Identification of a Novel Pleiotropic Transcriptional Regulator Involved in Sporulation and Secondary Metabolism Production in Chaetomium globosum
Abstract
:1. Introduction
2. Results
2.1. Identification of the CgXpp1 Gene in C. globosum W7
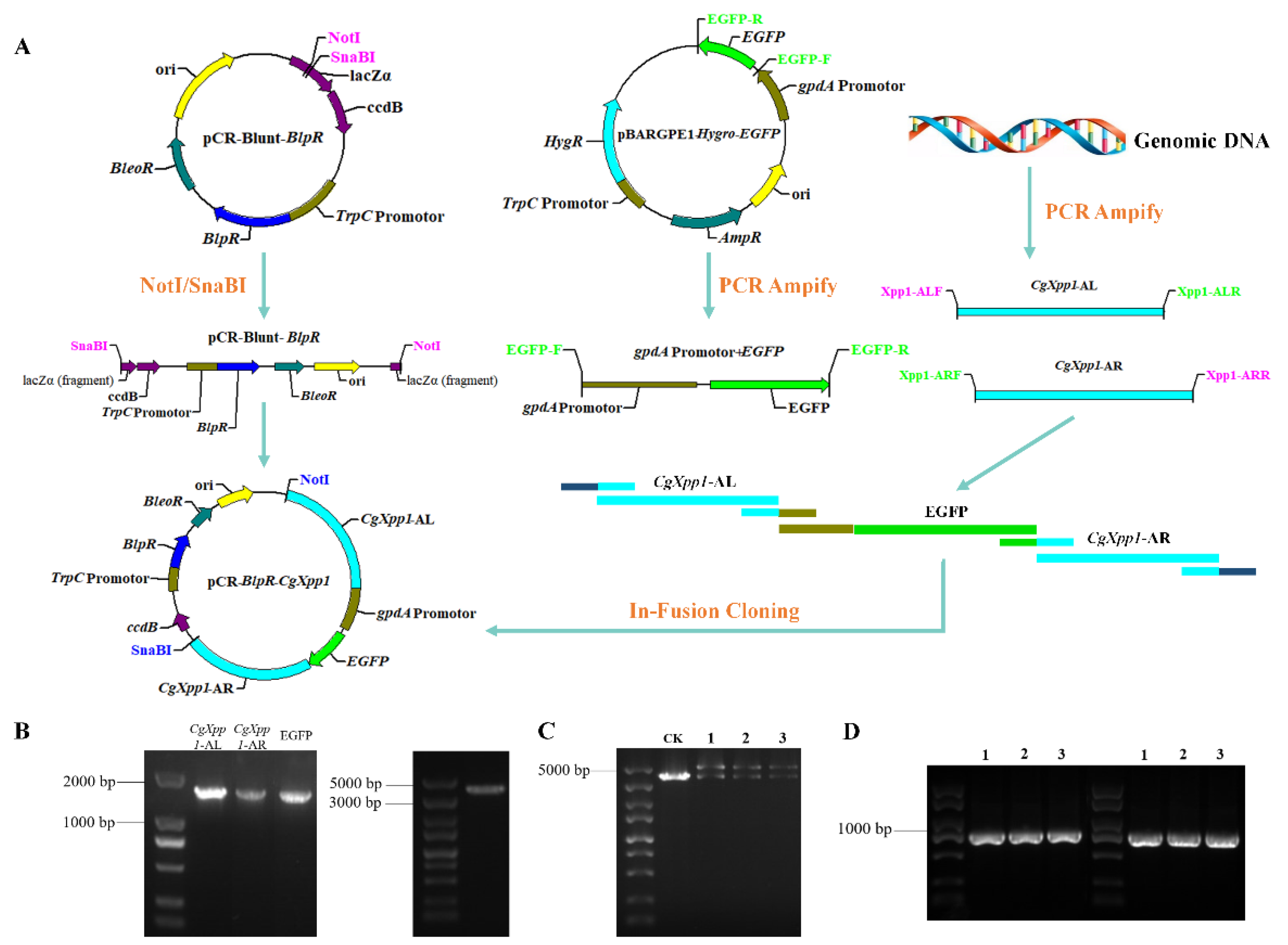
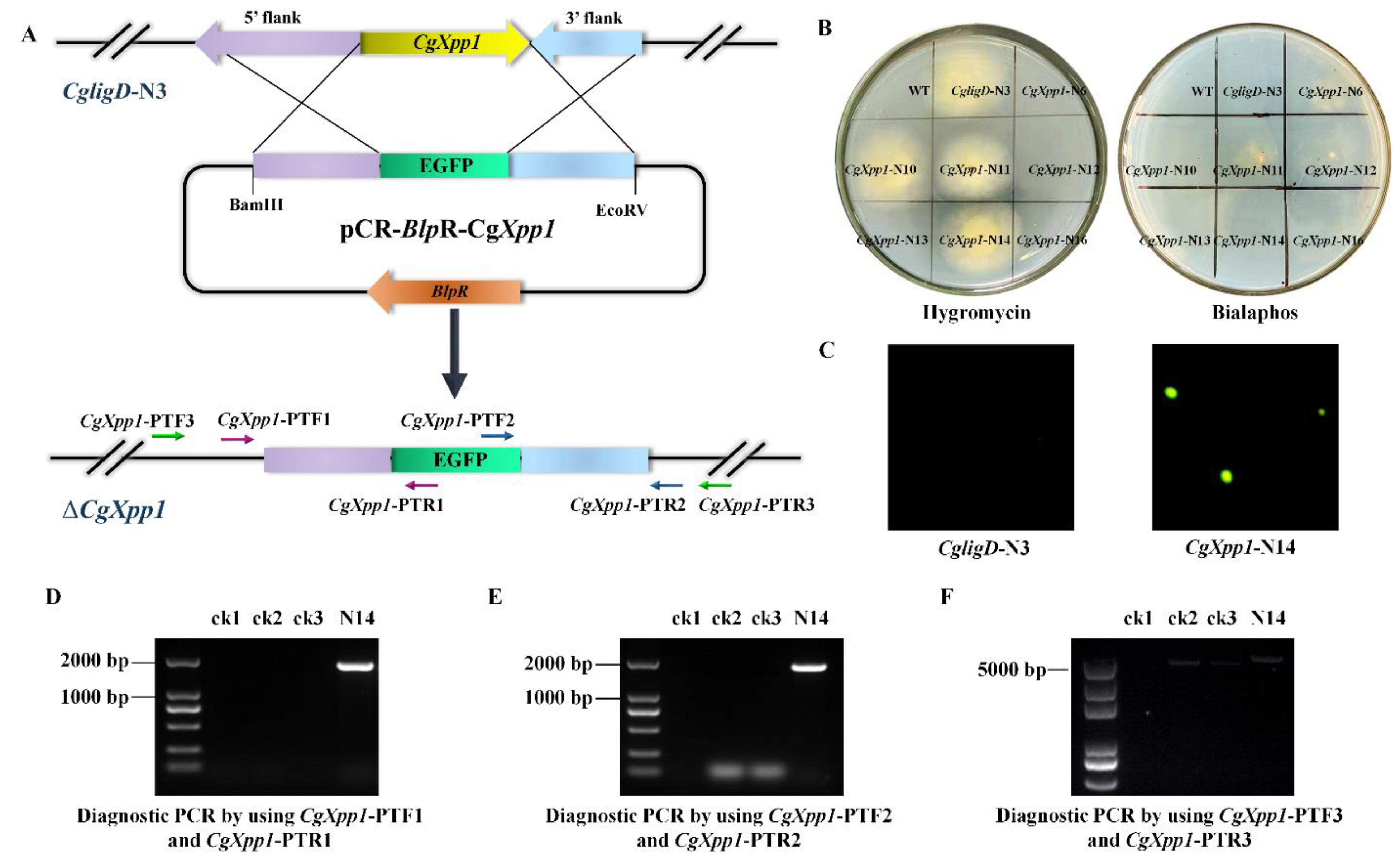
2.2. Construction of CgXpp1 Inactivation Mutants and Complementation
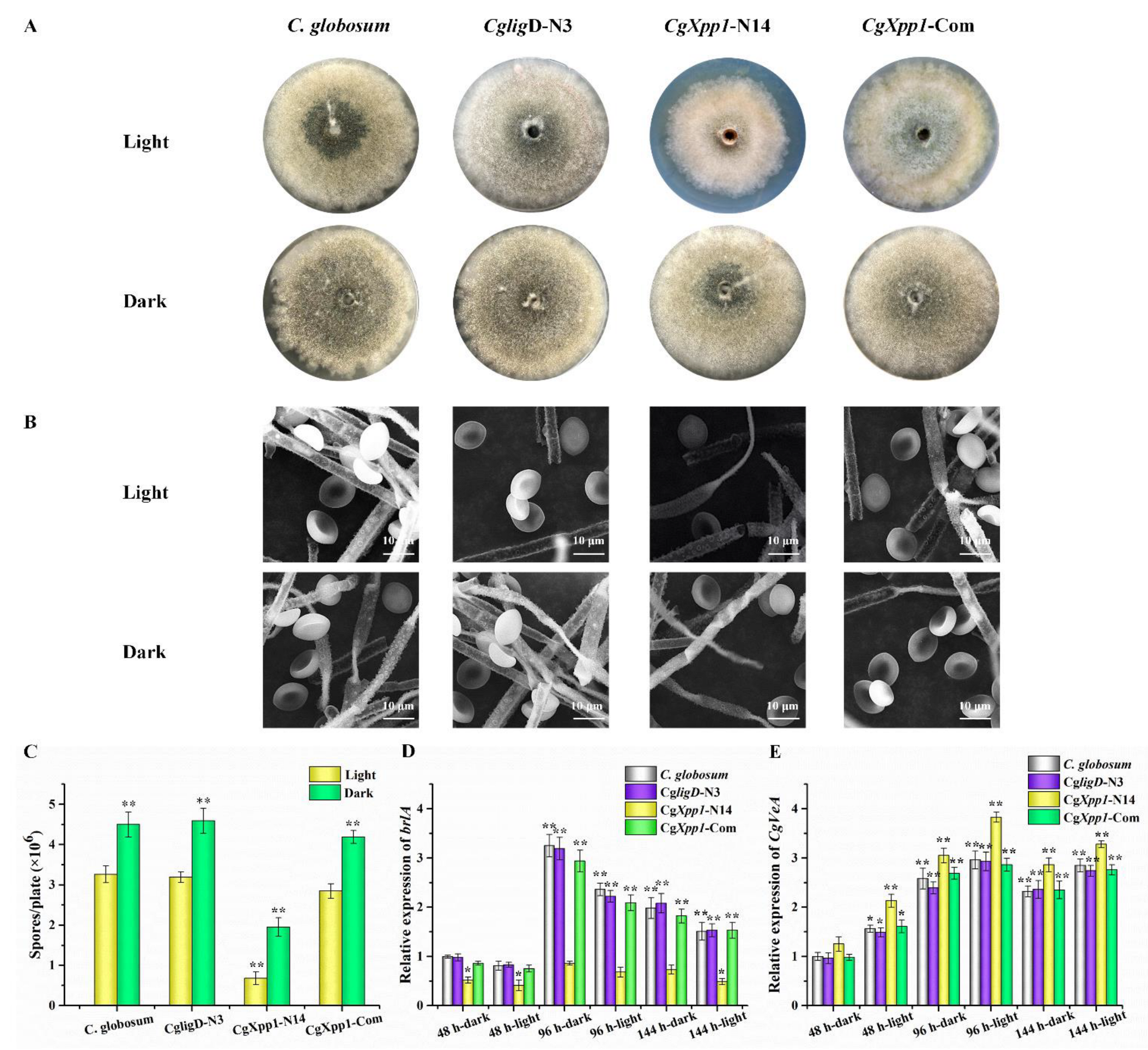
2.3. Effect of CgXpp1 on C. globosum W7 Development
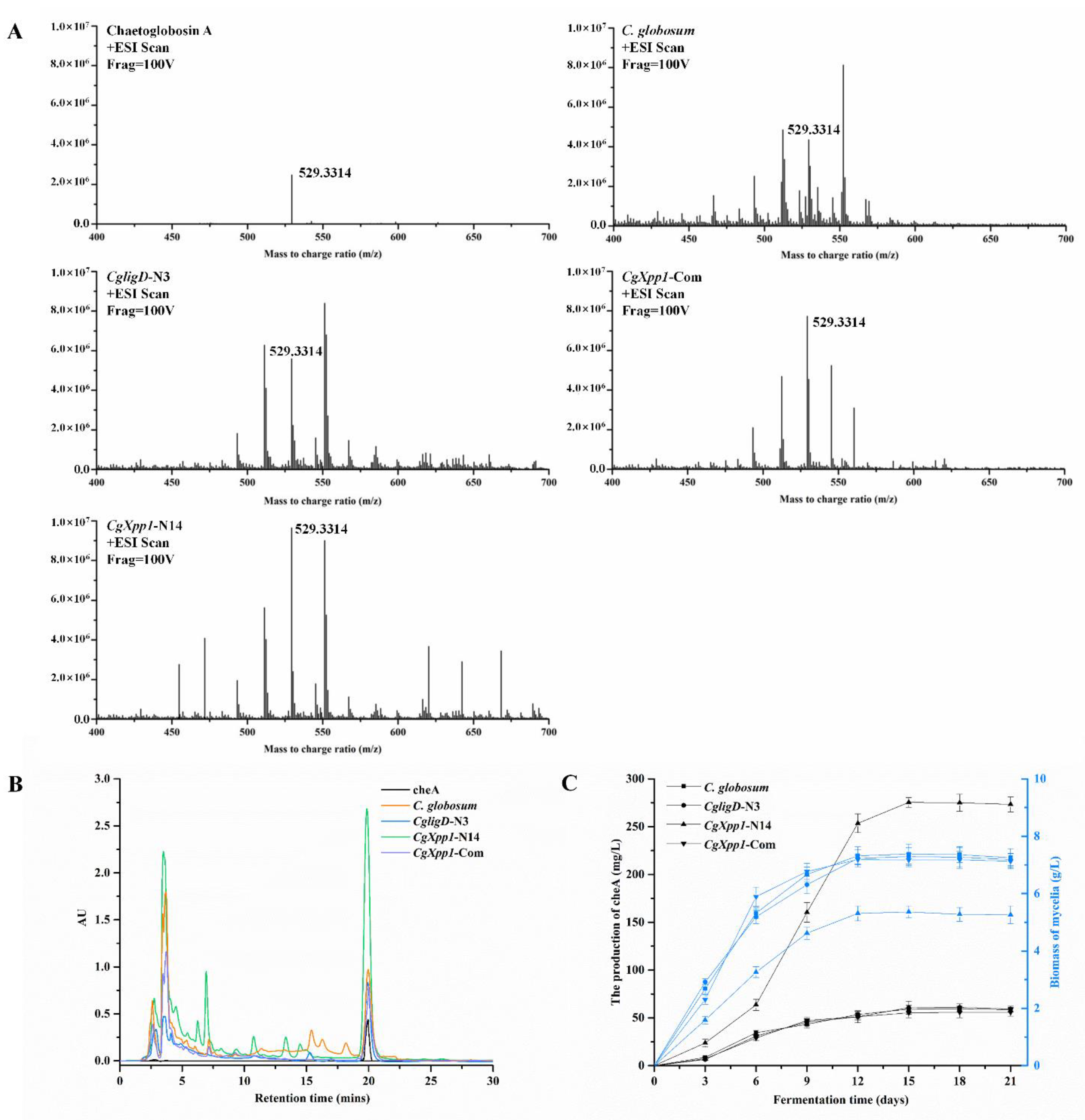
2.4. Effects of CgXpp1 Disruption on CheA Production
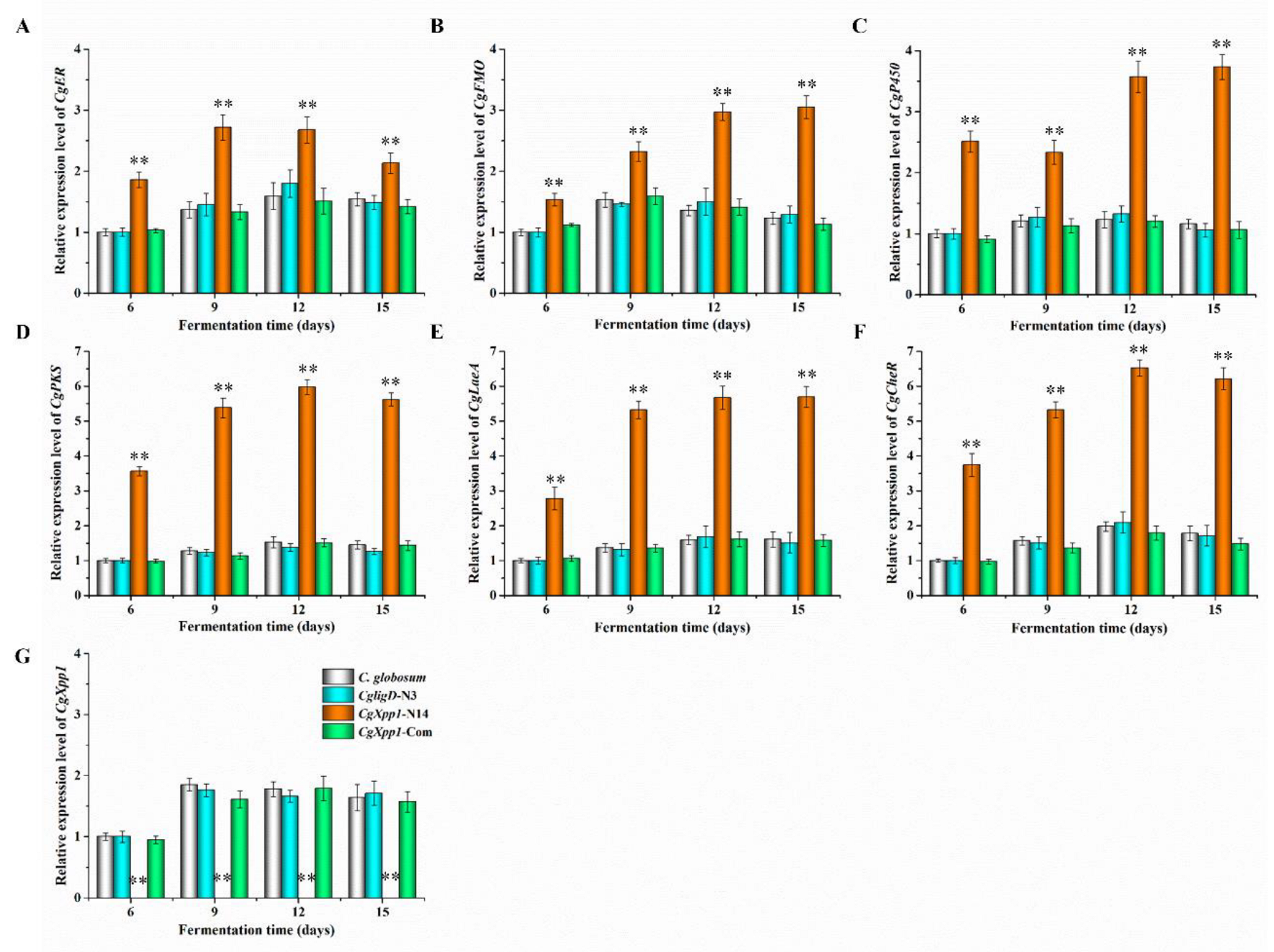
2.5. Effect of CgXpp1 on the Transcription of the CheA Biosynthetic Gene Cluster and Other Related Regulators
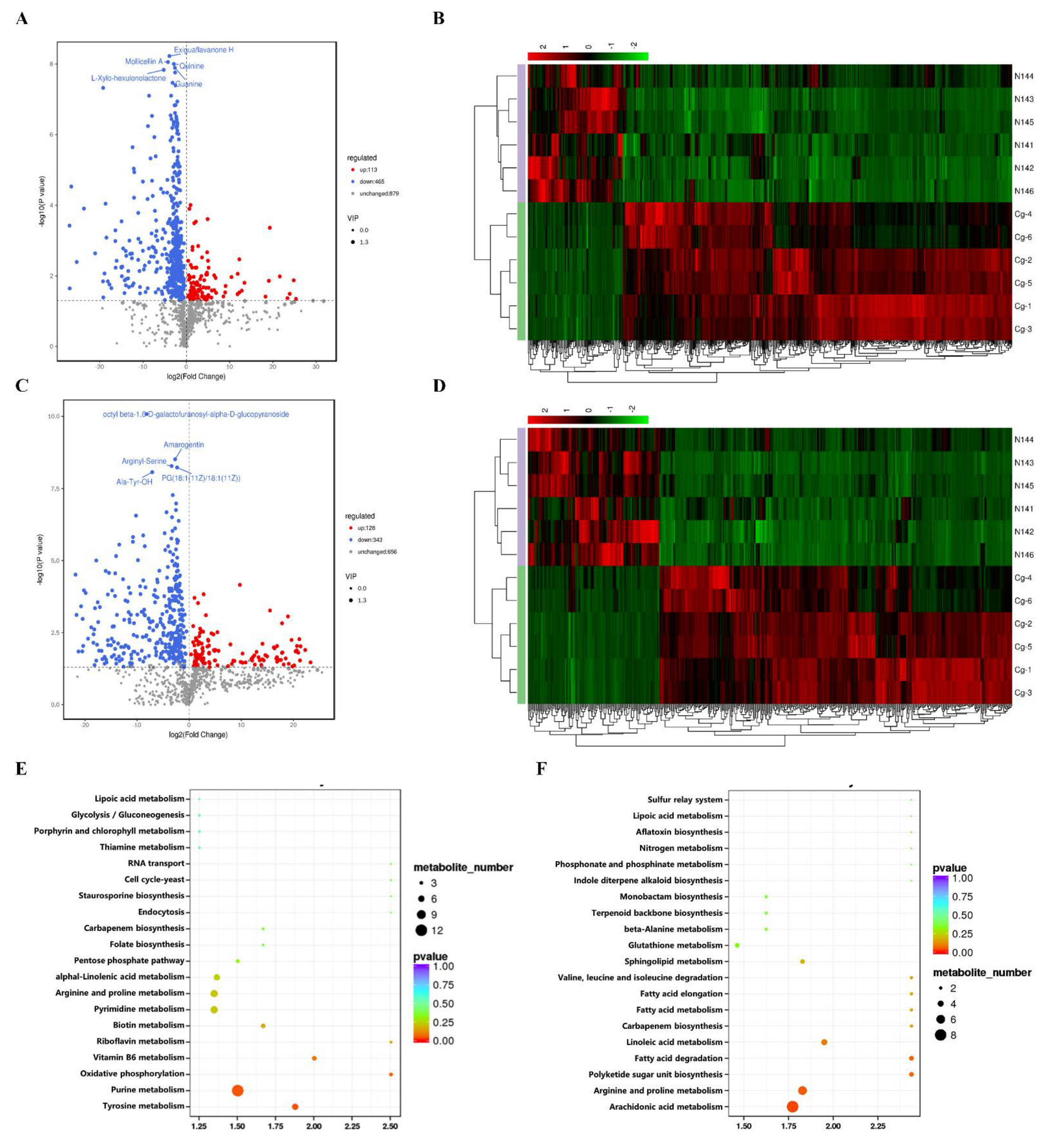
2.6. Analysis of the Metabolomics of C. globosum W7 and CgXpp1-N14
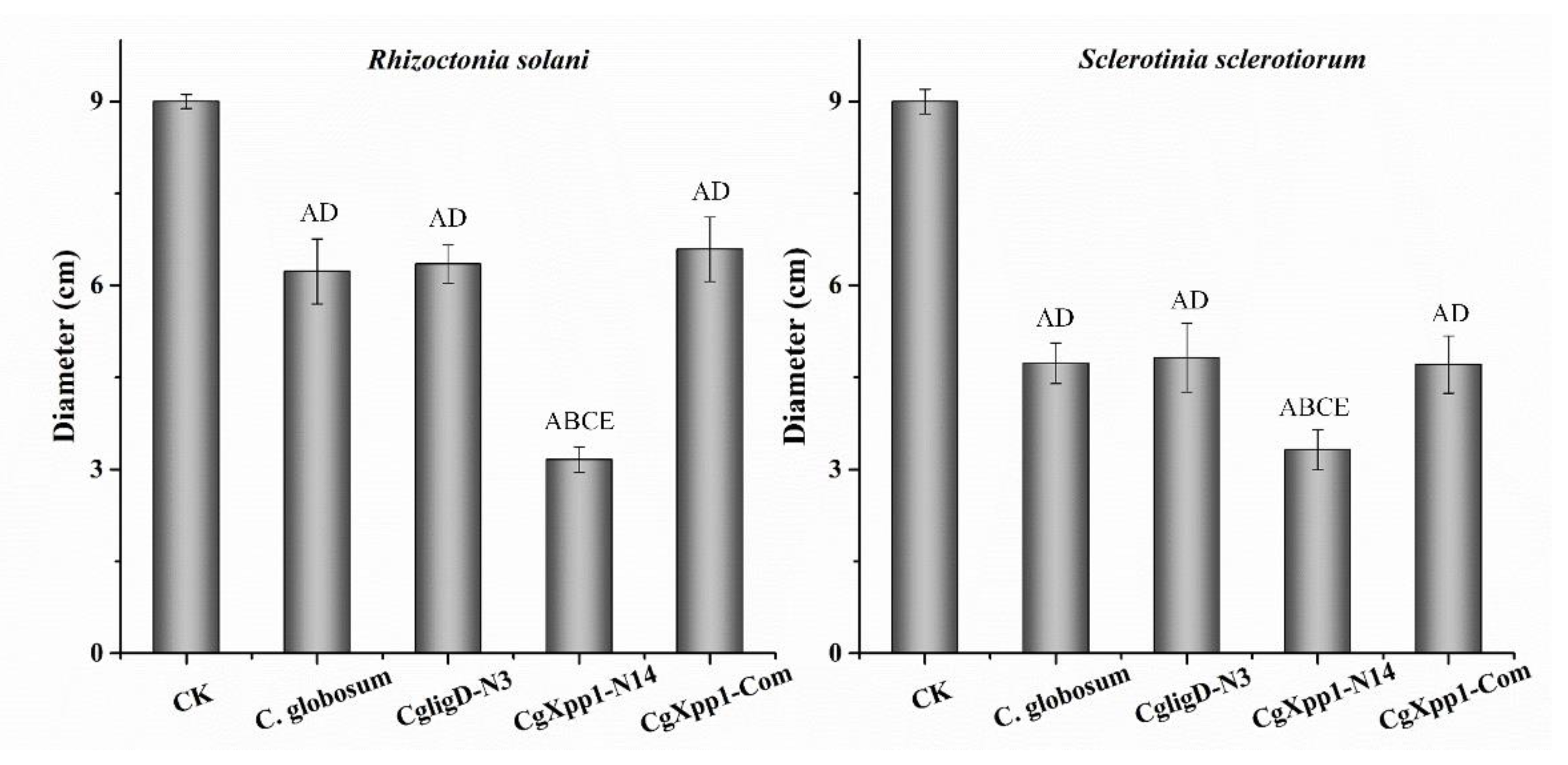
2.7. Impact of the CgXpp1 Gene in the Inhibitory Activities of C. globosum W7
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids, Primers, Culture Media, and Growth Conditions
4.2. In Silico Analysis of CgXpp1 and Construction of the Phylogenetic Tree
4.3. Construction of CgXpp1-Disruption Mutants and Their Complementation
4.4. Transformation of C. globosum W7 Protoplasts
4.5. Morphological Studies
4.6. Determination of Mycelial Biomass and CheA
4.7. RNA Isolation and Quantitative Real-Time PCR
4.8. Metabolome Analysis
4.9. Determination of the Antifungal Activity of Liquid Filtrates against Phytopathogenic Fungi
4.10. Expression and Purification of the CgXpp1 Protein
4.11. Electrophoretic Mobility Shift Assays (EMSAs)
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, R.; Kundu, A.; Dutta, A.; Saha, S.; Das, A. Profiling of volatile secondary metabolites of Chaetomium globosum for potential antifungal activity against soil borne fungi. Journal of Pharmacognosy and Phytochemistry 2020, 9, 922–927. [Google Scholar]
- Yan, W.; Ge, H.M.; Wang, G.; Jiang, N.; Mei, Y.N.; Jiang, R.; Li, S.J.; Chen, C.J.; Jiao, R.H.; Xu, Q.; et al. Pictet-Spengler reaction-based biosynthetic machinery in fungi. Proc. Natl. Acad. Sci. USA 2014, 111, 18138–18143. [Google Scholar] [CrossRef] [Green Version]
- Pontius, A.; Krick, A.; Kehraus, S.; Brun, R.; König, G.M. Antiprotozoal activities of heterocyclic-substituted xanthones from the marine-derived fungus Chaetomium sp. J. Nat. Prod. 2008, 71, 1579–1584. [Google Scholar] [CrossRef]
- Li, T.T.; Wang, Y.; Li, L.; Tang, M.Y.; Meng, Q.H.; Zhang, C.; Hua, E.B.; Pei, Y.H.; Sun, Y. New cytotoxic cytochalasans from a plant-associated fungus Chaetomium globosum kz-19. Mar. Drugs 2021, 19, 438. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Tan, X.M.; Yu, M.; Yang, J.; Sun, B.D.; Qin, J.C.; Guo, L.P.; Ding, G. Bioactive metabolites from the desert plant-associated endophytic fungus Chaetomium globosum (Chaetomiaceae). Phytochemistry 2021, 185, 112701. [Google Scholar] [CrossRef]
- Seklta, S.; Yoshlhira, K.; Natori, S. Structures of chaetoglobosin A and B, cytotoxic metabolites of Chaetomium globosum. Tetrahedron Lett. 1973, 23, 2109–2112. [Google Scholar] [CrossRef]
- Yan, W.; Zhao, S.S.; Ye, Y.H.; Zhang, Y.Y.; Zhang, Y.; Xu, J.Y.; Yin, S.M.; Tan, R.X. Generation of indoles with agrochemical significance through biotransformation by Chaetomium globosum. J. Nat. Prod. 2019, 82, 2132–2137. [Google Scholar] [CrossRef]
- Lösgen, S.; Schlörke, O.; Meindl, K.; Herbst-Irmer, R.; Zeeck, A. Structure and biosynthesis of chaetocyclinones, new polyketides produced by an endosymbiotic fungus. Eur. J. Org. Chem. 2007, 13, 2191–2196. [Google Scholar] [CrossRef]
- Jiang, C.; Song, J.Z.; Zhang, J.Z.; Yang, Q. Identification and characterization of the major antifungal substance against Fusarium sporotrichioides from Chaetomium globosum. World J. Microbiol. Biotechnol. 2017, 33, 108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.Z.; Wang, F.T.; Qin, J.C.; Wang, D.; Zhang, J.Y.; Zhang, Y.H.; Zhang, S.H.; Pan, H.Y. Efficacy assessment of antifungal metabolites from Chaetomium globosum No.05, a new biocontrol agent, against Setosphaeria turcica. Biol. Control 2013, 64, 90–98. [Google Scholar] [CrossRef]
- Schümann, J.; Hertweck, C. Molecular basis of cytochalasan biosynthesis in fungi: Gene cluster analysis and evidence for the involvement of a PKS-NRPS hybrid synthase by RNA silencing. J. Am. Chem. Soc. 2007, 129, 9564–9565. [Google Scholar] [CrossRef] [PubMed]
- Ishiuchi, K.; Nakazawa, T.; Yagishita, F.; Mino, T.; Noguchi, H.; Hotta, K.; Watanabe, K. Combinatorial generation of complexity by redox enzymes in the Chaetoglobosin A biosynthesis. J. Am. Chem. Soc. 2013, 135, 7371–7377. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.M.; Lee, K.H.; Sanchez, J.F.; Keller, N.P.; Wang, C.C. Unlocking fungal cryptic natural products. Nat. Prod. Commun. 2009, 4, 1505–1510. [Google Scholar] [CrossRef] [Green Version]
- Wiemann, P.; Keller, N.P. Strategies for mining fungal natural products. J. Ind. Microbiol. Biotechnol. 2014, 41, 301–313. [Google Scholar] [CrossRef]
- Estiarte, N.; Lawrence, C.B.; Sanchis, V.; Ramos, A.J.; Crespo-Sempere, A. LaeA and VeA are involved in growth morphology, asexual development, and mycotoxin production in Alternaria alternata. Int. J. Food Microbiol. 2016, 238, 153–164. [Google Scholar] [CrossRef] [Green Version]
- Jain, S.; Keller, N. Insights to fungal biology through LaeA sleuthing. Fungal Biol. Rev. 2013, 27, 51–59. [Google Scholar] [CrossRef]
- Chang, P.K.; Scharfenstein, L.L.; Ehrlich, K.C.; Wei, Q.; Bhatnagar, D.; Ingber, B.F. Effects of laeA deletion on Aspergillus flavus conidial development and hydrophobicity may contribute to loss of aflatoxin production. Fungal Biol. 2012, 116, 298–307. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, H.Y.; Wang, Y.L.; Liu, F.; Li, E.; Ma, J.N.; Yang, B.L.; Zhang, C.X.; Li, L.; Liu, Y. Requirement of LaeA, VeA, and VelB on asexual development, ochratoxin A biosynthesis, and fungal virulence in Aspergillus ochraceus. Front. Microbiol. 2019, 10, 2759. [Google Scholar] [CrossRef] [Green Version]
- Cheng, M.; Zhao, S.S.; Lin, C.Y.; Song, J.Z.; Yang, Q. Requirement of LaeA for sporulation, pigmentation and secondary metabolism in Chaetomium globosum. Fungal Biol. 2021, 125, 305–315. [Google Scholar] [CrossRef]
- Jiang, T.; Wang, M.H.; Li, L.; Si, J.G.; Song, B.; Zhou, C.; Yu, M.; Wang, X.W.; Zhang, Y.G.; Ding, G.; et al. Overexpression of the global regulator LaeA in Chaetomium globosum leads to the biosynthesis of Chaetoglobosin Z. J. Nat. Prod. 2016, 79, 2487–2494. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Wang, X.; Kong, F.D.; Huang, H.M.; Zhao, Y.N.; Liu, M.; Wang, Z.P.; Han, J. Overexpression of global regulator Talae1 leads to the discovery of new antifungal polyketides from endophytic fungus Trichoderma afroharzianum. Front Microbiol. 2020, 11, 622785. [Google Scholar] [CrossRef] [PubMed]
- Derntl, C.; Kluger, B.; Bueschl, C.; Schuhmacherb, R.; Mach, R.L.; Mach-Aigner, A.R. Transcription factor Xpp1 is a switch between primary and secondary fungal metabolism. Proc. Natl. Acad. Sci. USA 2017, 114, 560–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandit, S.S.; Lohmar, J.M.; Ahmed, S.; Etxebeste, O.; Espeso, E.A.; Calvo, A.M. UrdA controls secondary metabolite production and the balance between asexual and sexual development in Aspergillus nidulans. Genes 2018, 9, 570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, M.; Zhao, S.S.; Liu, H.; Liu, Y.T.; Lin, C.Y.; Song, J.Z.; Thawai, C.; Charoensettasilp, S.; Yang, Q. Functional analysis of a chaetoglobosin A biosynthetic regulator in Chaetomium globosum. Fungal Biol. 2021, 125, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Anthony, P.; Davey, M.R.; Power, J.B.; Washington, C.; Lowe, K.C. Synergistic enhancement of protoplast growth by oxygenated perfluorocarbon and Pluronic F-68. Plant Cell Rep. 1994, 13, 251–255. [Google Scholar] [CrossRef]
- Tamano, K.; Satoh, Y.; Ishii, T.; Terabayashi, Y.; Ohtaki, S.; Sano, M.; Takahashi, T.; Koyama, Y.; Mizutani, O.; Abe, K.; et al. The beta-1, 3-exoglucanase gene exgA (exg1) of Aspergillus oryzae is required to catabolize extracellular glucan, and is induced in growth on a solid surface. Biosci. Biotechnol. Biochem. 2007, 71, 926–934. [Google Scholar] [CrossRef] [Green Version]
- Won, D.C.; Kim, Y.J.; Kim, D.H.; Park, H.M.; Maeng, P.J. The putative C2H2 transcription factor RocA is a novel regulator of development and secondary metabolism in Aspergillus nidulans. J. Microbiol. 2020, 58, 574–587. [Google Scholar] [CrossRef]
- Hu, P.; Wang, Y.; Zhou, J.; Pan, Y.; Liu, G. AcstuA, which encodes an APSES transcription regulator, is involved in conidiation, cephalosporin biosynthesis and cell wall integrity of Acremonium chrysogenum. Fungal Genet. Biol. 2015, 83, 26–40. [Google Scholar] [CrossRef]
- Wang, Z.R.; Zhao, S.S.; Zhang, K.; Lin, C.Y.; Ru, X.; Yang, Q. CgVeA, a light signaling responsive regulator, is involved in regulation of chaetoglobosin A biosynthesis and conidia development in Chaetomium globosum. Synth. Syst. Biotechnol. 2022, 7, 1084–1094. [Google Scholar] [CrossRef]
- Lan, N.; Yue, Q.; An, Z.; Bills, G.F. Apc.LaeA and Apc.VeA of the velvet complex govern secondary metabolism and morphological development in the echinocandin-producing fungus Aspergillus pachycristatus. J. Ind. Microbiol. Biotechnol. 2020, 47, 155–168. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, X.; Bao, Y.; Wang, X.; Zhai, J.; Zhan, X.; Zhang, H. Characterization and anti-inflammation of a polysaccharide produced by Chaetomium globosum CGMCC 6882 on LPS-induced RAW 264.7 cells. Carbohydr. Polym. 2021, 251, 117129. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Min, X.T.; Huang, J.J.; Zhong, Y.; Wu, Y.H.; Li, X.X.; Deng, Y.Y.; Jiang, Z.D.; Shao, Z.Z.; Zhang, L.H.; et al. Cytoglobosins H and I, new antiproliferative cytochalasans from deep-sea-derived fungus Chaetomium globosum. Mar. Drugs 2016, 14, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, B.H.; Yu, Z.F.; Yang, X.Q.; Yang, Y.B.; Hu, M.; Zhang, Z.X.; Zhou, Q.Y.; Zhou, H.; Ding, Z.T. New bioactive compounds from aquatic endophyte Chaetomium globosum. Nat. Prod. Res. 2018, 32, 1050–1055. [Google Scholar] [CrossRef] [PubMed]
- Takanezawa, Y.; Nakamura, R.; Sone, Y.; Uraguchi, S.; Kobayashi, K.; Tomoda, H. Variation in the activity of distinct cytochalasins as autophagy inhibitiors in human lung A549 cells. Biochem. Biophys. Res. Commun. 2017, 494, 641–647. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, W.P.; Zhang, P.; Ruan, W.B.; Zhu, X.D. Nematicidal activity of chaetoglobosin A poduced by Chaetomium globosum NK102 against Meloidogyne incognita. J. Agric. Food Chem. 2013, 61, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Van, L.T.; Medema, M.H. Computational strategies for genome-based natural product discovery and engineering in fungi. Fungal Genet. Biol. 2016, 89, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Oakley, C.E.; Ahuja, M.; Sun, W.W.; Entwistle, R.; Akashi, T.; Yaegashi, J.; Guo, C.J.; Cerqueira, G.C.; Wortman, J.R.; Wang, C.C.; et al. Discovery of McrA, a master regulator of Aspergillus secondary metabolism. Mol. Microbiol. 2017, 103, 347–365. [Google Scholar] [CrossRef] [Green Version]
- Chiang, Y.M.; Ahuja, M.; Oakley, C.E.; Entwistle, R.; Zutz, C.; Wang, C.C.; Oakley, B.R. Development of genetic dereplication strains in Aspergillus nidulans results in the discovery of Aspercryptin. Angew. Chem. Int. Ed. 2016, 55, 1662–1665. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhu, P.H.; Wu, F.; Wang, X.F.; Zhang, J.F.; Ji, K.S. Identification and characterization of the basic helix-loop-helix transcription factor family in Pinus Massoniana. Forests 2020, 11, 1292. [Google Scholar] [CrossRef]
- Liu, J.; Yuan, Y.Z.; Wu, Z.; Li, N.; Chen, Y.L.; Qin, T.T.; Geng, H.; Xiong, L.; Liu, D.L. A novel sterol regulatory element-binding protein gene (sreA) identified in Penicillium digitatum is required for prochloraz resistance, full virulence and erg11 (cyp51) regulation. PLoS ONE 2015, 10, e0117115. [Google Scholar] [CrossRef] [Green Version]
- Jin, F.J.; Takahashi, T.; Matsushima, K.; Hara, S.; Shinohara, Y.; Maruyama, J.; Kitamoto, K.; Koyama, Y. SclR, a basic helix-loop-helix transcription factor, regulates hyphal morphology and promotes sclerotial formation in Aspergillus oryzae. Eukaryot. Cell. 2011, 10, 945–955. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.J.; Nishida, M.; Hara, S.; Koyama, Y. Identification and characterization of a putative basic helix-loop-helix transcription factor involved in the early stage of conidiophore development in Aspergillus oryzae. Fungal Genet. Biol. 2011, 48, 1108–1115. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xu, J.; Chang, P.K.; Liu, Z.; Kong, Q. New Insights of transcriptional regulator AflR in Aspergillus flavus physiology. Microbiol. Spectr. 2022, 10, e0079121. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.H.; Jun, S.C.; Lee, M.W.; Yu, J.H.; Shin, K.S. Characterization of the mbsA gene encoding a putative APSES transcription factor in Aspergillus fumigatus. Int. J. Mol. Sci. 2021, 22, 3777. [Google Scholar] [CrossRef]
- Caruso, M.L.; Litzka, O.; Martic, G.; Lottspeich, F.; Brakhage, A.A. Novel basic-region helix-loop-helix transcription factor (AnBH1) of Aspergillus nidulans counteracts the CCAAT-binding complex AnCF in the promoter of a penicillin biosynthesis gene. J. Mol. Biol. 2002, 323, 425–439. [Google Scholar] [CrossRef]
- Spröte, P.; Hynes, M.J.; Hortschansky, P.; Shelest, E.; Scharf, D.H.; Wolke, S.M.; Brakhage, A.A. Identification of the novel penicillin biosynthesis gene aatB of Aspergillus nidulans and its putative evolutionary relationship to this fungal secondary metabolism gene cluster. Mol. Microbiol. 2008, 70, 445–461. [Google Scholar] [CrossRef]
- He, H.R.; Ye, L.; Li, C.; Wang, H.Y.; Guo, X.W.; Wang, X.J.; Zhang, Y.Y.; Xiang, W.S. SbbR/SbbA, an important ArpA/AfsA-like system, regulates milbemycin production in Streptomyces bingchenggensis. Front. Microbiol. 2018, 9, 1064. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, M.; Zhang, Z.M.; Jin, L.; Wang, B.T.; Koyama, Y.; Jin, F.J. The basic-region Helix-Loop-Helix transcription factor DevR significantly affects polysaccharide metabolism in Aspergillus oryzae. Appl. Environ. Microbiol. 2019, 85, e00089-19. [Google Scholar] [CrossRef] [Green Version]
- Jin, F.J.; Han, P.; Zhuang, M.; Zhang, Z.M.; Jin, L.; Koyama, Y. Comparative proteomic analysis: SclR is importantly involved in carbohydrate metabolism in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2018, 102, 319–332. [Google Scholar] [CrossRef]
- Liu, H.; Cheng, M.; Zhao, S.S.; Lin, C.Y.; Song, J.Z.; Yang, Q. ATP-binding cassette transporter regulates N, N’-diacetylchitobiose transportation and chitinase production in Trichoderma asperellum T4. Int. J. Mol. Sci. 2019, 20, 2412. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.06. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.C.; Li, K.; Susko, E.; Roger, A.J. A class frequency mixture model that adjusts for site-specific amino acid frequencies and improves inference of protein phylogeny. BMC Evol. Biol. 2008, 8, 331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.J.; Liu, C.X.; Zhao, J.W.; Fang, B.Z.; Zhang, Y.J.; Li, L.J.; Jin, P.J.; Wang, X.J.; Xiang, W.S. Streptomyces maoxianensis sp. nov., a novel actinomycete isolated from soil in Maoxian, China. Antonie van Leeuwenhoek 2015, 107, 1119–1126. [Google Scholar] [CrossRef]
- Long, L.K.; Yang, J.; An, Y.; Liu, G. Disruption of a glutathione reductase encoding gene in Acremonium chrysogenum leads to reduction of its growth, cephalosporin production and antioxidative ability which is recovered by exogenous methionine. Fungal Genet. Biol. 2012, 49, 114–122. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; He, H.R.; Liu, H.; Wang, H.Y.; Wang, X.J.; Xiang, W.S. Characterization of a pathway-specific activator of milbemycin biosynthesis and improved milbemycin production by its overexpression in Streptomyces bingchenggensis. Microb. Cell Fact. 2016, 15, 152. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.J.; Tian, Z.H.; Xu, Y.; Zhang, J.H.; Liu, W.; Tan, H.R. Coordinative modulation of chlorothricin biosynthesis by binding of the glycosylated intermediates and end product to a responsive regulator ChlF1. J. Biol. Chem. 2016, 291, 5406–5417. [Google Scholar] [CrossRef]
| Strain or Plasmid | Description | Source or Reference |
|---|---|---|
| C. globosum W7 | Parental strain; chaetoglobosin A producer | This study |
| CgligD-N3 | CgligD deletion mutant | This study |
| CgXpp1-N14 | CgXpp1-deletion mutant | This study |
| CgXpp1-Com | CgXpp1-complemented mutant | This study |
| DH5α | Host strain for cloning | Vazyme Biotech Co., Ltd, Nanjing, China |
| TransDB3.1 | ccdB gene survival competent cell, used to propagate and maintain vectors containing the ccdB gene | TransGen Biotech, Beijing, China |
| E. coli BL21 (DE3) | Heterologous expression host | Comate Biosciences Co., Ltd, Changchun, China |
| pCR-Blunt | Routine DNA cloning contains bleomycin (BleoR) and kanamycin (KanR) resistance gene, the vector involvement of a ccdB lethality gene | Thermo Fisher Scientific, Carlsbad, CA, USA |
| pET-28a | Prokaryotic expression vector | Comate Biosciences Co., Ltd, Changchun, China |
| pBARGPE1-mCherry | Obtaining bialaphos resistance gene (BlpR) | Miaoling company, Wuhan, China |
| pCR-Blunt-BlpR | Skeleton vector for gene knockout, contains bleomycin (BleoR) and bialaphos (BlpR) resistance gene based on pCR-Blunt | This study |
| pBARGPE1-EGFP | Obtaining green fluorescent protein EGFP selectable marker | Miaoling company, Wuhan, China |
| pET-28a-CgXpp1 | Heterologous expression vector of CgXpp1 gene | This study |
| pCR-BlpR-CgXpp1 | CgXpp1-deletion vector based on pCR-Blunt-BlpR | This study |
| pBlpR-CgXpp1-Com | CgXpp1-complemented vector based on pCR-Blunt-BlpR | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.; Zhang, K.; Lin, C.; Cheng, M.; Song, J.; Ru, X.; Wang, Z.; Wang, W.; Yang, Q. Identification of a Novel Pleiotropic Transcriptional Regulator Involved in Sporulation and Secondary Metabolism Production in Chaetomium globosum. Int. J. Mol. Sci. 2022, 23, 14849. https://doi.org/10.3390/ijms232314849
Zhao S, Zhang K, Lin C, Cheng M, Song J, Ru X, Wang Z, Wang W, Yang Q. Identification of a Novel Pleiotropic Transcriptional Regulator Involved in Sporulation and Secondary Metabolism Production in Chaetomium globosum. International Journal of Molecular Sciences. 2022; 23(23):14849. https://doi.org/10.3390/ijms232314849
Chicago/Turabian StyleZhao, Shanshan, Kai Zhang, Congyu Lin, Ming Cheng, Jinzhu Song, Xin Ru, Zhengran Wang, Wan Wang, and Qian Yang. 2022. "Identification of a Novel Pleiotropic Transcriptional Regulator Involved in Sporulation and Secondary Metabolism Production in Chaetomium globosum" International Journal of Molecular Sciences 23, no. 23: 14849. https://doi.org/10.3390/ijms232314849
APA StyleZhao, S., Zhang, K., Lin, C., Cheng, M., Song, J., Ru, X., Wang, Z., Wang, W., & Yang, Q. (2022). Identification of a Novel Pleiotropic Transcriptional Regulator Involved in Sporulation and Secondary Metabolism Production in Chaetomium globosum. International Journal of Molecular Sciences, 23(23), 14849. https://doi.org/10.3390/ijms232314849








