Host Cell Binding Mediated by Leptospira interrogans Adhesins
Abstract
1. Introduction
2. Results
2.1. Evaluation of Leptospira Binding to Mammalian Cells by ELISA
2.2. Evaluation of Leptospiral Membrane Protein Binding to Mammalian Cells
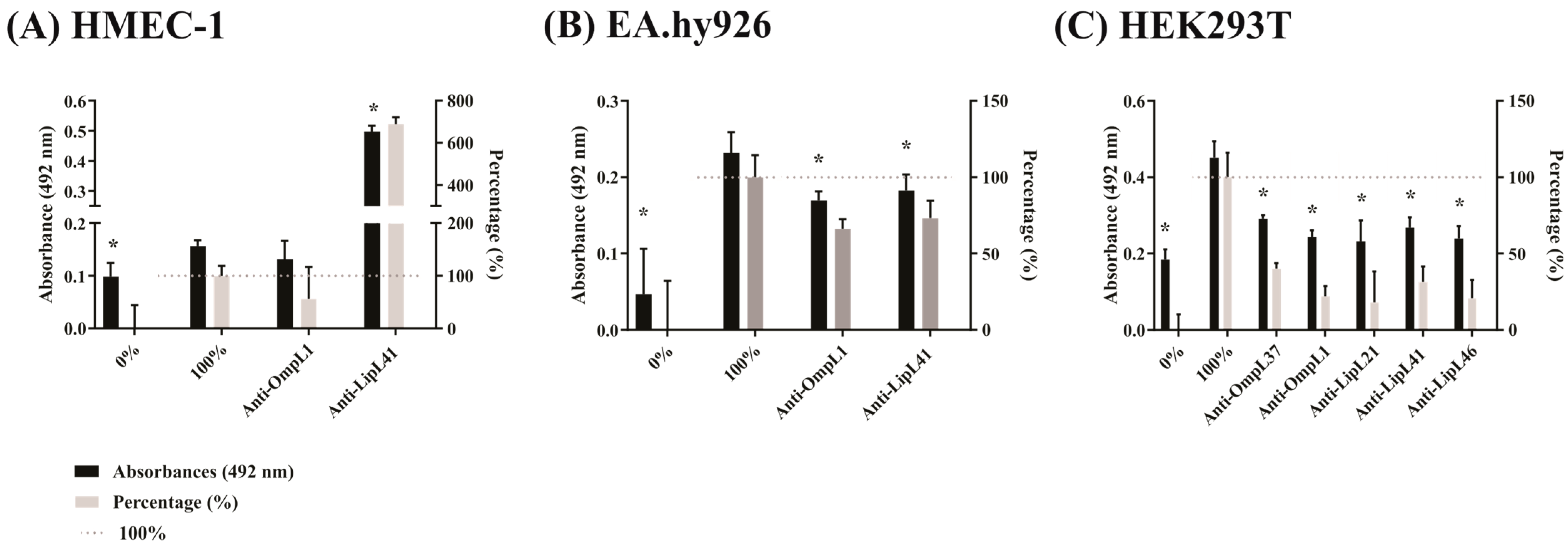
2.3. Effect of Antiserum against the Recombinant Proteins on the Adhesion of L. interrogans to Mammalian Cells
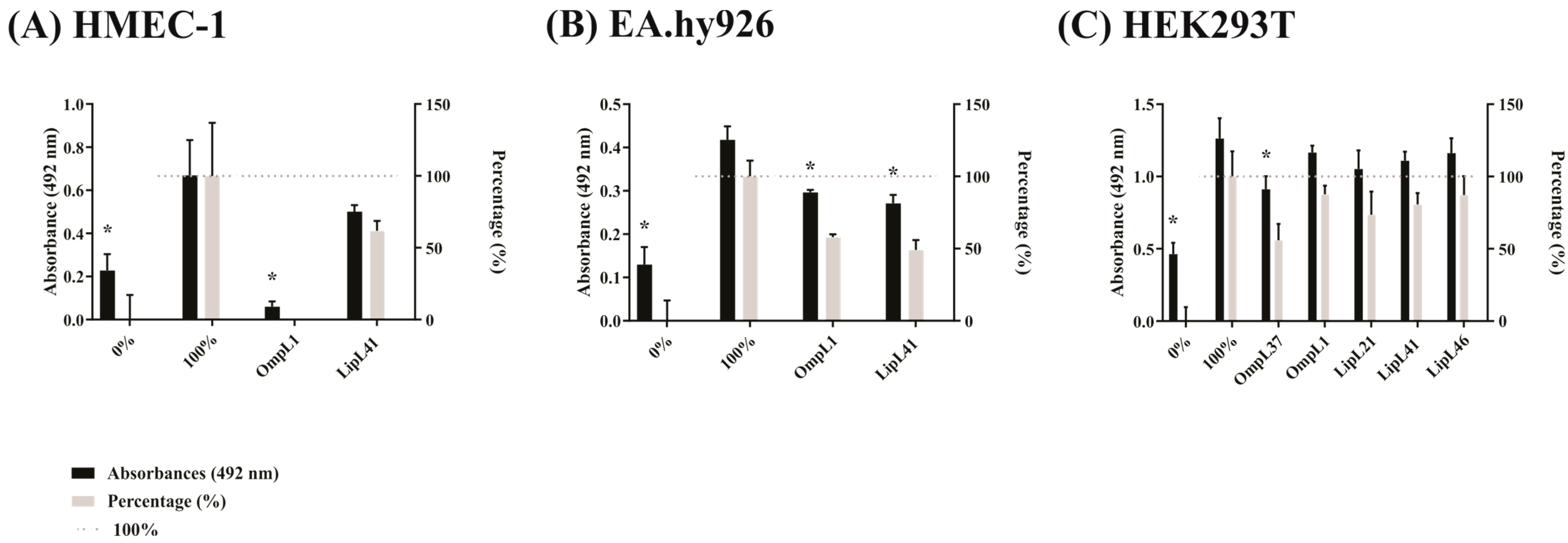
2.4. Effect of Recombinant Proteins on the Adhesion of L. interrogans to Mammalian Cells
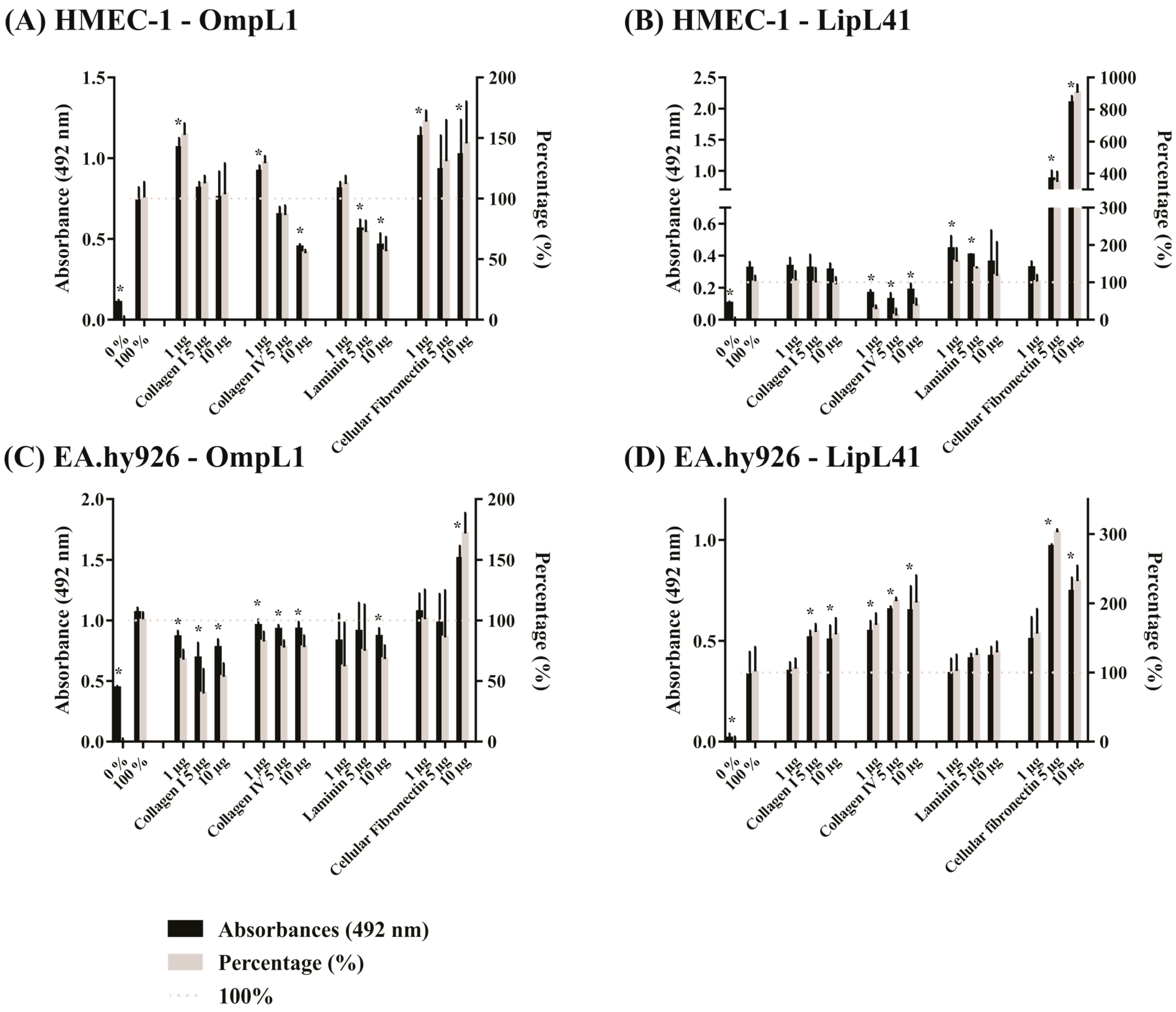
2.5. Effect of Extracellular Matrix Components on Leptospiral Protein Adhesion to Mammalian Cells
3. Discussion
4. Materials and Methods
4.1. Leptospira Strains and Mammalian Cell Lines
4.2. Evaluation of Leptospire Binding to Mammalian Cells by ELISA
4.3. Determination of Leptospiral Membrane Protein Binding to Mammalian Cells
4.4. Effect of Leptospiral Membrane Proteins on Bacterial Adhesion to Mammalian Cells
4.5. Evaluation of the Effect of Host Components on Adhesion of Leptospiral Membrane Proteins to Mammalian Cells
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Patel, S.; Mathivanan, N.; Goyal, A. Bacterial Adhesins, the Pathogenic Weapons to Trick Host Defense Arsenal. Biomed. Pharmacother. 2017, 93, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Paulsson, M.; Riesbeck, K. How Bacteria Hack the Matrix and Dodge the Bullets of Immunity. Eur. Respir. Rev. 2018, 27, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Coburn, J.; Leong, J.; Chaconas, G. Illuminating the Roles of the Borrelia burgdorferi Adhesins. Trends Microbiol. 2013, 21, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Daroz, B.B.; Fernandes, L.G.V.; Cavenague, M.F.; Kochi, L.T.; Passalia, F.J.; Takahashi, M.B.; Nascimento Filho, E.G.; Teixeira, A.F.; Nascimento, A.L.T.O. A Review on Host-Leptospira Interactions: What We Know and Future Expectations. Front. Cell. Infect. Microbiol. 2021, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Vieira, M.L.; Fernandes, L.G.; Domingos, R.F.; Oliveira, R.; Siqueira, G.H.; Souza, N.M.; Teixeira, A.R.F.; Atzingen, M.v.; Nascimento, A.L.T.O. Leptospiral Extracellular Matrix Adhesins as Mediators of Pathogen-Host Interactions. FEMS Microbiol. Lett. 2014, 352, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, L.G.V.; Vieira, M.L.; Kirchgatter, K.; Alves, I.J.; de Morais, Z.M.; Vasconcellos, S.A.; Romero, E.C.; Nascimento, A.L.T.O. Ompl1 Is an Extracellular Matrix- and Plasminogen-Interacting Protein of Leptospira spp. Infect. Immun. 2012, 80, 3679–3692. [Google Scholar] [CrossRef]
- Robbins, G.T.; Hahn, B.L.; Evangelista, K.V.; Padmore, L.; Aranda, P.S.; Coburn, J. Evaluation of Cell Binding Activities of Leptospira ECM Adhesins. PLoS Negl. Trop. Dis. 2015, 9, e0003712. [Google Scholar] [CrossRef]
- Pinne, M.; Choy, H.A.; Haake, D.A. The OmpL37 Surface-Exposed Protein Is Expressed by Pathogenic Leptospira during Infection and Binds Skin and Vascular Elastin. PLoS Negl. Trop. Dis. 2010, 4, e815. [Google Scholar] [CrossRef]
- Santos, J.v.; Pereira, P.R.M.; Fernandes, L.G.v.; Siqueira, G.H.; de Souza, G.O.; Souza Filho, A.; Vasconcellos, S.A.; Heinemann, M.B.; Chapola, E.G.B.; Nascimento, A.L.T.O. Binding of Human Plasminogen by the Lipoprotein LipL46 of Leptospira interrogans. Mol. Cell. Probes 2018, 37, 12–21. [Google Scholar] [CrossRef]
- Takahashi, M.B.; Teixeira, A.F.; Nascimento, A.L.T.O. The Leptospiral LipL21 and LipL41 Proteins Exhibit a Broad Spectrum of Interactions with Host Cell Components. Virulence 2021, 12, 2798–2813. [Google Scholar] [CrossRef]
- Breiner, D.D.; Fahey, M.; Salvador, R.; Novakova, J.; Coburn, J. Leptospira Interrogans Binds to Human Cell Surface Receptors Including Proteoglycans. Infect. Immun. 2009, 77, 5528–5536. [Google Scholar] [CrossRef] [PubMed]
- Cinco, M.; Domenis, R.; Perticarari, S.; Presani, G.; Marangoni, A.; Blasi, E. Interaction of Leptospires with Murine Microglial Cells. New Microbiol. 2006, 29, 193–199. [Google Scholar]
- Evangelista, K.V.; Hahn, B.; Wunder, E.A.; Ko, A.I.; Haake, D.A.; Coburn, J. Identification of Cell-Binding Adhesins of Leptospira Interrogans. PLoS Negl. Trop. Dis. 2014, 8, e3215. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, K.; Franco, R.; Schwab, A.; Coburn, J. Leptospira interrogans Binds to Cadherins. PLoS Negl. Trop. Dis. 2014, 8, e2672. [Google Scholar] [CrossRef] [PubMed]
- Merien, F.; Truccolo, J.; Rougier, Y.; Baranton, G.; Perolat, P. In Vivo Apoptosis of Hepatocytes in Guinea Pigs Infected with Leptospira interrogans Serovar Icterohaemorrhagiae. FEMS Microbiol. Lett. 1998, 169, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Merien, F.; Baranton, G.; Perolat, P. Invasion of Vero Cells and Induction of Apoptosis in Macrophages by Pathogenic Leptospira interrogans Are Correlated with Virulence. Infect. Immun. 1997, 65, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.D.; Higbie, L.M. In Vitro Association of Leptospires with Host Cells. Infect Immun. 1990, 58, 581–585. [Google Scholar] [CrossRef]
- Eshghi, A.; Gaultney, R.A.; England, P.; Brûlé, S.; Miras, I.; Sato, H.; Coburn, J.; Bellalou, J.; Moriarty, T.J.; Haouz, A.; et al. An Extracellular Leptospira interrogans Leucine-Rich Repeat Protein Binds Human E- and VE-Cadherins. Cell. Microbiol. 2019, 21, e12949. [Google Scholar] [CrossRef]
- Hsieh, C.L.; Tseng, A.; He, H.; Kuo, C.J.; Wang, X.; Chang, Y.F. Leptospira Immunoglobulin-like Protein B Interacts with the 20th Exon of Human Tropoelastin Contributing to Leptospiral Adhesion to Human Lung Cells. Front. Cell. Infect. Microbiol. 2017, 7, 163. [Google Scholar] [CrossRef]
- Lima, S.S.; Ching, A.T.; Fávaro, R.D.; da Silva, J.B.; Oliveira, M.L.; Carvalho, E.; Abreu, P.A.; Vasconcellos, S.A.; Ho, P.L. Adhesin Activity of Leptospira Interrogans Lipoprotein Identified by In Vivo and In Vitro Shotgun Phage Display. Biochem. Biophys. Res. Commun. 2013, 431, 342–347. [Google Scholar] [CrossRef]
- Lin, Y.P.; Mcdonough, S.P.; Sharma, Y.; Chang, Y.F. The Terminal Immunoglobulin-like Repeats of LigA and LigB of Leptospira Enhance Their Binding to Gelatin Binding Domain of Fibronectin and Host Cells. PLoS ONE 2010, 5, e11301. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, A.S.; Abreu, P.A.; Neves, F.O.; Atzingen, M.V.; Watanabe, M.M.; Vieira, M.L.; Morais, Z.M.; Vasconcellos, S.A.; Nascimento, A.L. A Newly Identified Leptospiral Adhesin Mediates Attachment to Laminin. Infect. Immun. 2006, 74, 6356–6364. [Google Scholar] [CrossRef] [PubMed]
- Adler, B. Pathogenesis of Leptospirosis: Cellular and Molecular Aspects. Vet. Microbiol. 2014, 172, 353–358. [Google Scholar] [CrossRef]
- Haake, D.A.; Levett, P.N. Leptospirosis in Humans. Curr. Top. Microbiol. Immunol. 2015, 387, 65–97. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lopez, D.G.; Fahey, M.; Coburn, J. Responses of Human Endothelial Cells to Pathogenic and Non-Pathogenic Leptospira Species. PLoS Negl. Trop. Dis. 2010, 4, e918. [Google Scholar] [CrossRef]
- Croda, J.; Figueira, C.P.; Wunder, E.A.; Santos, C.S.; Reis, M.G.; Ko, A.I.; Picardeau, M. Targeted Mutagenesis in Pathogenic Leptospira Species: Disruption of the LigB Gene Does Not Affect Virulence in Animal Models of Leptospirosis. Infect. Immun. 2008, 76, 5826–5833. [Google Scholar] [CrossRef]
- Schubert, A.; Zakikhany, K.; Pietrocola, G.; Meinke, A.; Speziale, P.; Eikmanns, B.J.; Reinscheid, D.J. The Fibrinogen Receptor FbsA Promotes Adherence of Streptococcus agalactiae to Human Epithelial Cells. Infect. Immun. 2004, 72, 6197–6205. [Google Scholar] [CrossRef]
- Chen, T.; Duncan, M.J. Gingipain Adhesin Domains Mediate Porphyromonas gingivalis Adherence to Epithelial Cells. Microb. Pathog. 2004, 36, 205–209. [Google Scholar] [CrossRef]
- Posadas, D.M.; Ruiz-Ranwez, V.; Bonomi, H.R.; Martín, F.A.; Zorreguieta, A. BmaC, a Novel Autotransporter of Brucella Suis, Is Involved in Bacterial Adhesion to Host Cells. Cell. Microbiol. 2012, 14, 965–982. [Google Scholar] [CrossRef]
- Girón, J.A.; Torres, A.G.; Freer, E.; Kaper, J.B. The Flagella of Enteropathogenic Escherichia Coli Mediate Adherence to Epithelial Cells. Mol. Microbiol. 2002, 44, 361–379. [Google Scholar] [CrossRef]
- Dibb-Fuller, M.P.; Allen-Vercoe, E.; Thorns, C.J.; Woodward, M.J. Fimbriae- and Flagella-Mediated Association with and Invasion of Cultured Epithelial Cells by Salmonella enteritidis. Microbiology 1999, 145, 1023–1031. [Google Scholar] [CrossRef]
- Speziale, P.; Joh, D.; Visai, L.; Bozzini, S.; House-Pompeo, K.; Lindberg, M.; Höök, M. A Monoclonal Antibody Enhances Ligand Binding of Fibronectin MSCRAMM (Adhesin) from Streptococcus dysgalactiae. J. Biol. Chem. 1996, 271, 1371–1378. [Google Scholar] [CrossRef]
- Meenan, N.A.G.; Visai, L.; Valtulina, V.; Schwarz-Linek, U.; Norris, N.C.; Gurusiddappa, S.; Höök, M.; Speziale, P.; Potts, J.R. The Tandem β-Zipper Model Defines High Affinity Fibronectin-Binding Repeats within Staphylococcus Aureus FnBPA. J. Biol. Chem. 2007, 282, 25893–25902. [Google Scholar] [CrossRef] [PubMed]
- Courtney, H.S.; Dale, J.B.; Hasty, D.L. Differential Effects of the Streptococcal Fibronectin-Binding Protein, FBP54, on Adhesion of Group A Streptococci to Human Buccal Cells and HEp-2 Tissue Culture Cells. Infect. Immun. 1996, 64, 2415–2419. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Fleury, C.; Jalalvand, F.; Riesbeck, K. Human Pathogens Utilize Host Extracellular Matrix Proteins Laminin and Collagen for Adhesion and Invasion of the Host. FEMS Microbiol. Rev. 2012, 36, 1122–1180. [Google Scholar] [CrossRef] [PubMed]
- Anderton, J.M.; Rajam, G.; Romero-Steiner, S.; Summer, S.; Kowalczyk, A.P.; Carlone, G.M.; Sampson, J.S.; Ades, E.W. E-Cadherin Is a Receptor for the Common Protein Pneumococcal Surface Adhesin A (PsaA) of Streptococcus Pneumoniae. Microb. Pathog. 2007, 42, 225–236. [Google Scholar] [CrossRef]

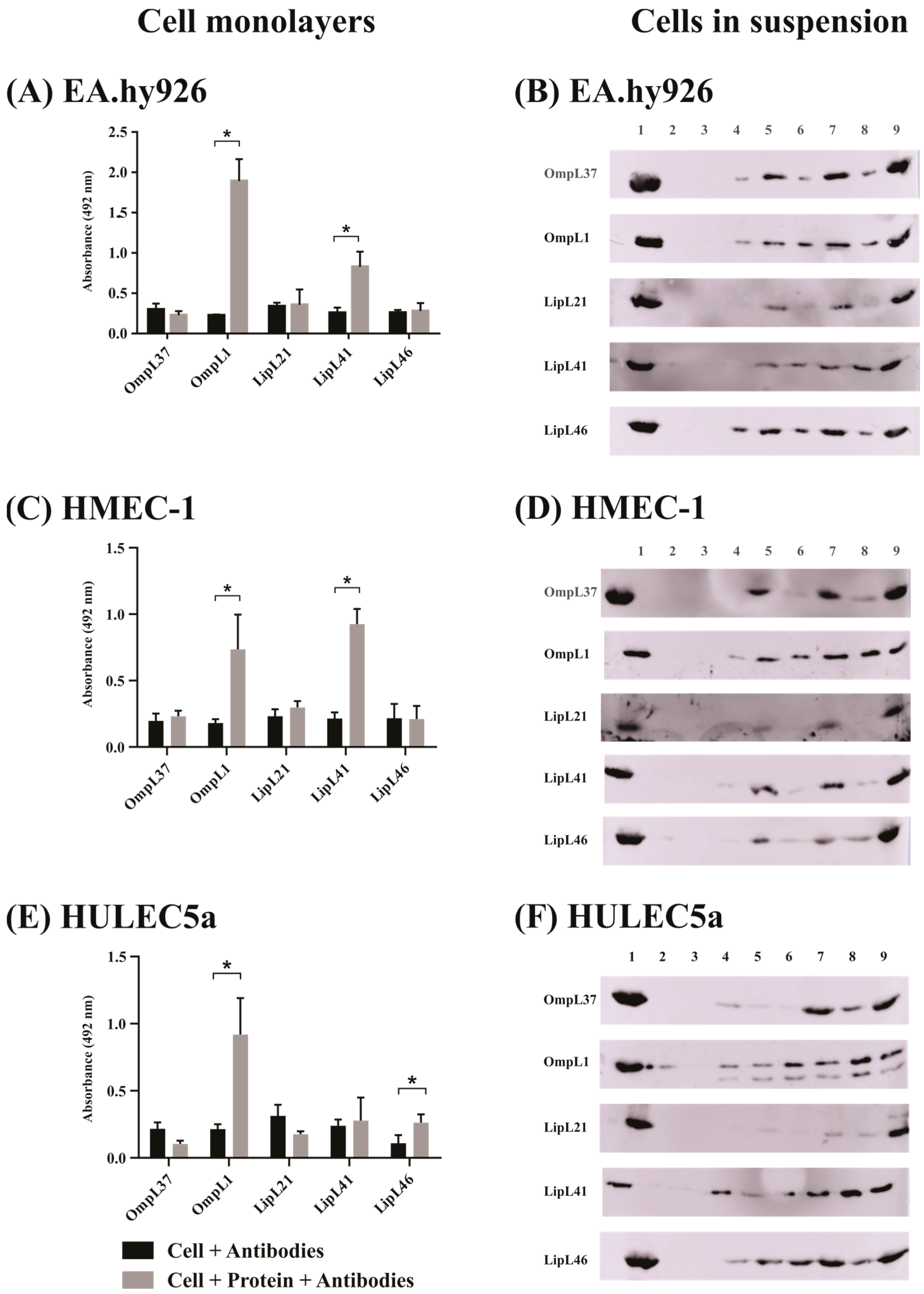
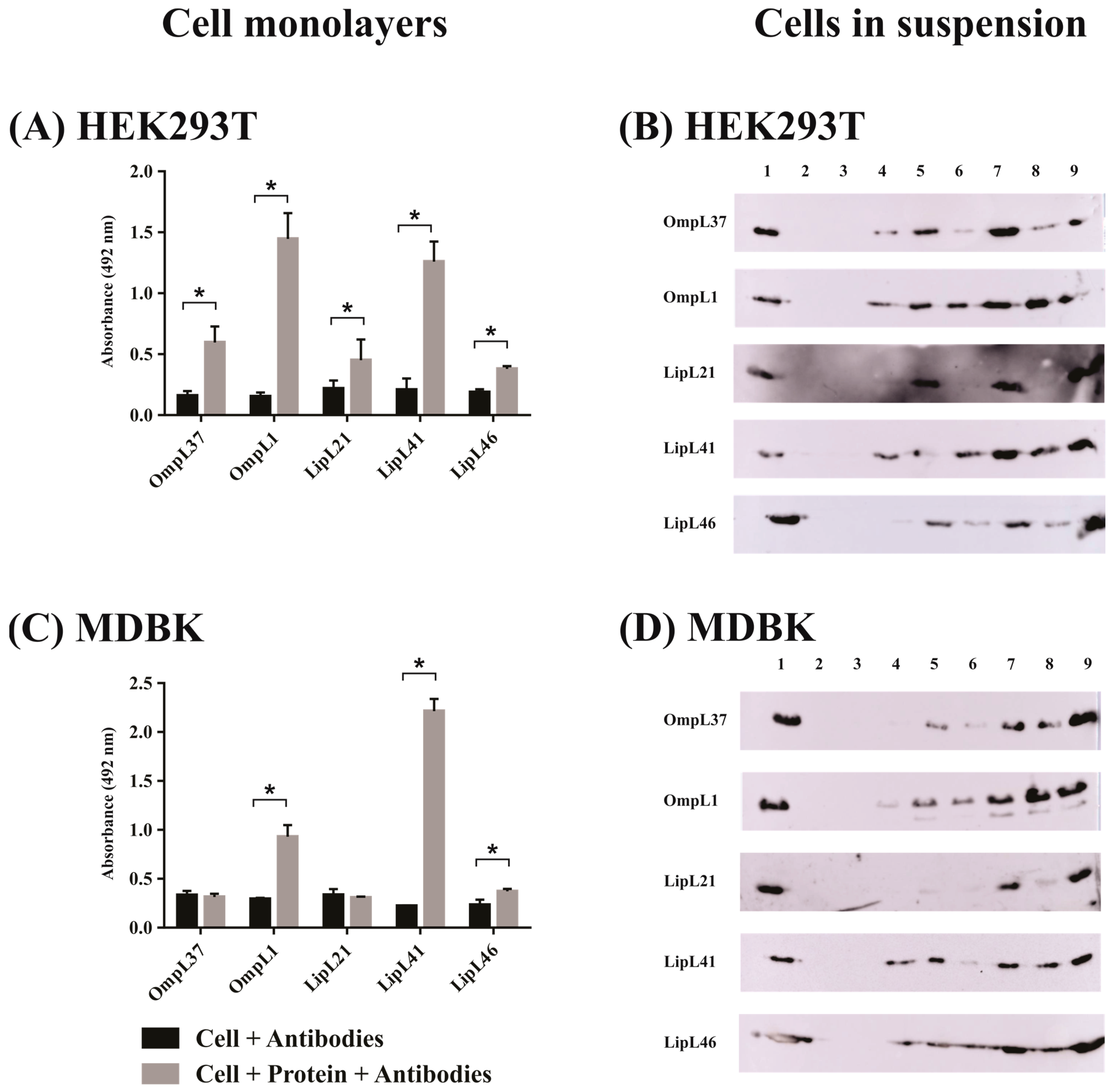

| Protein | Mammalian Endothelial Cells | ||
|---|---|---|---|
| EA.hy926 | HMEC-1 | HULEC5a | |
| OmpL1 | 40.8 ± 11.9 | 174.8 ± 136.1 | 55.2 ± 54.5 |
| LipL41 | - | 11.7 ± 6.0 | - |
| LipL46 | - | - | - |
| Protein | Mammalian Epithelial Cells | ||
| HEK293T | MDBK | ||
| OmpL37 | 606.5 ± 223.4 | - | |
| OmpL1 | - | 40.9 ± 19.4 | |
| LipL21 | 28.7 ± 14.5 | - | |
| LipL41 | 280.1 ± 108.8 | 53.2 ± 34.2 | |
| LipL46 | - | 1576 ± 1325 | |
| Protein | Mammalian Fibroblasts | ||
| E. Derm | BHK-21 | ||
| OmpL1 | 20.6 ± 9.9 | - | |
| LipL41 | 3.6 ± 2.6 | - | |
| Protein | Cell Line | |||||
|---|---|---|---|---|---|---|
| HMEC | EA.hy926 | HEK293T | ||||
| (nM) | (μg) | (nM) | (μg) | (nM) | (μg) | |
| OmpL37 | 4070 | 17.5 | 507 | 2.2 | 507 | 2.2 |
| OmpL1 | 793 | 2.5 | 198 | 0.6 | 198 | 0.6 |
| LipL21 | 2750 | 5.0 | 343 | 0.6 | 343 | 0.6 |
| LipL41 | 672 | 2.5 | 1340 | 5.0 | 672 | 2.5 |
| LipL46 | 8140 | 35.0 | 8140 | 35.0 | 8140 | 35.0 |
| Host Component | Molecular Weight | Concentration (nM) | ||
|---|---|---|---|---|
| (kDa) | 1 μg | 5 μg | 10 μg | |
| Collagen I | 695 | 14.4 | 71.9 | 143.8 |
| Collagen IV | 492.5 | 20.3 | 101.5 | 203.1 |
| Laminin | 810 | 12.4 | 61.7 | 123.5 |
| Cellular fibronectin | 550 | 18.2 | 90.9 | 181.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, M.B.; Teixeira, A.F.; Nascimento, A.L.T.O. Host Cell Binding Mediated by Leptospira interrogans Adhesins. Int. J. Mol. Sci. 2022, 23, 15550. https://doi.org/10.3390/ijms232415550
Takahashi MB, Teixeira AF, Nascimento ALTO. Host Cell Binding Mediated by Leptospira interrogans Adhesins. International Journal of Molecular Sciences. 2022; 23(24):15550. https://doi.org/10.3390/ijms232415550
Chicago/Turabian StyleTakahashi, Maria Beatriz, Aline Florencio Teixeira, and Ana Lucia Tabet Oller Nascimento. 2022. "Host Cell Binding Mediated by Leptospira interrogans Adhesins" International Journal of Molecular Sciences 23, no. 24: 15550. https://doi.org/10.3390/ijms232415550
APA StyleTakahashi, M. B., Teixeira, A. F., & Nascimento, A. L. T. O. (2022). Host Cell Binding Mediated by Leptospira interrogans Adhesins. International Journal of Molecular Sciences, 23(24), 15550. https://doi.org/10.3390/ijms232415550








