Controlling Macrophage Polarization to Modulate Inflammatory Cues Using Immune-Switch Nanoparticles
Abstract
:1. Introduction
2. Results and Discussion
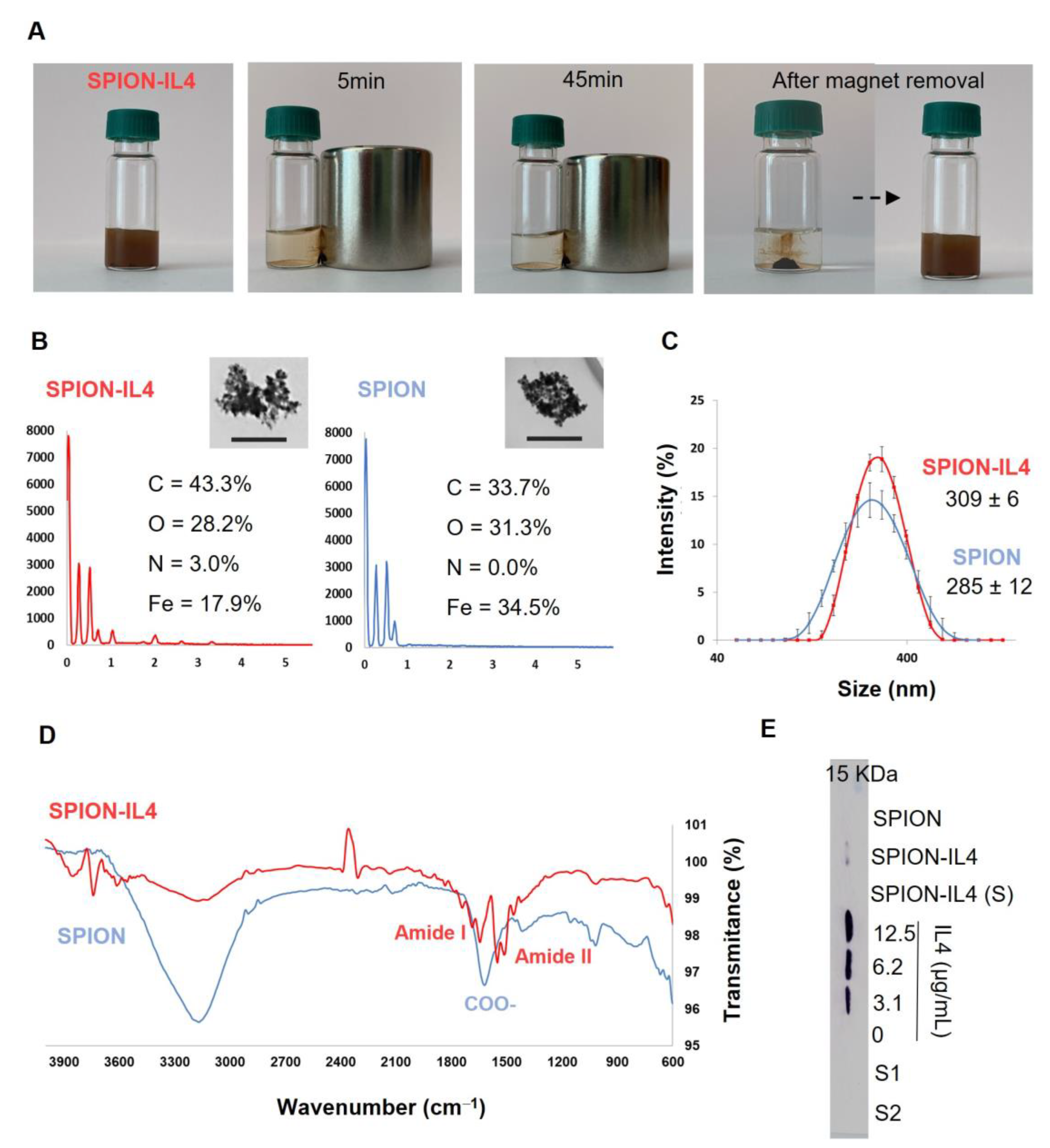
2.1. Production and Characterization of IL4-Functionalized SPION
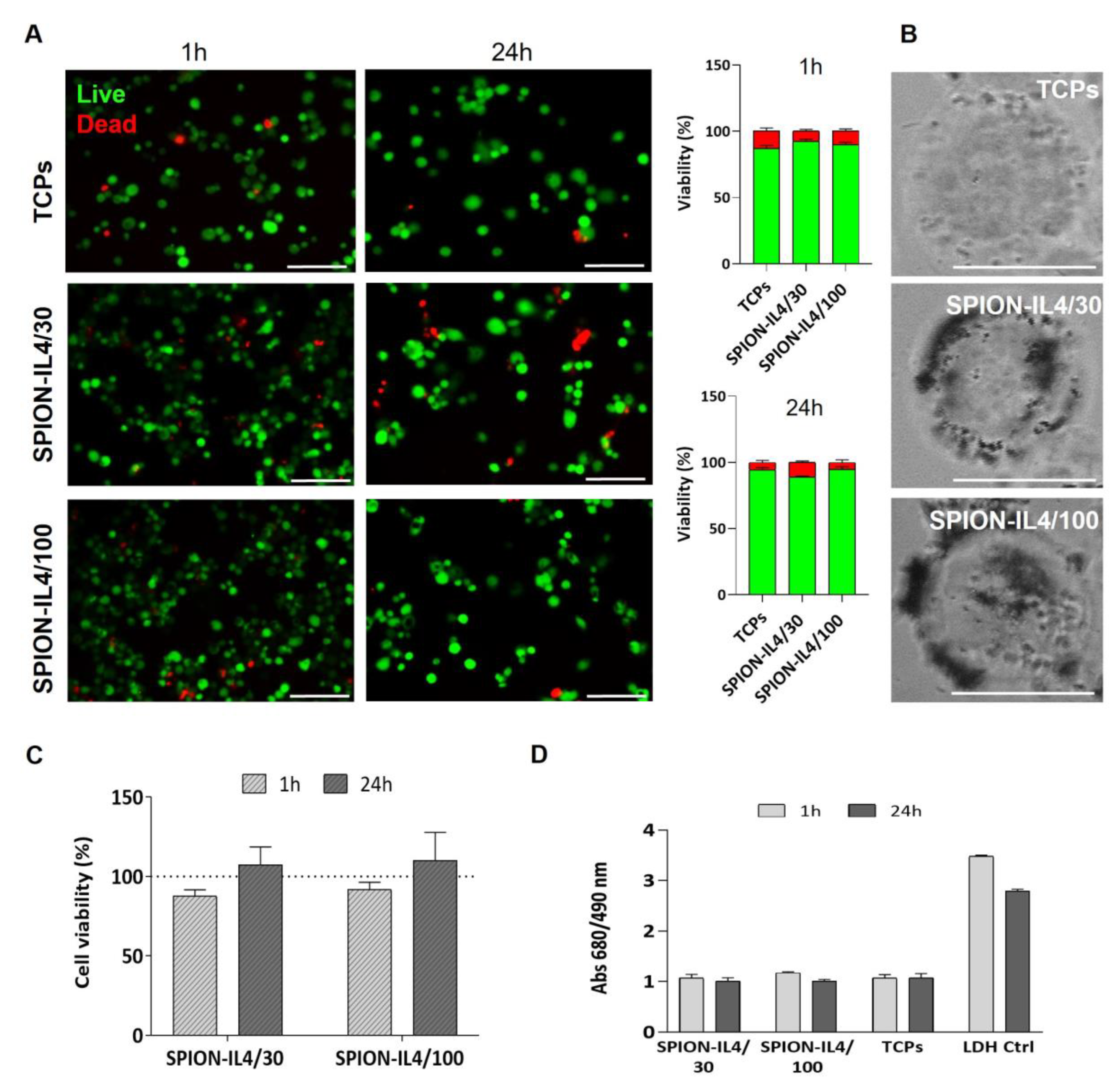
2.2. In Vitro Cytotoxicity Assessment of SPION-IL4
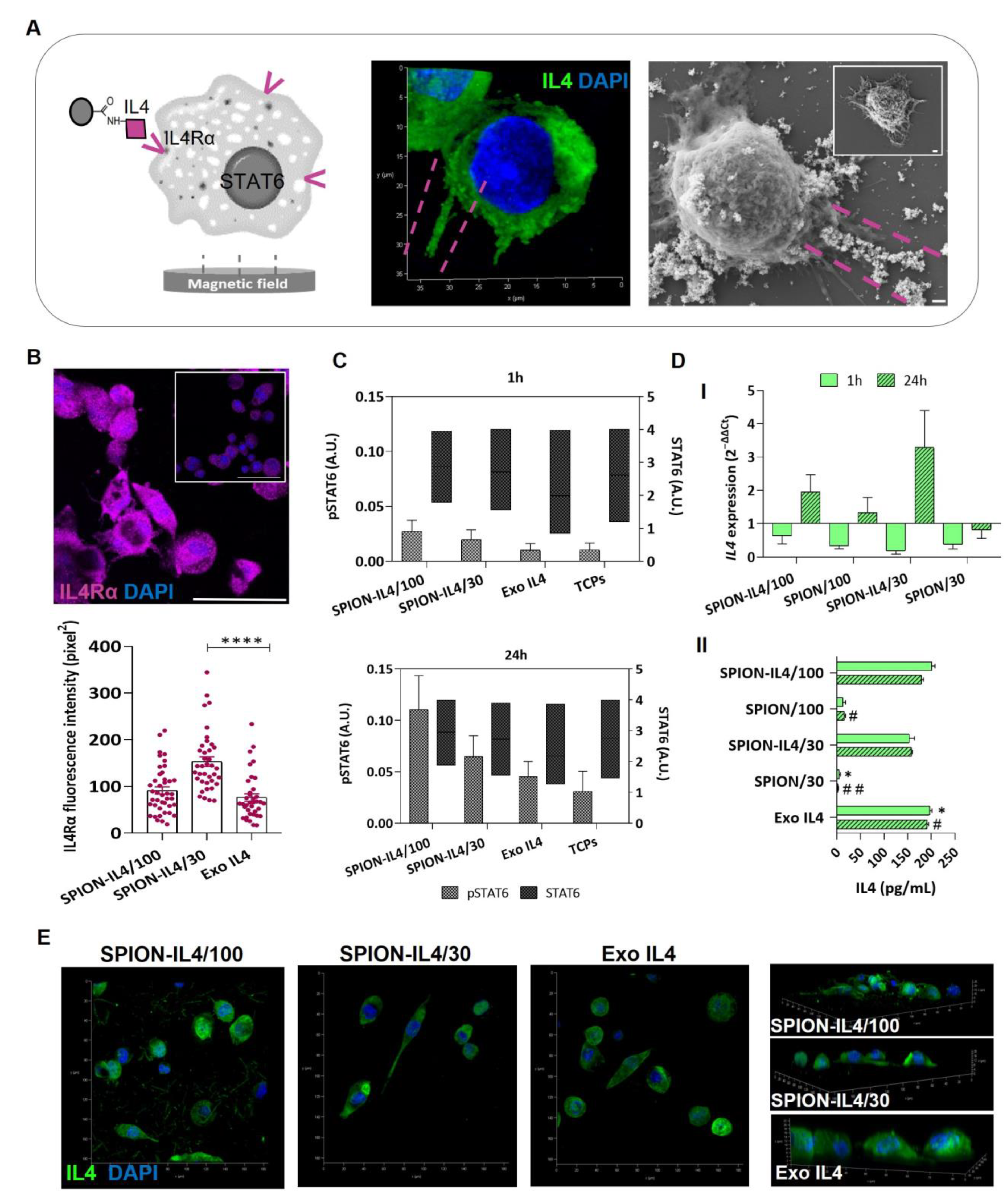
2.3. SPION-IL4 Incentivizes the Phosphorylation of STAT6 and Influences the Synthesis and Expression of IL4 and IL4Rα
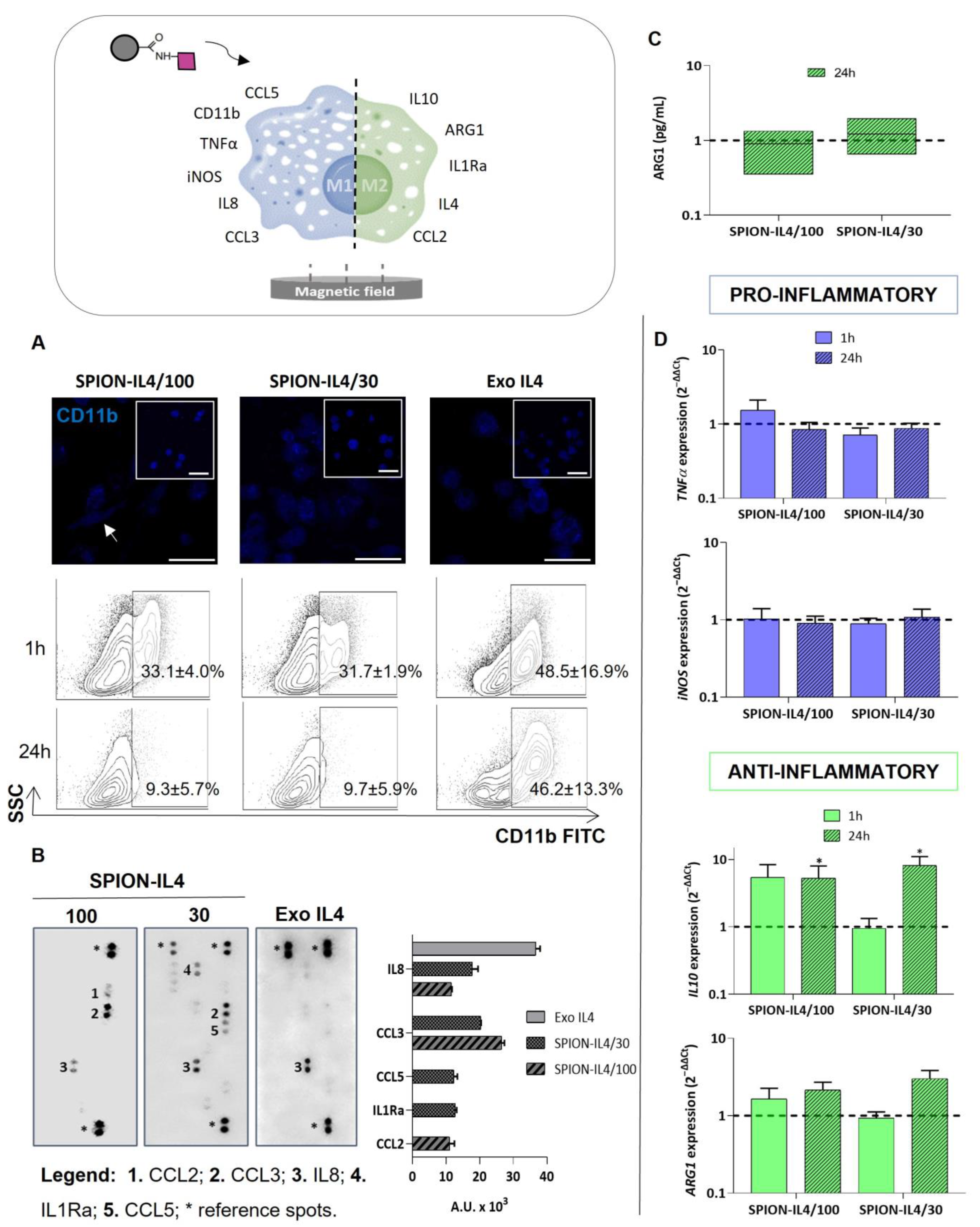
2.4. Profiling Immune-Modulatory Genes and Inflammatory Mediators upon Treatment with SPION-IL4
3. Materials and Methods
3.1. Superparamagnetic Iron Oxide Nanoparticles (SPION) Functionalization with IL4
3.2. Characterization of SPION-IL4
3.2.1. Physico-Chemical Characterization of SPION-IL4
3.2.2. Functionalization Efficiency
3.3. Macrophage Assays
3.4. Cytotoxicity Assessment
3.5. Characterization of Macrophages upon Contact with SPION-IL4
3.5.1. RNA Isolation and Gene Expression Analysis
3.5.2. Determining the Phosphorylation of STAT6
3.5.3. Immunodetection of IL4Rα, IL4, and CD11b
3.5.4. SPION-IL4/Cells Surface Interaction Analysis by SEM
3.5.5. Cytokine Production Assays
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, Q.; Gong, H.; Gao, W.; Zhang, L. Recent Progress in Capturing and Neutralizing Inflammatory Cytokines. CCS Chem. 2020, 2, 376–389. [Google Scholar] [CrossRef]
- Chen, Z.; Bozec, A.; Ramming, A.; Schett, G. Anti-inflammatory and immune-regulatory cytokines in rheumatoid arthritis. Nat. Rev. Rheumatol. 2019, 15, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Guzman-Martinez, L.; Maccioni, R.B.; Andrade, V.; Navarrete, L.P.; Pastor, M.G.; Ramos-Escobar, N. Neuroinflammation as a Common Feature of Neurodegenerative Disorders. Front. Pharmacol. 2019, 10, 1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasheed, A.; Rayner, K.J. Macrophage Responses to Environmental Stimuli During Homeostasis and Disease. Endocr. Rev. 2021, 42, 407–435. [Google Scholar] [CrossRef]
- Stein, M.; Keshav, S.; Harris, N.; Gordon, S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: A marker of alternative immunologic macrophage activation. J. Exp. Med. 1992, 176, 287–292. [Google Scholar] [CrossRef] [Green Version]
- Hart, P.H.; Vitti, G.F.; Burgess, D.R.; Whitty, G.A.; Piccoli, D.S.; Hamilton, J.A. Potential antiinflammatory effects of interleukin 4: Suppression of human monocyte tumor necrosis factor alpha, interleukin 1, and prostaglandin E2. Proc. Natl. Acad. Sci. USA 1989, 86, 3803–3807. [Google Scholar] [CrossRef] [Green Version]
- Linehan, S.A.; Coulson, P.S.; Wilson, R.A.; Mountford, A.P.; Brombacher, F.; Martínez-Pomares, L.; Gordon, S. IL-4 receptor signaling is required for mannose receptor expression by macrophages recruited to granulomata but not resident cells in mice infected with Schistosoma mansoni. Lab. Investig. J. Tech. Methods Pathol. 2003, 83, 1223–1231. [Google Scholar] [CrossRef] [Green Version]
- Heller, N.M.; Qi, X.; Junttila, I.S.; Shirey, K.A.; Vogel, S.N.; Paul, W.E.; Keegan, A.D. Type I IL-4Rs selectively activate IRS-2 to induce target gene expression in macrophages. Sci. Signal. 2008, 1, ra17. [Google Scholar] [CrossRef] [Green Version]
- Czimmerer, Z.; Daniel, B.; Horvath, A.; Rückerl, D.; Nagy, G.; Kiss, M.; Peloquin, M.; Budai, M.M.; Cuaranta-Monroy, I.; Simandi, Z.; et al. The Transcription Factor STAT6 Mediates Direct Repression of Inflammatory Enhancers and Limits Activation of Alternatively Polarized Macrophages. Immunity 2018, 48, 75–90.e6. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Brombacher, F.; Tunyogi-Csapo, M.; Glant, T.T.; Finnegan, A. Interleukin-4 regulates proteoglycan-induced arthritis by specifically suppressing the innate immune response. Arthritis Rheum. 2007, 56, 861–870. [Google Scholar] [CrossRef]
- Haikal, S.M.; Abdeltawab, N.F.; Rashed, L.A.; Abd El-Galil, T.I.; Elmalt, H.A.; Amin, M.A. Combination Therapy of Mesenchymal Stromal Cells and Interleukin-4 Attenuates Rheumatoid Arthritis in a Collagen-Induced Murine Model. Cells 2019, 8, 823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghoreschi, K.; Thomas, P.; Breit, S.; Dugas, M.; Mailhammer, R.; van Eden, W.; van der Zee, R.; Biedermann, T.; Prinz, J.; Mack, M.; et al. Interleukin-4 therapy of psoriasis induces Th2 responses and improves human autoimmune disease. Nat. Med. 2003, 9, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Mejías, R.; Costo, R.; Roca, A.G.; Arias, C.F.; Veintemillas-Verdaguer, S.; González-Carreño, T.; del Puerto Morales, M.; Serna, C.J.; Mañes, S.; Barber, D.F. Cytokine adsorption/release on uniform magnetic nanoparticles for localized drug delivery. J. Control. Release 2008, 130, 168–174. [Google Scholar] [CrossRef]
- Almeida, A.F.; Vinhas, A.; Gonçalves, A.I.; Miranda, M.S.; Rodrigues, M.T.; Gomes, M.E. Magnetic triggers in biomedical applications—Prospects for contact free cell sensing and guidance. J. Mater. Chem. B 2021, 9, 1259–1271. [Google Scholar] [CrossRef]
- Gonçalves, A.I.; Rotherham, M.; Markides, H.; Rodrigues, M.T.; Reis, R.L.; Gomes, M.E.; El Haj, A.J. Triggering the activation of Activin A type II receptor in human adipose stem cells towards tenogenic commitment using mechanomagnetic stimulation. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1149–1159. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, S.; Jiang, S. Dual-Functional Biomimetic Materials: Nonfouling Poly(carboxybetaine) with Active Functional Groups for Protein Immobilization. Biomacromolecules 2006, 7, 3311–3315. [Google Scholar] [CrossRef]
- Bartczak, D.; Kanaras, A.G. Preparation of Peptide-Functionalized Gold Nanoparticles Using One Pot EDC/Sulfo-NHS Coupling. Langmuir 2011, 27, 10119–10123. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, B.; Sun, S. Dumbbell-like Au−Fe3O4 Nanoparticles for Target-Specific Platin Delivery. J. Am. Chem. Soc. 2009, 131, 4216–4217. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, C.V.; Delzeit, L.; Cassell, A.M.; Li, J.; Han, J.; Meyyappan, M. Preparation of Nucleic Acid Functionalized Carbon Nanotube Arrays. Nano Lett. 2002, 2, 1079–1081. [Google Scholar] [CrossRef]
- Wang, Y.; Qi, H.; Miron, R.; Zhang, Y. Modulating macrophage polarization on titanium implant surface by poly(dopamine)-assisted immobilization of IL4. Clin. Implant Dent. Relat. Res. 2019, 21, 977–986. [Google Scholar] [CrossRef]
- Alric, C.; Aubrey, N.; Allard-Vannier, É.; di Tommaso, A.; Blondy, T.; Dimier-Poisson, I.; Chourpa, I.; Hervé-Aubert, K. Covalent conjugation of cysteine-engineered scFv to PEGylated magnetic nanoprobes for immunotargeting of breast cancer cells. RSC Adv. 2016, 6, 37099–37109. [Google Scholar] [CrossRef]
- Schlachter, E.K.; Widmer, H.R.; Bregy, A.; Lönnfors-Weitzel, T.; Vajtai, I.; Corazza, N.; Bernau, V.J.; Weitzel, T.; Mordasini, P.; Slotboom, J.; et al. Metabolic pathway and distribution of superparamagnetic iron oxide nanoparticles: In vivo study. Int. J. Nanomed. 2011, 6, 1793–1800. [Google Scholar]
- Singh, N.; Jenkins, G.J.S.; Asadi, R.; Doak, S.H. Potential toxicity of superparamagnetic iron oxide nanoparticles (SPION). Nano Rev. 2010, 1, 5358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elias, A.; Tsourkas, A. Imaging circulating cells and lymphoid tissues with iron oxide nanoparticles. Hematology 2009, 2009, 720–726. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, H.L.; Cronholm, P.; Gustafsson, J.; Möller, L. Copper Oxide Nanoparticles Are Highly Toxic: A Comparison between Metal Oxide Nanoparticles and Carbon Nanotubes. Chem. Res. Toxicol. 2008, 21, 1726–1732. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.M.; Hess, K.L.; Gearhart, J.M.; Geiss, K.T.; Schlager, J.J. In vitro toxicity of nanoparticles in BRL 3A rat liver cells. Toxicol. In Vitro 2005, 19, 975–983. [Google Scholar] [CrossRef]
- Waqas, S.F.H.; Ampem, G.; Röszer, T. Analysis of IL-4/STAT6 Signaling in Macrophages. Methods Mol. Biol. 2019, 1966, 211–224. [Google Scholar]
- Kaplan, M.H.; Schindler, U.; Smiley, S.T.; Grusby, M.J. Stat6 Is Required for Mediating Responses to IL-4 and for the Development of Th2 Cells. Immunity 1996, 4, 313–319. [Google Scholar] [CrossRef] [Green Version]
- Takeda, K.; Tanaka, T.; Shi, W.; Matsumoto, M.; Minami, M.; Kashiwamura, S.-I.; Nakanishi, K.; Yoshida, N.; Kishimoto, T.; Akira, S. Essential role of Stat6 in IL-4 signalling. Nature 1996, 380, 627–630. [Google Scholar] [CrossRef]
- Xu, Y.; Li, Y.; Liu, X.; Pan, Y.; Sun, Z.; Xue, Y.; Wang, T.; Dou, H.; Hou, Y. SPIONs enhances IL-10-producing macrophages to relieve sepsis via Cav1-Notch1/HES1-mediated autophagy. Int. J. Nanomed. 2019, 14, 6779–6797. [Google Scholar] [CrossRef] [Green Version]
- Schmid, M.C.; Khan, S.Q.; Kaneda, M.M.; Pathria, P.; Shepard, R.; Louis, T.L.; Anand, S.; Woo, G.; Leem, C.; Faridi, M.H.; et al. Integrin CD11b activation drives anti-tumor innate immunity. Nat. Commun. 2018, 9, 5379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, S.T.; Edgington, T.S. Integrin regulation of leukocyte inflammatory functions. CD11b/CD18 enhancement of the tumor necrosis factor-alpha responses of monocytes. J. Immunol. Baltim. Md. 1950 1993, 150, 2972–2980. [Google Scholar]
- Ling, G.S.; Bennett, J.; Woollard, K.J.; Szajna, M.; Fossati-Jimack, L.; Taylor, P.R.; Scott, D.; Franzoso, G.; Cook, H.T.; Botto, M. Integrin CD11b positively regulates TLR4-induced signalling pathways in dendritic cells but not in macrophages. Nat. Commun. 2014, 5, 3039. [Google Scholar] [CrossRef] [PubMed]
- McWhorter, F.Y.; Wang, T.; Nguyen, P.; Chung, T.; Liu, W.F. Modulation of macrophage phenotype by cell shape. Proc. Natl. Acad. Sci. USA 2013, 110, 17253–17258. [Google Scholar] [CrossRef] [Green Version]
- Chan, L.P.; Liu, C.; Chiang, F.Y.; Wang, L.F.; Lee, K.W.; Chen, W.T.; Kuo, P.L.; Liang, C.H. IL-8 promotes inflammatory mediators and stimulates activation of p38 MAPK/ERK-NF-κB pathway and reduction of JNK in HNSCC. Oncotarget 2017, 8, 56375–56388. [Google Scholar] [CrossRef] [Green Version]
- Mahmoud, L.; Al-Enezi, F.; Al-Saif, M.; Warsy, A.; Khabar, K.S.A.; Hitti, E.G. Sustained stabilization of Interleukin-8 mRNA in human macrophages. RNA Biol. 2014, 11, 124–133. [Google Scholar] [CrossRef] [Green Version]
- Maurer, M.; von Stebut, E. Macrophage inflammatory protein-1. Int. J. Biochem. Cell Biol. 2004, 36, 1882–1886. [Google Scholar] [CrossRef]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef]
- Deci, M.B.; Ferguson, S.W.; Scatigno, S.L.; Nguyen, J. Modulating Macrophage Polarization through CCR2 Inhibition and Multivalent Engagement. Mol. Pharm. 2018, 15, 2721–2731. [Google Scholar] [CrossRef]
- Chen, H.; Sun, H.; You, F.; Sun, W.; Zhou, X.; Chen, L.; Yang, J.; Wang, Y.; Tang, H.; Guan, Y.; et al. Activation of STAT6 by STING Is Critical for Antiviral Innate Immunity. Cell 2011, 147, 436–446. [Google Scholar] [CrossRef] [Green Version]
- Madej, M.P.; Töpfer, E.; Boraschi, D.; Italiani, P. Different Regulation of Interleukin-1 Production and Activity in Monocytes and Macrophages: Innate Memory as an Endogenous Mechanism of IL-1 Inhibition. Front. Pharmacol. 2017, 8, 335. [Google Scholar] [CrossRef] [PubMed]
- Harrell, C.R.; Markovic, B.S.; Fellabaum, C.; Arsenijevic, N.; Djonov, V.; Volarevic, V. The role of Interleukin 1 receptor antagonist in mesenchymal stem cell-based tissue repair and regeneration. BioFactors 2020, 46, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Campbell, L.; Saville, C.R.; Murray, P.J.; Cruickshank, S.M.; Hardman, M.J. Local Arginase 1 Activity Is Required for Cutaneous Wound Healing. J. Investig. Dermatol. 2013, 133, 2461–2470. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Dai, X.; Chen, J.; Zhao, J.; Xu, M.; Zhang, L.; Yang, B.; Zhang, W.; Rocha, M.; Nakao, T.; et al. STAT6/Arg1 promotes microglia/macrophage efferocytosis and inflammation resolution in stroke mice. JCI Insight 2019, 4, e131355. [Google Scholar] [CrossRef] [PubMed]
- Richards, D.M.; Endres, R.G. The mechanism of phagocytosis: Two stages of engulfment. Biophys. J. 2014, 107, 1542–1553. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almeida, A.F.; Miranda, M.S.; Vinhas, A.; Gonçalves, A.I.; Gomes, M.E.; Rodrigues, M.T. Controlling Macrophage Polarization to Modulate Inflammatory Cues Using Immune-Switch Nanoparticles. Int. J. Mol. Sci. 2022, 23, 15125. https://doi.org/10.3390/ijms232315125
Almeida AF, Miranda MS, Vinhas A, Gonçalves AI, Gomes ME, Rodrigues MT. Controlling Macrophage Polarization to Modulate Inflammatory Cues Using Immune-Switch Nanoparticles. International Journal of Molecular Sciences. 2022; 23(23):15125. https://doi.org/10.3390/ijms232315125
Chicago/Turabian StyleAlmeida, Ana F., Margarida S. Miranda, Adriana Vinhas, Ana I. Gonçalves, Manuela E. Gomes, and Márcia T. Rodrigues. 2022. "Controlling Macrophage Polarization to Modulate Inflammatory Cues Using Immune-Switch Nanoparticles" International Journal of Molecular Sciences 23, no. 23: 15125. https://doi.org/10.3390/ijms232315125
APA StyleAlmeida, A. F., Miranda, M. S., Vinhas, A., Gonçalves, A. I., Gomes, M. E., & Rodrigues, M. T. (2022). Controlling Macrophage Polarization to Modulate Inflammatory Cues Using Immune-Switch Nanoparticles. International Journal of Molecular Sciences, 23(23), 15125. https://doi.org/10.3390/ijms232315125







