cellular Uptake, DNA Interaction, and Biological Evaluation of Cobaltabisdicarbollide [COSAN]. Chemistry 2018, 24, 17239–17254. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Chou, F.I.; Yang, B.H.; Chang, C.W.; Chen, Y.W.; Hwang, J.J. Similar T/N ratio between 18F-FBPA diagnostic and BPA therapeutic dosages for boron neutron capture therapy in orthotropic tongue cancer model. Ann. Nucl. Med. 2020, 34, 58–64. [Google Scholar] [CrossRef]
- Fontanilles, M.; Marguet, F.; Ruminy, P.; Basset, C.; Noel, A.; Beaussire, L.; Viennot, M.; Viailly, P.J.; Cassinari, K.; Chambon, P.; et al. Simultaneous detection of EGFR amplification and EGFRvIII variant using digital PCR-based method in glioblastoma. Acta Neuropathol. Commun. 2020, 8, 52. [Google Scholar] [CrossRef]
- Ohta, K.; Iijima, T.; Kawachi, E.; Kagechika, H.; Endo, Y. Novel retinoid X receptor (RXR) antagonists having a dicarba-closo-dodecaborane as a hydrophobic moiety. Bioorg. Med. Chem. Lett. 2004, 14, 5913–5918. [Google Scholar] [CrossRef]
- Fujii, S.; Goto, T.; Ohta, K.; Hashimoto, Y.; Suzuki, T.; Ohta, S.; Endo, Y. Potent Androgen Antagonists Based on Carborane as a Hydrophobic Core Structure. J. Med. Chem. 2005, 48, 4654–4662. [Google Scholar] [CrossRef] [PubMed]
- Otero, R.; Seoane, S.; Sigüeiro, R.; Belorusova, A.Y.; Maestro, M.A.; Pérez-Fernández, R.; Rochel, N.; Mouriño, A. Carborane-based design of a potent vitamin D receptor agonist. Chem. Sci. 2016, 7, 1033–1037. [Google Scholar] [CrossRef] [Green Version]
- Worm, D.J.; Els-Heindl, S.; Kellert, M.; Kuhnert, R.; Saretz, S.; Koebberling, J.; Riedl, B.; Hey-Hawkins, E.; Beck-Sickinger, A.G. A stable meta-carborane enables the generation of boron-rich peptide agonists targeting the ghrelin receptor. J. Pept. Sci. 2018, 24, e3119. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, V.M.; Frank, R.; Boehnke, S.; Schütz, C.L.; Hampel, G.; Iffland, D.S.; Bings, N.H.; Hey-Hawkins, E.; Beck-Sickinger, A.G. Receptor-mediated uptake of boron-rich neuropeptide y analogues for boron neutron capture therapy. ChemMedChem 2015, 10, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Bednarska-Szczepaniak, K.; Mieczkowski, A.; Kierozalska, A.; Pavlović Saftić, D.; Głąbała, K.; Przygodzki, T.; Stańczyk, L.; Karolczak, K.; Watała, C.; Rao, H.; et al. Synthesis and evaluation of adenosine derivatives as A(1), A(2A), A(2B) and A(3) adenosine receptor ligands containing boron clusters as phenyl isosteres and selective A(3) agonists. Eur. J. Med. Chem. 2021, 223, 113607. [Google Scholar] [CrossRef]
- Mori, S.; Tsuemoto, N.; Kasagawa, T.; Nakano, E.; Fujii, S.; Kagechika, H. Development of Boron-Cluster-Based Progesterone Receptor Antagonists Bearing a Pentafluorosulfanyl (SF5) Group. Chem. Pharm. Bull. 2019, 67, 1278–1283. [Google Scholar] [CrossRef] [Green Version]
- Chou, F.I.; Chung, H.P.; Liu, H.M.; Chi, C.W.; Lui, W.Y. Suitability of boron carriers for BNCT: Accumulation of boron in malignant and normal liver cells after treatment with BPA, BSH and BA. Appl. Radiat. Isot. Incl. Data Instrum. Methods Use Agric. Ind. Med. 2009, 67, S105–S108. [Google Scholar] [CrossRef]
- Fukuo, Y.; Hattori, Y.; Kawabata, S.; Kashiwagi, H.; Kanemitsu, T.; Takeuchi, K.; Futamura, G.; Hiramatsu, R.; Watanabe, T.; Hu, N.; et al. The Therapeutic Effects of Dodecaborate Containing Boronophenylalanine for Boron Neutron Capture Therapy in a Rat Brain Tumor Model. Biology 2020, 12, 437. [Google Scholar] [CrossRef]
- Hillaireau, H.; Couvreur, P. Nanocarriers’ entry into the cell: Relevance to drug delivery. Cell. Mol. Life Sci. 2009, 66, 2873–2896. [Google Scholar] [CrossRef]
- Xie, X.; Liao, J.; Shao, X.; Li, Q.; Lin, Y. The Effect of shape on Cellular Uptake of Gold Nanoparticles in the forms of Stars, Rods, and Triangles. Sci. Rep. 2017, 7, 3827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.Y.; Hsieh, H.H.; Chang, T.Y.; Lin, J.J.; Wu, C.C.; Hsu, M.H.; Lin, M.C.; Peng, S.L. Development of MRI-Detectable Boron-Containing Gold Nanoparticle-Encapsulated Biodegradable Polymeric Matrix for Boron Neutron Capture Therapy (BNCT). Int. J. Mol. Sci. 2021, 22, 8050. [Google Scholar] [CrossRef] [PubMed]
- Nakase, I.; Aoki, A.; Sakai, Y.; Hirase, S.; Ishimura, M.; Takatani-Nakase, T.; Hattori, Y.; Kirihata, M. Antibody-Based Receptor Targeting Using an Fc-Binding Peptide-Dodecaborate Conjugate and Macropinocytosis Induction for Boron Neutron Capture Therapy. ACS Omega 2020, 5, 22731–22738. [Google Scholar] [CrossRef]
- Assaf, K.I.; Begaj, B.; Frank, A.; Nilam, M.; Mougharbel, A.S.; Kortz, U.; Nekvinda, J.; Grüner, B.; Gabel, D.; Nau, W.M. High-Affinity Binding of Metallacarborane Cobalt Bis(dicarbollide) Anions to Cyclodextrins and Application to Membrane Translocation. J. Org. Chem. 2019, 84, 11790–11798. [Google Scholar] [CrossRef]
- Grüner, B.; Brynda, J.; Das, V.; Šícha, V.; Štěpánková, J.; Nekvinda, J.; Holub, J.; Pospíšilová, K.; Fábry, M.; Pachl, P.; et al. Metallacarborane Sulfamides: Unconventional, Specific, and Highly Selective Inhibitors of Carbonic Anhydrase IX. J. Med. Chem. 2019, 62, 9560–9575. [Google Scholar] [CrossRef] [PubMed]
- Viñas, C.; Núñez, R.; Bennour, I.; Teixidor, F. Periphery Decorated and Core Initiated Neutral and Polyanionic Borane large molecules: Forthcoming and Promising properties for medicinal applications. Curr. Med. Chem. 2019, 21, 9560–9575. [Google Scholar] [CrossRef]
- Chaari, M.; Gaztelumendi, N.; Cabrera-González, J.; Peixoto-Moledo, P.; Viñas, C.; Xochitiotzi-Flores, E.; Farfán, N.; Ben Salah, A.; Nogués, C.; Núñez, R. Fluorescent BODIPY-Anionic Boron Cluster Conjugates as Potential Agents for Cell Tracking. Bioconjug. Chem. 2018, 29, 1763–1773. [Google Scholar] [CrossRef]
- Wang, S.; Allen, N.; Vickers, T.A.; Revenko, A.S.; Sun, H.; Liang, X.H.; Crooke, S.T. Cellular uptake mediated by epidermal growth factor receptor facilitates the intracellular activity of phosphorothioate-modified antisense oligonucleotides. Nucleic Acids Res. 2018, 46, 3579–3594. [Google Scholar] [CrossRef] [Green Version]
- Crooke, S.T.; Wang, S.; Vickers, T.A.; Shen, W.; Liang, X.H. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017, 35, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; de Bruijn, H.S.; Farrell, E.; Sioud, M.; Mashayekhi, V.; Oliveira, S.; van Dam, G.M.; Roodenburg, J.L.N.; Witjes, M.J.H.; Robinson, D.J. Epidermal growth factor receptor (EGFR) density may not be the only determinant for the efficacy of EGFR-targeted photoimmunotherapy in human head and neck cancer cell lines. Lasers Surg. Med. 2018, 50, 513–522. [Google Scholar] [CrossRef]
- Bramsen, J.; Kjems, J. Development of Therapeutic-Grade Small Interfering RNAs by Chemical Engineering. Front. Genet. 2012, 3, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pugh, R.A.; Honda, M.; Spies, M. Ensemble and single-molecule fluorescence-based assays to monitor DNA binding, translocation, and unwinding by iron-sulfur cluster containing helicases. Methods 2010, 51, 313–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fink, K.; Kobak, K.; Kasztura, M.; Boratyński, J.; Goszczyński, T.M. Synthesis and Biological Activity of Thymosin β4-Anionic Boron Cluster Conjugates. Bioconjug. Chem. 2018, 29, 3509–3515. [Google Scholar] [CrossRef] [PubMed]
- Vorobyeva, M.A.; Dymova, M.A.; Novopashina, D.S.; Kuligina, E.V.; Timoshenko, V.V.; Kolesnikov, I.A.; Taskaev, S.Y.; Richter, V.A.; Venyaminova, A.G. Tumor Cell-Specific 2′-Fluoro RNA Aptamer Conjugated with Closo-Dodecaborate as A Potential Agent for Boron Neutron Capture Therapy. Int. J. Mol. Sci. 2021, 22, 7326. [Google Scholar] [CrossRef]
- Li, J.; Sun, Q.; Lu, C.; Xiao, H.; Guo, Z.; Duan, D.; Zhang, Z.; Liu, T.; Liu, Z. Boron encapsulated in a liposome can be used for combinational neutron capture therapy. Nat. Commun. 2022, 13, 2143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, G.; Li, S.; Wang, Z.; He, D.; Wang, Y.; Zhang, J.; Li, J.; Bai, Z.; Zhang, Q.; et al. Asialoglycoprotein receptor targeted micelles containing carborane clusters for effective boron neutron capture therapy of hepatocellular carcinoma. Colloids Surf. B Biointerfaces 2019, 182, 110397. [Google Scholar] [CrossRef]
- Bhupathiraju, N.V.; Hu, X.; Zhou, Z.; Fronczek, F.R.; Couraud, P.O.; Romero, I.A.; Weksler, B.; Vicente, M.G. Synthesis and in vitro evaluation of BBB permeability, tumor cell uptake, and cytotoxicity of a series of carboranylporphyrin conjugates. J. Med. Chem. 2014, 15, 6718–6728. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, P.M.; Jelliss, P.A.; Nonaka, N.; Shi, X.; Banks, W.A. Permeability of the blood-brain barrier to a rhenacarborane. J. Pharmacol. Exp. Ther. 2009, 2, 608–614. [Google Scholar] [CrossRef] [Green Version]
- Glenresearch. Available online: http://www.glenresearch.com/GlenReports/GR25-24.html (accessed on 18 May 2017).
- Pirhonen, J.; Sareneva, T.; Kurimoto, M.; Julkunen, I.; Matikainen, S. Virus infection activates IL-1 beta and IL-18 production in human macrophages by a caspase-1-dependent pathway. J. Immunol. 1999, 162, 7322–7329. [Google Scholar]
- Cypryk, W.; Ohman, T.; Eskelinen, E.L.; Matikainen, S.; Nyman, T.A. Quantitative proteomics of extracellular vesicles released from human monocyte-derived macrophages upon β-glucan stimulation. J. Proteome Res. 2014, 13, 2468–2477. [Google Scholar] [CrossRef]


| No. | Sequences | MW calc. (g/mol) | MALDI-TOF MS (m/z) | RP-HPLC (Rt, min) |
|---|---|---|---|---|
| BIOT-1-FESAN | 5′-biotin-2′OMe(GAUB.1UC)-3′ | 2507.4 | 2509.8 | 20.3 * |
| BIOT-1-1,2-DCDDB | 5′-biotin-2′OMe(GAUB.2UC)-3′ | 2283.6 | 2286.7 | 17.7 * |
| BIOT-1 | 5′-biotin-2′OMe(GAUUC)-3′ | 2034.5 | 1016.2 1/ 2032.4 2 | 15.5 |
| FAM-1-FESAN | 5′-FAM-2′OMe(GAUB.1UC)-3′ | 2610.9 | 2611.8 | 20.2 * |
| ASO anti-EGFR | 5′-d(TTT CTT TTC CTC CAG AGC CCGA)-3′ | 6612 | 6612 | 16.0 |
| ASO-C | 5′-d(ATG AAG GTT CAA TCT GAT TTT)-3′ | 6450.3 | 6450.12 | 12.8 |
| B-ASO | 5′-d(UB.1TUB.1 CUB.1TTUB.1CCUB.1C CAG AGC CCGA)-3′ | 9063.0 | 9065.8 | 24.7 * |
| FAM-B-ASO | 5′-FAM-d(UB.1TUB.1CUB.1TTUB.1CCUB.1C CAG AGC CCGA)-3′ | 9601.7 | 9603.0 | 24.0 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaniowski, D.; Suwara, J.; Ebenryter-Olbińska, K.; Jakóbik-Kolon, A.; Nawrot, B. EGFR-Targeted Cellular Delivery of Therapeutic Nucleic Acids Mediated by Boron Clusters. Int. J. Mol. Sci. 2022, 23, 14793. https://doi.org/10.3390/ijms232314793
Kaniowski D, Suwara J, Ebenryter-Olbińska K, Jakóbik-Kolon A, Nawrot B. EGFR-Targeted Cellular Delivery of Therapeutic Nucleic Acids Mediated by Boron Clusters. International Journal of Molecular Sciences. 2022; 23(23):14793. https://doi.org/10.3390/ijms232314793
Chicago/Turabian StyleKaniowski, Damian, Justyna Suwara, Katarzyna Ebenryter-Olbińska, Agata Jakóbik-Kolon, and Barbara Nawrot. 2022. "EGFR-Targeted Cellular Delivery of Therapeutic Nucleic Acids Mediated by Boron Clusters" International Journal of Molecular Sciences 23, no. 23: 14793. https://doi.org/10.3390/ijms232314793





