A Protective Role of Phenylalanine Ammonia-Lyase from Astragalus membranaceus against Saline-Alkali Stress
Abstract
:1. Introduction
2. Results
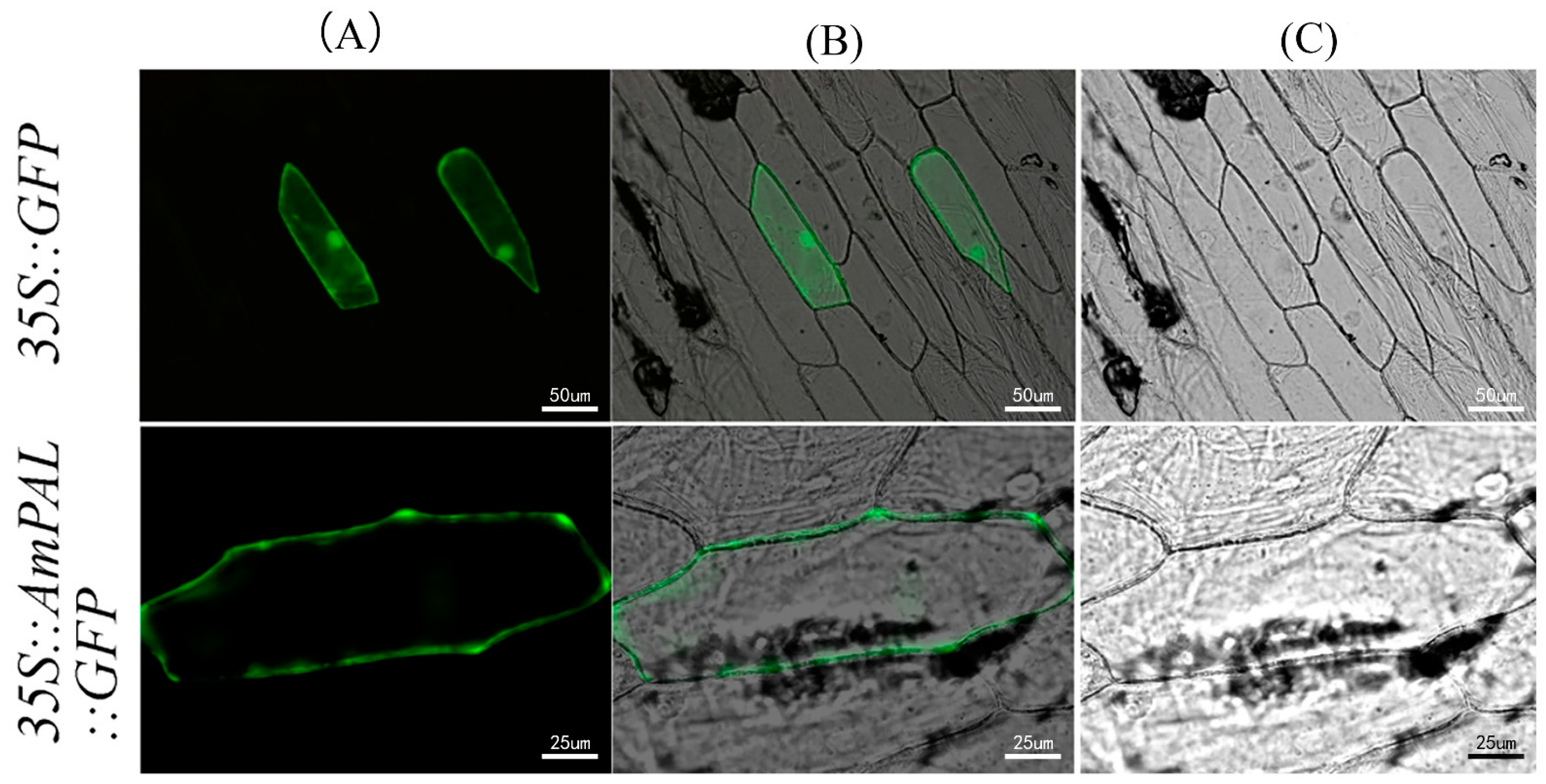
2.1. Bioinformatics and Subcellular Localization of AmPAL
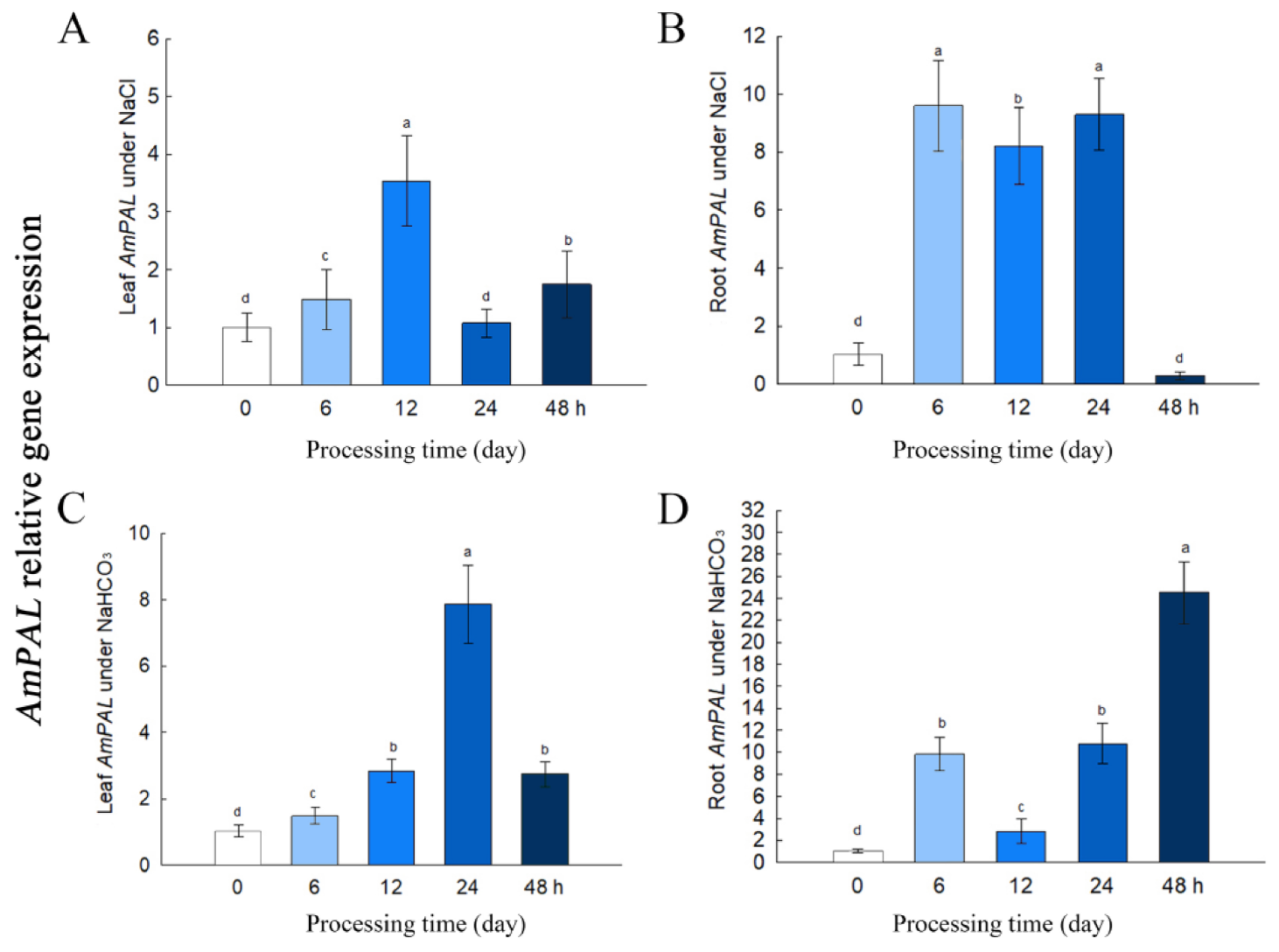
2.2. Expression of AmPAL Is Induced by Saline-Alkaline Stress
2.3. Overexpressing of AmPAL in N. tabacum
2.4. Impact on Root Growth under Saline-Alkali Stress
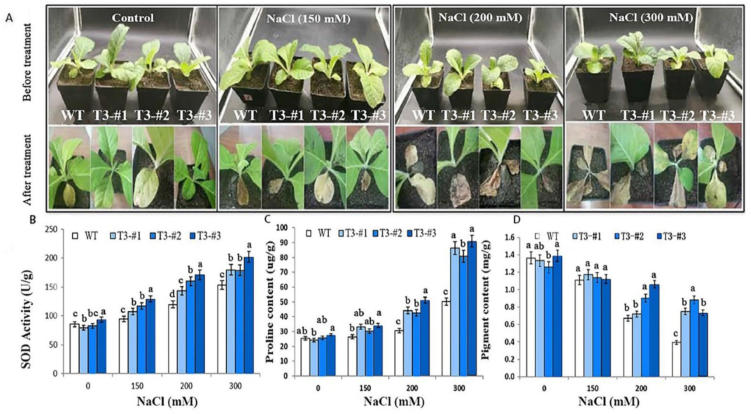
2.5. Enhanced Salt Tolerance in Soil-Grown Plants by Overexpressing AmPAL
2.6. Enhanced Alkali Tolerance in Soil-Grown Plants by Overexpressing AmPAL
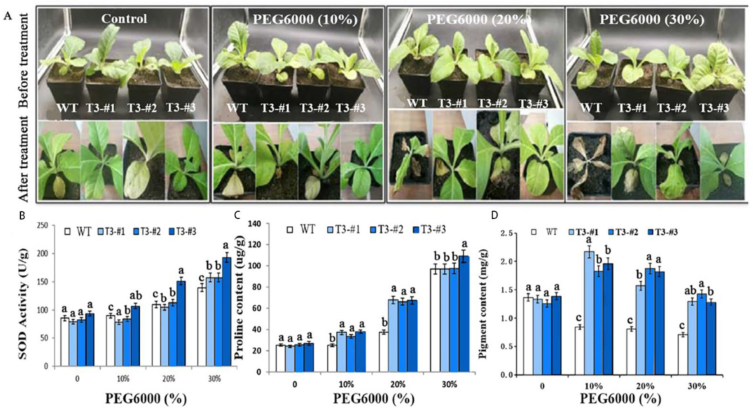
2.7. Enhanced Drought Tolerance in Soil-Grown Plants by Overexpressing AmPAL
3. Discussions
4. Conclusions
5. Materials and Methods
5.1. Plant Materials and Reagents
5.2. Bioinformatics
5.3. Subcellular Localization
5.4. Gene Expression Analysis
5.5. Vector Construction and Plant Transformation
5.6. Identification of Transgenic Plants
5.7. Root Growth, PAL Activity, and MDA of Seedlings
5.8. Abiotic Stress in Soil-Grown Plants
5.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koukol, J.; Conn, E.E. The Metabolism of Aromatic Compounds in Higher Plants: IV. purification and properties of the phenylalanine deaminase of hordeum vulgare. J. Biol. Chem. 1961, 236, 2692–2698. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraser, C.M.; Chapple, C. The phenylpropanoid pathway in Arabidopsis. Arab. Book 2011, 9, e152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauch-Mani, B.; Slusarenko, A. Production of Salicylic Acid Precursors Is a Major Function of Phenylalanine Ammonia-Lyase in the Resistance of Arabidopsis to Peronospora parasitica. Plant Cell 1996, 8, 203–212. [Google Scholar] [CrossRef]
- Minami, E.; Ozeki, Y.; Matsuoka, M.; Koizuka, N.; Tanaka, Y. Structure and some characterization of the gene for phenylalanine ammonia-lyase from rice plants. Eur. J. Biochem. 1989, 185, 19–25. [Google Scholar] [CrossRef]
- Fukasawa-Akada, T.; Kung, S.D.; Watson, J.C. Phenylalanine ammonia-lyase gene structure, expression, and evolution in Nicotiana. Plant Mol. Biol. 1996, 30, 711–722. [Google Scholar] [CrossRef]
- Cochrane, F.C.; Davin, L.B.; Lewis, N.G. The Arabidopsis phenylalanine ammonia lyase gene family: Kinetic characterization of the four PAL isoforms. Phytochemistry 2004, 65, 1557–1564. [Google Scholar] [CrossRef]
- Engelsma, G. On the Mechanism of the Changes in Phenylalanine Ammonia-lyase Activity Induced by Ultraviolet and Blue Light in Gherkin Hypocotyls. Plant Physiol. 1974, 54, 702–705. [Google Scholar] [CrossRef] [Green Version]
- Loschke, D.C.; Hadwiger, L.A. Effects of Light and of Fusarium solani on Synthesis and Activity of Phenylalanine Ammonia-Lyase in Peas. Plant Physiol. 1981, 68, 680–685. [Google Scholar] [CrossRef] [Green Version]
- Smith, B.G.; Rubery, P.H. Modifications of wound-induced changes in phenylalanine ammonia-lyase activity in potato tuber tissue. Plant Sci. Lett. 1979, 15, 29–33. [Google Scholar] [CrossRef]
- Brown, G.E. Changes in Phenylalanine Ammonia-Lyase, Soluble Phenolics and Lignin in Injured Orange Exocarp. Proc. Fla. State Hort. Soc. 2015, 103, 234–237. [Google Scholar]
- Yuan, L.; Yanqun, Z.; Jianjun, C.; Haiyan, C.; Jilong, Y.; Zhide, H. Intraspecific differences in physiological response of 20 wheat cultivars to enhanced ultraviolet-B radiation under field conditions. Environ. Exp. Bot. 2000, 44, 95–103. [Google Scholar] [CrossRef]
- Zhao, Z.; Qiu, Z.; Chen, X.; Huang, D.; Zhu, D.; Qiu, Y.; Zheng, S. Study on the activity of phenylalanine ammonia-lyase and flavonoid content in Houlijuan climbing type and dwarf type Tetranyl. Chin. J. Ecol. Environ. 2011, 20, 1670–1674. [Google Scholar] [CrossRef]
- He, J.; Liu, Y.; Yuan, D.; Duan, M.; Liu, Y.; Shen, Z.; Yang, C.; Qiu, Z.; Liu, D.; Wen, P.; et al. An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc. Natl. Acad. Sci. USA 2020, 117, 271–277. [Google Scholar] [CrossRef]
- Fu, J.; Xiao, Y.; Wang, Y.F.; Liu, Z.H.; Yang, K. Saline-alkaline stress in growing maize seedlings is alleviated by Trichoderma asperellum through regulation of the soil environment. Sci. Rep. 2021, 11, 11152. [Google Scholar] [CrossRef]
- Jia, X.; Sun, C.; Zuo, Y.; Li, G.; Li, G.; Ren, L.; Chen, G. Integrating transcriptomics and metabolomics to characterise the response of Astragalus membranaceus Bge. var. mongolicus (Bge.) to progressive drought stress. BMC Genom. 2016, 17, 188. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Sun, Y.; Huang, J.; Wang, B.; Gong, Y.; Fang, Y.; Liu, Y.; Wang, S.; Guo, Y.; Wang, H.; et al. Anti-tumor effects and mechanisms of Astragalus membranaceus (AM) and its specific immunopotentiation: Status and prospect. J. Ethnopharmacol. 2020, 258, 112797. [Google Scholar] [CrossRef]
- Abla, M.; Sun, H.; Li, Z.; Wei, C.; Gao, F.; Zhou, Y.; Feng, J. Identification of miRNAs and Their Response to Cold Stress in Astragalus Membranaceus. Biomolecules 2019, 9, 182. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Luan, Y.; Liu, Z. Overexpression ofSpWRKY1 promotes resistance toPhytophthora nicotianae and tolerance to salt and drought stress in transgenic tobacco. Physiol. Plant. 2015, 155, 248–266. [Google Scholar] [CrossRef]
- Jones, D.H. Phenylalanine ammonia-lyase: Regulation of its induction, and its role in plant development. Phytochemistry 1984, 23, 1349–1359. [Google Scholar] [CrossRef]
- Hanson, K.R.; Havir, E.A. L-phenylalanine ammonia-lyase. IV. Evidence that the prosthetic group contains a dehydroalanyl residue and mechanism of action. Arch. Biochem. Biophys. 1970, 141, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.W.; Dron, M.; Cramer, C.L.; Dixon, R.A.; Lamb, C.J. Differential regulation of phenylalanine ammonia-lyase genes during plant development and by environmental cues. J. Biol. Chem. 1989, 264, 14486–14492. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wang, Z. RNAi-mediated suppression of the phenylalanine ammonia-lyase gene in Salvia miltiorrhiza causes abnormal phenotypes and a reduction in rosmarinic acid biosynthesis. J. Plant Res. 2011, 124, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Chaoui, A.; Mazhoudi, S.; Ghorbal, M.H.; El Ferjani, E. Cadmium and zinc induction of lipid peroxidation and effects on antioxidant enzyme activities in bean (Phaseolus vulgaris L.). Plant Sci. 1997, 127, 139–147. [Google Scholar] [CrossRef]
- Chen, J.Y.; He, L.H.; Jiang, Y.M.; Wang, Y.; Joyce, D.C.; Ji, Z.L.; Lu, W.J. Role of phenylalanine ammonia-lyase in heat pretreatment-induced chilling tolerance in banana fruit. Physiol. Plant. 2008, 132, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, X.; Zhang, F.; Dong, L.; Wu, J.; Cheng, Q.; Qi, D.; Yan, X.; Jiang, L.; Fan, S.; et al. Phenylalanine ammonia-lyase2.1 contributes to the soybean response towards Phytophthora sojae infection. Sci. Rep. 2017, 7, 7242. [Google Scholar] [CrossRef] [Green Version]
- Cass, C.L.; Peraldi, A.; Dowd, P.F.; Mottiar, Y.; Santoro, N.; Karlen, S.D.; Bukhman, Y.V.; Foster, C.E.; Thrower, N.; Bruno, L.C.; et al. Effects of PHENYLALANINE AMMONIA LYASE (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J. Exp. Bot. 2015, 66, 4317–4335. [Google Scholar] [CrossRef] [Green Version]
- Gho, Y.S.; Kim, S.J.; Jung, K.H. Phenylalanine ammonia-lyase family is closely associated with response to phosphate deficiency in rice. Genes Genom. 2020, 42, 67–76. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Luo, W.; Tanaka, G.; Konishi, Y.; Matsuura, H.; Takahashi, K. Wounding stress induces phenylalanine ammonia lyases, leading to the accumulation of phenylpropanoids in the model liverwort Marchantia polymorpha. Phytochemistry 2018, 155, 30–36. [Google Scholar] [CrossRef]
- Yamada, T.; Sriprasertsak, P.; Kato, H.; Hashimoto, T.; Shimizu, H.; Shiraishi, T. Functional analysis of the promoters of phenylalanine ammonia-lyase genes in pea. Plant Cell Physiol. 1994, 35, 917–926. [Google Scholar]
- Yin, S. Comparison of stress effects of NaCl and Na2CO3 on hydroponic wheat seedlings. J. Zhejiang Wanli Univ. 2002, 15, 59–62. [Google Scholar]
- Yang, Y.H.; Wang, C.J.; Li, R.F.; Zhang, Z.Y.; Yang, H.; Chu, C.Y.; Li, J.T. Overexpression of RgPAL family genes involved in phenolic biosynthesis promotes the replanting disease development in Rehmannia glutinosa. J. Plant Physiol. 2021, 257, 153339. [Google Scholar] [CrossRef]
- Yu, H.; Li, D.; Yang, D.; Xue, Z.; Li, J.; Xing, B.; Yan, K.; Han, R.; Liang, Z. SmKFB5 protein regulates phenolic acid biosynthesis by controlling the degradation of phenylalanine ammonia-lyase in Salvia miltiorrhiza. J. Exp. Bot. 2021, 72, 4915–4929. [Google Scholar] [CrossRef]
- Kim, J.I.; Zhang, X.; Pascuzzi, P.E.; Liu, C.J.; Chapple, C. Glucosinolate and phenylpropanoid biosynthesis are linked by proteasome-dependent degradation of PAL. New Phytol. 2020, 225, 154–168. [Google Scholar] [CrossRef] [Green Version]
- Wei, J.; Liao, S.; Li, M.; Zhu, B.; Wang, H.; Gu, L.; Yin, H.; Du, X. AetSRG1 contributes to the inhibition of wheat Cd accumulation by stabilizing phenylalanine ammonia lyase. J. Hazard. Mater. 2022, 428, 128226. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, C. Multifaceted Regulations of Gateway Enzyme Phenylalanine Ammonia-Lyase in the Biosynthesis of Phenylpropanoids. Mol. Plant. 2015, 8, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Pang, Q.; Zhang, T.; Wang, Y.; Kong, W.; Guan, Q.; Yan, X.; Chen, S. Metabolomics of Early Stage Plant Cell-Microbe Interaction Using Stable Isotope Labeling. Front. Plant Sci. 2018, 9, 760. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Meng, L.; Kong, W.; Yin, Z.; Wang, Y.; Schneider, J.D.; Chen, S. Quantitative proteomics reveals a role of JAZ7 in plant defense response to Pseudomonas syringae DC3000. J. Proteom. 2018, 175, 114–126. [Google Scholar] [CrossRef]
- Meng, L.; Zhang, T.; Geng, S.; Scott, P.B.; Li, H.; Chen, S. Comparative proteomics and metabolomics of JAZ7-mediated drought tolerance in Arabidopsis. J. Proteom. 2019, 196, 81–91. [Google Scholar] [CrossRef]
- Chen, S.; Songkumarn, P.; Liu, J.; Wang, G.L. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 2009, 150, 1111–1121. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Zu, Y.; Guan, Q.; Wu, J. Cloning and Sequence Analysis of a Novel Phenylalanine Ammonia-Lyase Gene from Astragalus Membranaceus. Chin. Herb. Med. 2010, 41, 456–460. [Google Scholar]
- Alinsug, M.V.; Chen, F.F.; Luo, M.; Tai, R.; Jiang, L.; Wu, K. Subcellular localization of class II HDAs in Arabidopsis thaliana: Nucleocytoplasmic shuttling of HDA15 is driven by light. PLoS ONE 2012, 7, e30846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Jing, R.; Mao, X.; Chang, X.; Li, A. TaABC1, a member of the activity of bc1 complex protein kinase family from common wheat, confers enhanced tolerance to abiotic stresses in Arabidopsis. J. Exp. Bot. 2011, 62, 1299–1311. [Google Scholar] [CrossRef] [PubMed]
- Gallois, P.; Marinho, P. Leaf disk transformation using Agrobacterium tumefaciens-expression of heterologous genes in tobacco. Methods Mol. Biol. 1995, 49, 39–48. [Google Scholar] [CrossRef]
- Howland, J.L. Short Protocols in Molecular Biology, 3rd ed.; Ausubel, F., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K., Eds.; John Wiley & Sons: New York, NY, USA, 1995; p. 836. ISBN 0-471-13781-2. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; Volume 6, pp. 4–6. [Google Scholar]
- Hodges, D.M.; DeLong, J.M.; Forney, C.F.; Prange, R.K. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 1999, 207, 604–611. [Google Scholar] [CrossRef]
- Abraham, E.; Hourton-Cabassa, C.; Erdei, L.; Szabados, L. Methods for determination of proline in plants. Methods Mol. Biol. 2010, 639, 317–331. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Kim, D.; Paek, N.C. Salt Treatments and Induction of Senescence. Methods Mol. Biol. 2018, 1744, 141–149. [Google Scholar] [CrossRef]
- Yao, J.; Sun, D.; Cen, H.; Xu, H.; Weng, H.; Yuan, F.; He, Y. Phenotyping of Arabidopsis Drought Stress Response Using Kinetic Chlorophyll Fluorescence and Multicolor Fluorescence Imaging. Front. Plant Sci. 2018, 9, 603. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, L.; Shi, G.; Yang, J.; Liu, G.; Niu, Z.; Ye, W.; Wu, S.; Wang, L.; Guan, Q. A Protective Role of Phenylalanine Ammonia-Lyase from Astragalus membranaceus against Saline-Alkali Stress. Int. J. Mol. Sci. 2022, 23, 15686. https://doi.org/10.3390/ijms232415686
Fan L, Shi G, Yang J, Liu G, Niu Z, Ye W, Wu S, Wang L, Guan Q. A Protective Role of Phenylalanine Ammonia-Lyase from Astragalus membranaceus against Saline-Alkali Stress. International Journal of Molecular Sciences. 2022; 23(24):15686. https://doi.org/10.3390/ijms232415686
Chicago/Turabian StyleFan, Lijuan, Gongfa Shi, Juan Yang, Guiling Liu, Zhaoqian Niu, Wangbin Ye, Songquan Wu, Ling Wang, and Qingjie Guan. 2022. "A Protective Role of Phenylalanine Ammonia-Lyase from Astragalus membranaceus against Saline-Alkali Stress" International Journal of Molecular Sciences 23, no. 24: 15686. https://doi.org/10.3390/ijms232415686
APA StyleFan, L., Shi, G., Yang, J., Liu, G., Niu, Z., Ye, W., Wu, S., Wang, L., & Guan, Q. (2022). A Protective Role of Phenylalanine Ammonia-Lyase from Astragalus membranaceus against Saline-Alkali Stress. International Journal of Molecular Sciences, 23(24), 15686. https://doi.org/10.3390/ijms232415686






