Screening Key Genes and Biological Pathways in Nasopharyngeal Carcinoma by Integrated Bioinformatics Analysis
Abstract
:1. Introduction
2. Results
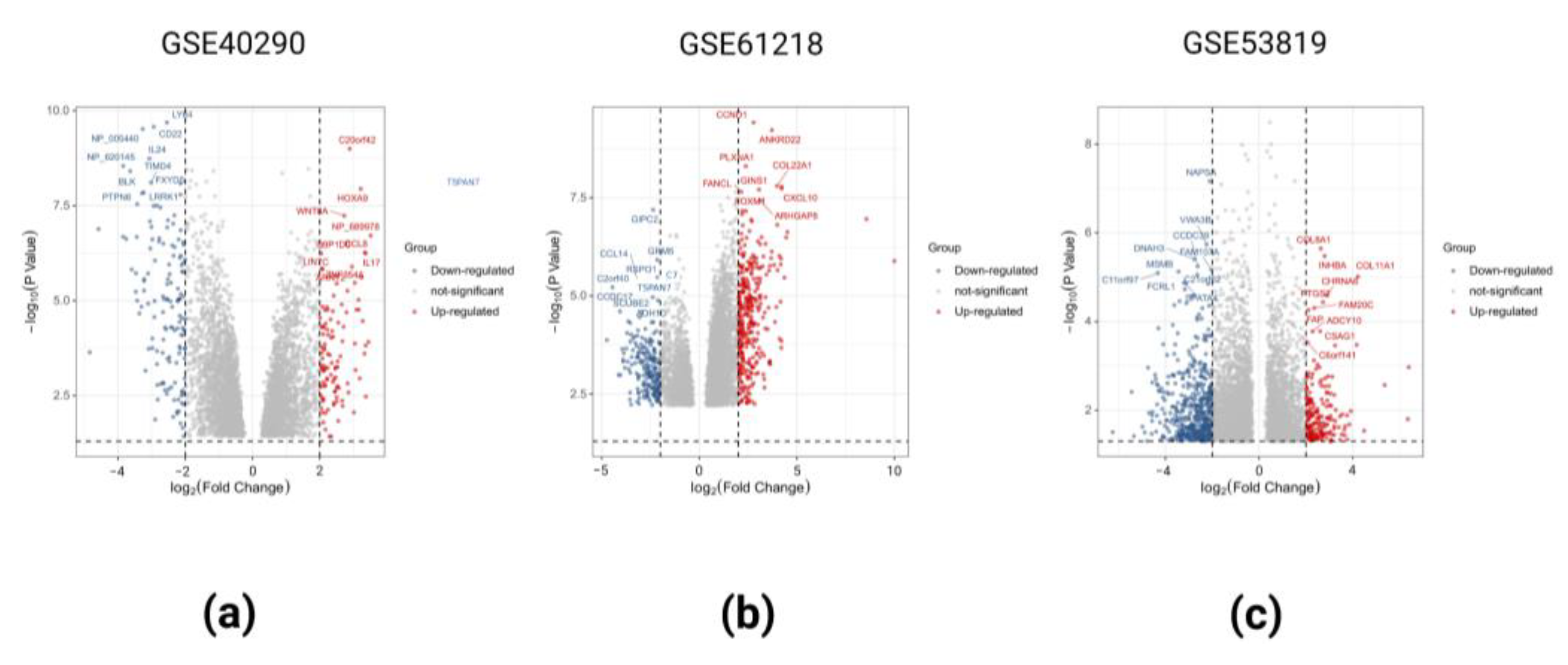
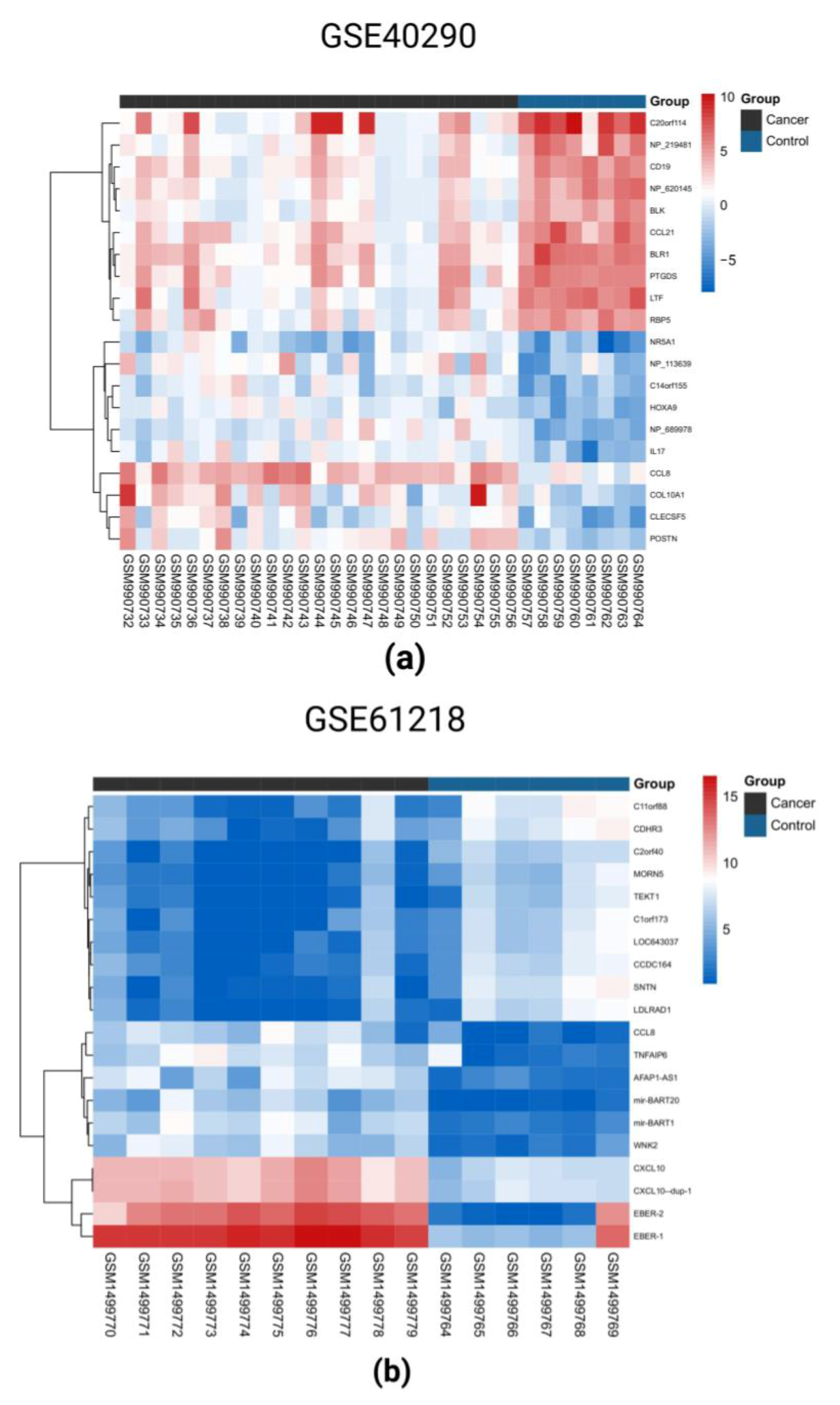
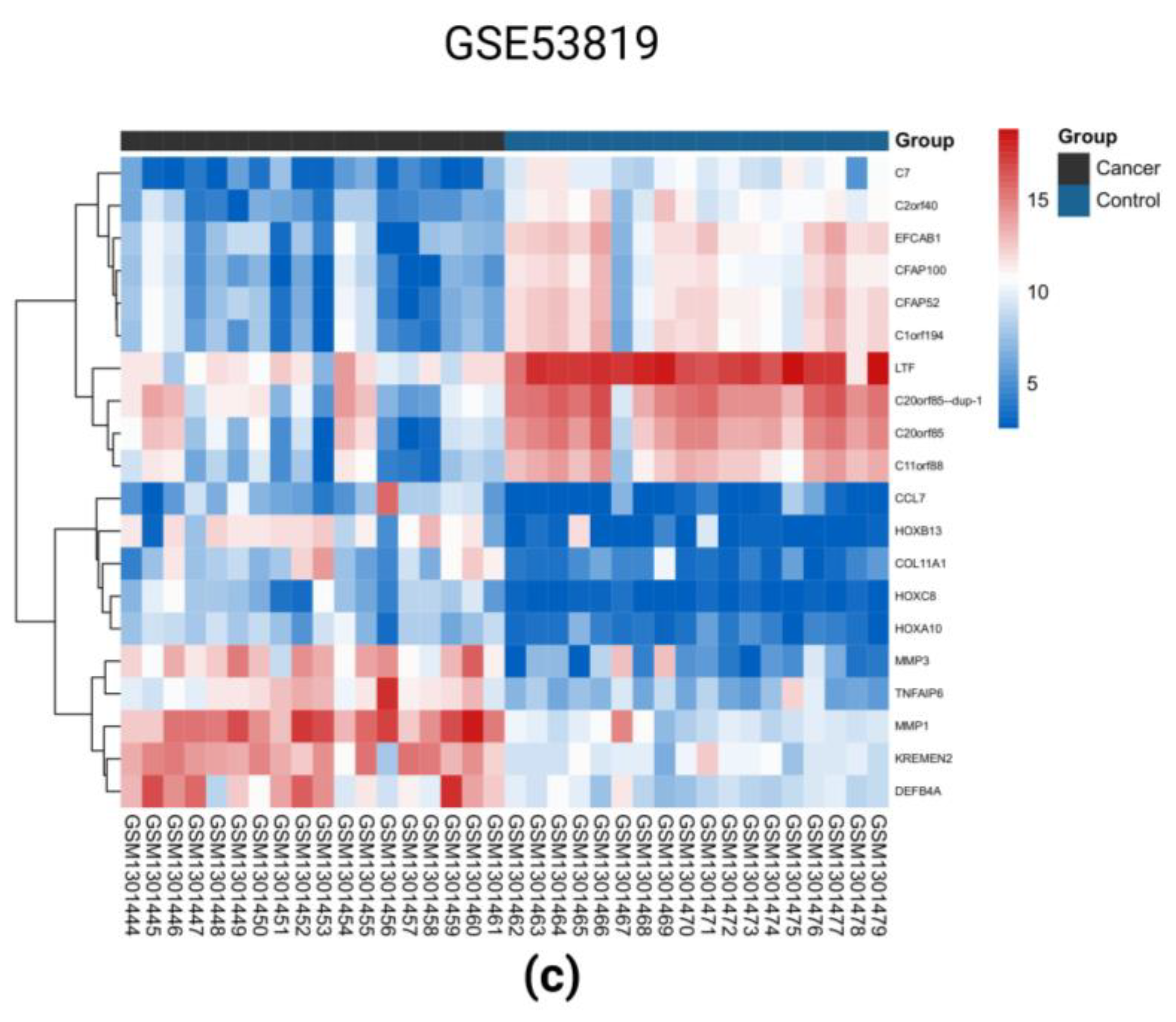
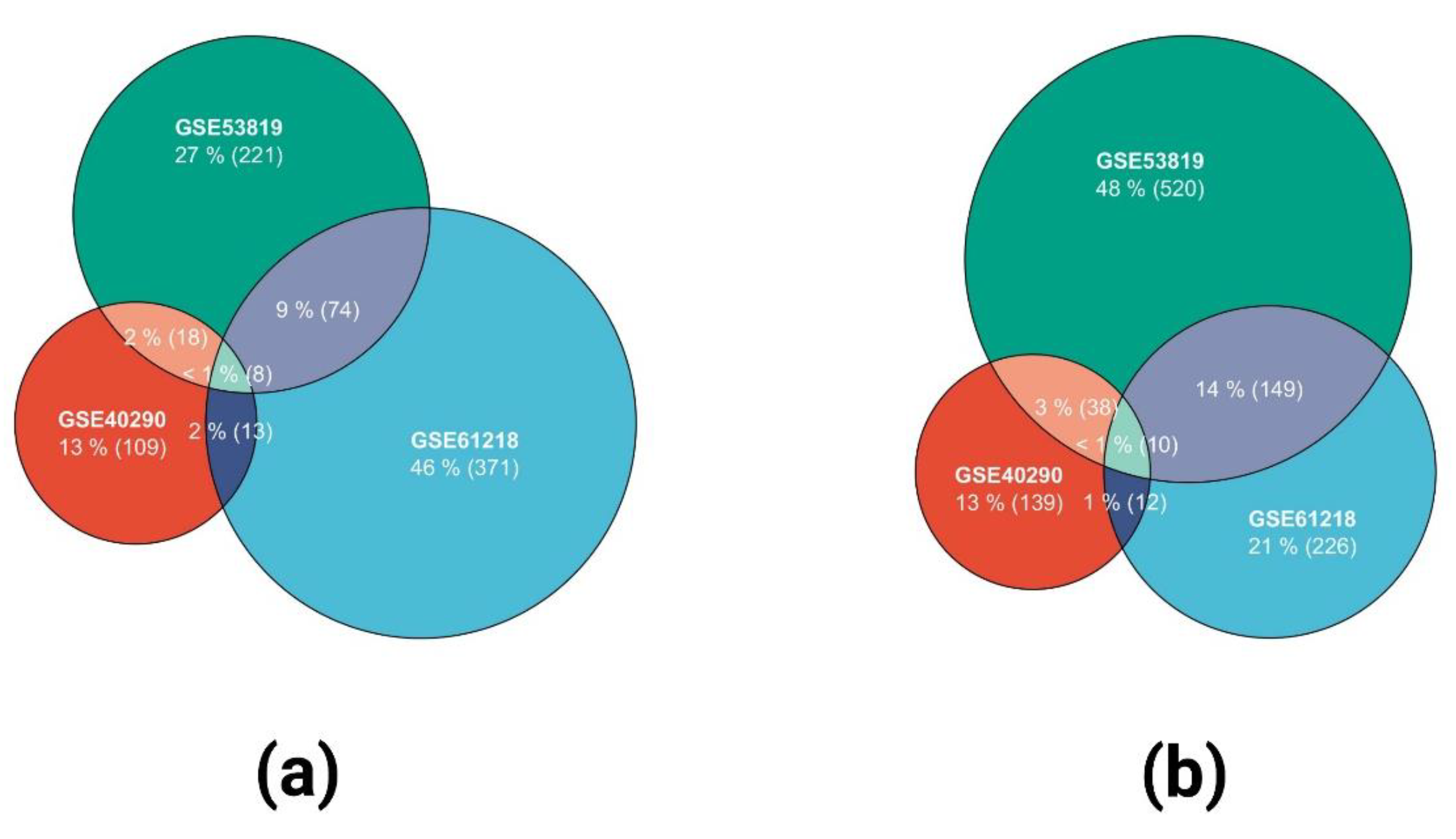
2.1. Identification of DEGs in the Three GEO Data Sets
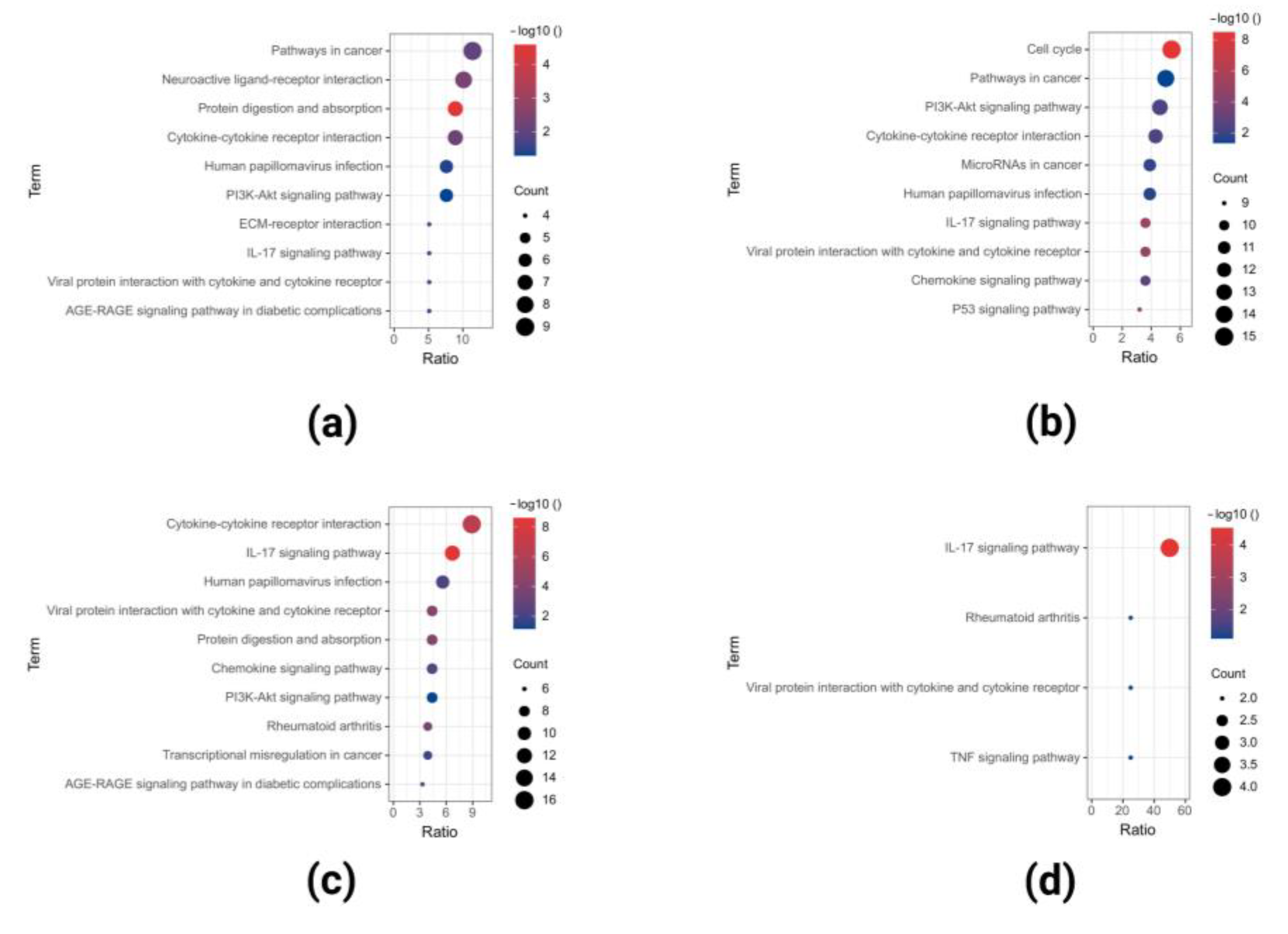
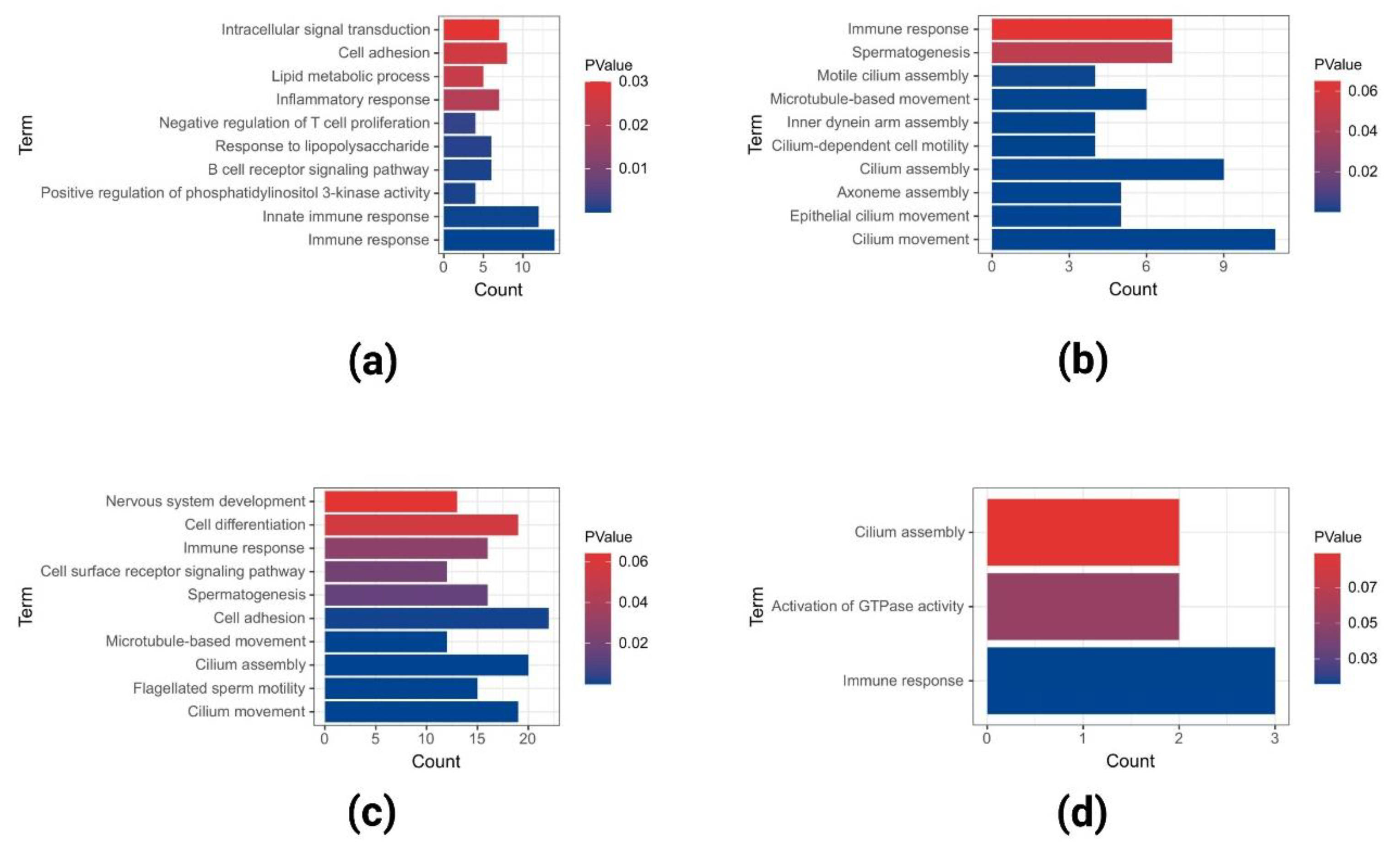
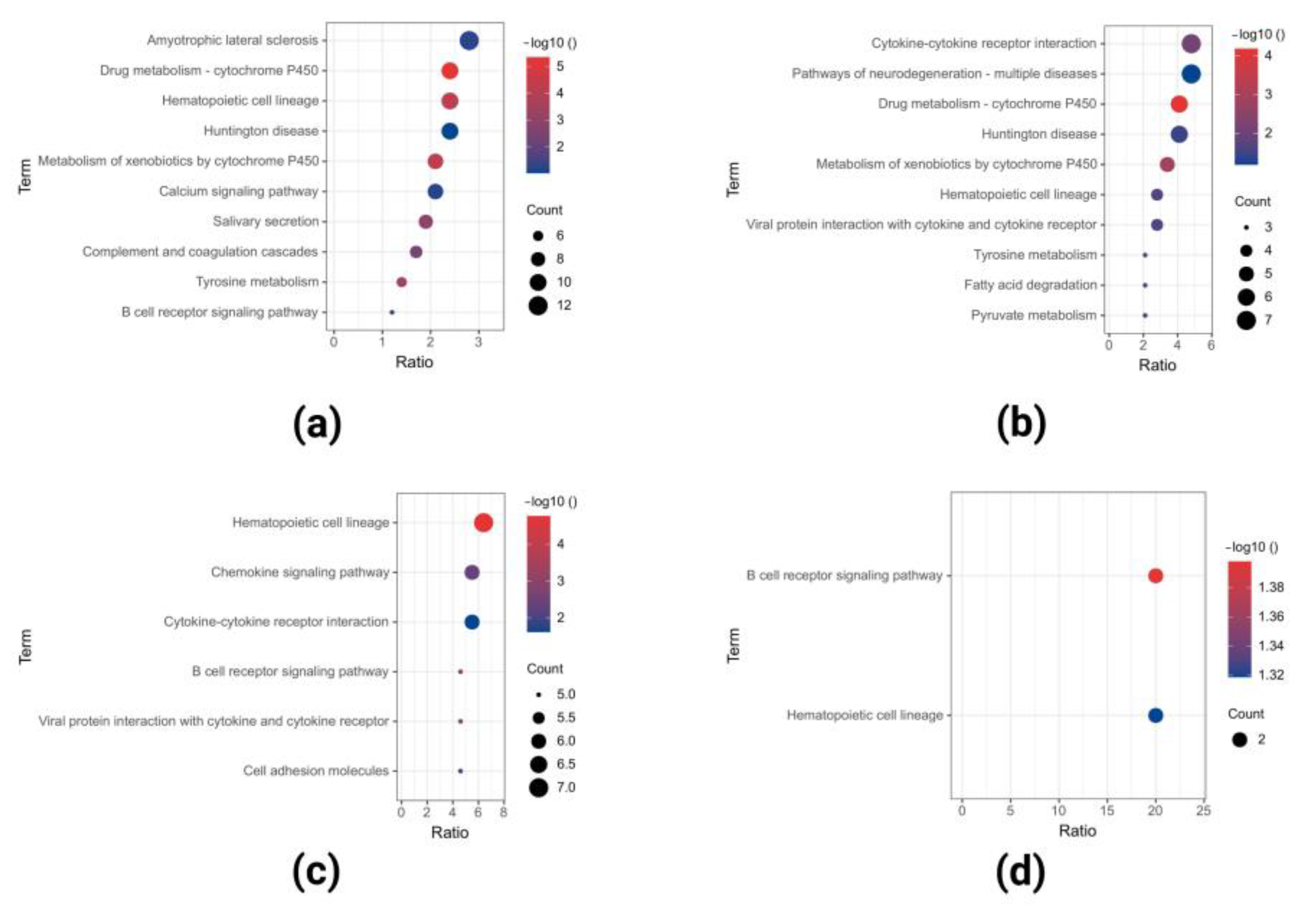
2.2. GO and KEGG Pathway Enrichment Analysis of Upregulated DEGs
2.3. GO and KEGG Pathway Enrichment Analyses of Downregulated DEGs
2.4. PPI Network Construction of Co-DEGs
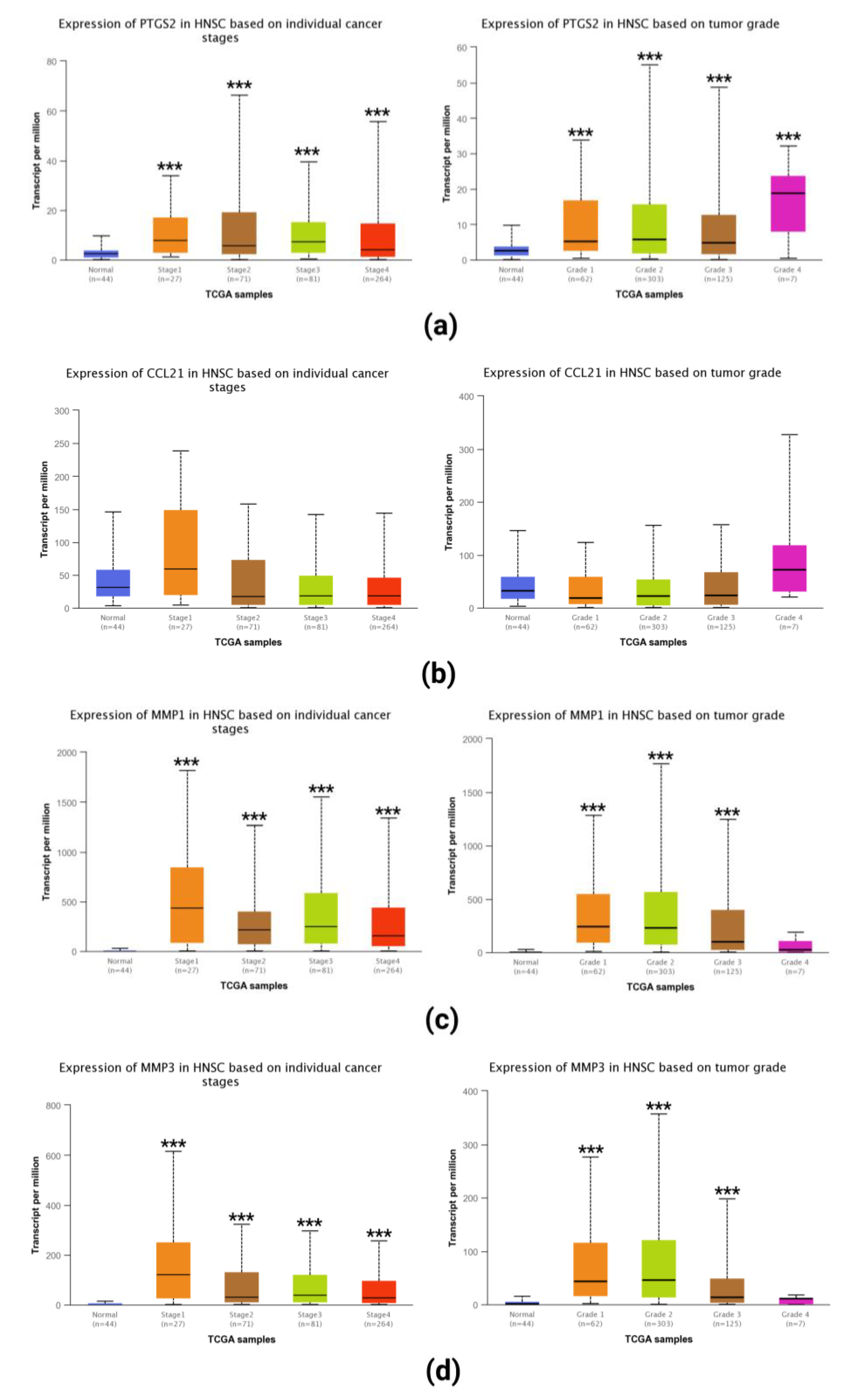
2.5. Expression of Selected Hub Genes in Tumor Tissues
3. Discussion
4. Materials and Methods
4.1. Microarray Data
4.2. Identification of DEGs and Data Visualization
4.3. GO and KEGG Pathway Enrichment Analysis of Up- and Downregulated DEGs
4.4. PPI Network Construction of Up- and Downregulated DEGs
4.5. Analyzing the Expression of Hub Genes in Tumor
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.P.; Chan, A.T.C.; Le, Q.T.; Blanchard, P.; Sun, Y.; Ma, J. Nasopharyngeal carcinoma. Lancet 2019, 394, 64–80. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.M.; Okuda, K.S.; González, F.E.; Patel, V. Current Perspectives on Nasopharyngeal Carcinoma. Adv. Exp. Med. Biol. 2019, 1164, 11–34. [Google Scholar] [CrossRef] [PubMed]
- Chua, M.L.K.; Wee, J.T.S.; Hui, E.P.; Chan, A.T.C. Nasopharyngeal carcinoma. Lancet 2016, 387, 1012–1024. [Google Scholar] [CrossRef]
- Chua, M.L.; Sun, Y.; Supiot, S. Advances in nasopharyngeal carcinoma-"West meets East". Br. J. Radiol. 2019, 92, 20199004. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Wei, J.; Huang, L.; Wu, L. Chemotherapy and chemo-resistance in nasopharyngeal carcinoma. Eur. J. Med. Chem. 2020, 207, 112758. [Google Scholar] [CrossRef]
- Tao, Z.; Shi, A.; Li, R.; Wang, Y.; Wang, X.; Zhao, J. Microarray bioinformatics in cancer—A review. J. Buon. 2017, 22, 838–843. [Google Scholar]
- Song, Y.; Feng, T.; Cao, W.; Yu, H.; Zhang, Z. Identification of Key Genes in Nasopharyngeal Carcinoma Based on Bioinformatics Analysis. Comput. Intell. Neurosci. 2022, 2022, 9022700. [Google Scholar] [CrossRef]
- Yue, H.; Zhu, H.; Luo, D.; Du, Q.; Xie, Y.; Huang, S.; Liu, W. Differentially Expressed Genes in Nasopharyngeal Carcinoma Tissues and Their Correlation with Recurrence and Metastasis of Nasopharyngeal Carcinoma. Comput. Math. Methods Med. 2022, 2022, 1941412. [Google Scholar] [CrossRef]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. Methods Mol. Biol. 2016, 1418, 93–110. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Ling, Z.; Arabnia, H.; Deng, Y. Current trend and development in bioinformatics research. BMC Bioinform. 2020, 21, 538. [Google Scholar] [CrossRef]
- Li, Z.; Tsai, M.H.; Shumilov, A.; Baccianti, F.; Tsao, S.W.; Poirey, R.; Delecluse, H.J. Epstein-Barr virus ncRNA from a nasopharyngeal carcinoma induces an inflammatory response that promotes virus production. Nat. Microbiol. 2019, 4, 2475–2486. [Google Scholar] [CrossRef] [PubMed]
- El Badry, A.A.; El-Fadle, A.A.; El-Balshy, A.L. Tissue inhibitor of matrix metalloproteinase-2 in nasopharyngeal carcinoma. MedGenMed 2007, 9, 3. [Google Scholar] [PubMed]
- Lu, J.; Liu, Q.H.; Wang, F.; Tan, J.J.; Deng, Y.Q.; Peng, X.H.; Liu, X.; Zhang, B.; Xu, X.; Li, X.P. Exosomal miR-9 inhibits angiogenesis by targeting MDK and regulating PDK/AKT pathway in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 147. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Ma, R.; Kang, Z.; Zhang, Y.; Ding, H.; Guo, W.; Gao, Q.; Xu, M. Effect of IL-17A on the migration and invasion of NPC cells and related mechanisms. PLoS ONE 2014, 9, e108060. [Google Scholar] [CrossRef]
- Hasler, F. Recent aspects of Epstein-Barr virus infections. Schweiz. Med. Wochenschr. 1984, 114, 520–523. [Google Scholar] [PubMed]
- Tsang, C.M.; Lui, V.W.Y.; Bruce, J.P.; Pugh, T.J.; Lo, K.W. Translational genomics of nasopharyngeal cancer. Semin. Cancer Biol. 2020, 61, 84–100. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Li, L.; Xiang, Y.Q.; Lung, M.L.; Zeng, T.; Lu, J.; Tsao, S.W.; Zeng, M.S.; Yun, J.P.; Kwong, D.L.W.; et al. Epstein-Barr Virus miRNA BART2-5p Promotes Metastasis of Nasopharyngeal Carcinoma by Suppressing RND3. Cancer Res. 2020, 80, 1957–1969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, Y.M.; Tan, L.P.; Abdul Aziz, N.; Mushiroda, T.; Kubo, M.; Mohd Kornain, N.K.; Tan, G.W.; Khoo, A.S.; Krishnan, G.; Pua, K.C.; et al. Integrated pathway analysis of nasopharyngeal carcinoma implicates the axonemal dynein complex in the Malaysian cohort. Int. J. Cancer 2016, 139, 1731–1739. [Google Scholar] [CrossRef] [Green Version]
- Morrison, J.A.; Raab-Traub, N. Roles of the ITAM and PY motifs of Epstein-Barr virus latent membrane protein 2A in the inhibition of epithelial cell differentiation and activation of {beta}-catenin signaling. J. Virol. 2005, 79, 2375–2382. [Google Scholar] [CrossRef] [Green Version]
- Han, B.; Yang, X.; Zhang, P.; Zhang, Y.; Tu, Y.; He, Z.; Li, Y.; Yuan, J.; Dong, Y.; Hosseini, D.K.; et al. DNA methylation biomarkers for nasopharyngeal carcinoma. PLoS ONE 2020, 15, e0230524. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.F.; Liou, M.H.; Huang, Y.J.; Chao, P.J.; Ting, H.M.; Lee, H.Y.; Fang, F.M. LASSO NTCP predictors for the incidence of xerostomia in patients with head and neck squamous cell carcinoma and nasopharyngeal carcinoma. Sci. Rep. 2014, 4, 6217. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.Z.; Aljabban, J.; Silverman, D.; McDermott, S.; Wanner, R.A.; Rohr, M.; Hadley, D.; Panahiazar, M. Meta-Analysis illustrates possible role of lipopolysaccharide (LPS)-induced tissue injury in nasopharyngeal carcinoma (NPC) pathogenesis. PLoS ONE 2021, 16, e0258187. [Google Scholar] [CrossRef] [PubMed]
- Sim, C.C.; Sim, E.U. Over-expression of cyclo-oxygenase-2 predicts poor survival of patients with nasopharyngeal carcinoma: A meta-analysis. J. Laryngol. Otol. 2020, 134, 338–343. [Google Scholar] [CrossRef]
- Zhu, Y.; Shi, C.; Zeng, L.; Liu, G.; Jiang, W.; Zhang, X.; Chen, S.; Guo, J.; Jian, X.; Ouyang, J.; et al. High COX-2 expression in cancer-associated fibiroblasts contributes to poor survival and promotes migration and invasiveness in nasopharyngeal carcinoma. Mol. Carcinog. 2020, 59, 265–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, N.; Wang, X.Q. Correlation between COX-2 gene polymorphism and susceptibility to nasopharyngeal carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5770–5778. [Google Scholar] [CrossRef] [PubMed]
- Ben Nasr, H.; Chahed, K.; Remadi, S.; Zakhama, A.; Chouchane, L. Expression and clinical significance of latent membrane protein-1, matrix metalloproteinase-1 and Ets-1 transcription factor in tunisian nasopharyngeal carcinoma patients. Arch. Med. Res. 2009, 40, 196–203. [Google Scholar] [CrossRef]
- Tsai, C.W.; Chang, W.S.; Gong, C.L.; Shih, L.C.; Chen, L.Y.; Lin, E.Y.; Li, H.T.; Yen, S.T.; Wu, C.N.; Bau, D.T. Contribution of Matrix Metallopeptidase-1 Genotypes, Smoking, Alcohol Drinking and Areca Chewing to Nasopharyngeal Carcinoma Susceptibility. Anticancer Res. 2016, 36, 3335–3340. [Google Scholar]
- Song, L.; Liu, H.; Liu, Q. Matrix metalloproteinase 1 promotes tumorigenesis and inhibits the sensitivity to 5-fluorouracil of nasopharyngeal carcinoma. Biomed. Pharmacother. 2019, 118, 109120. [Google Scholar] [CrossRef]
- Li, Y.; Feng, Z.; Xing, S.; Liu, W.; Zhang, G. Combination of serum matrix metalloproteinase-3 activity and EBV antibodies improves the diagnostic performance of nasopharyngeal carcinoma. J. Cancer 2020, 11, 6009–6018. [Google Scholar] [CrossRef]
- Lan, Y.Y.; Yeh, T.H.; Lin, W.H.; Wu, S.Y.; Lai, H.C.; Chang, F.H.; Takada, K.; Chang, Y. Epstein-Barr virus Zta upregulates matrix metalloproteinases 3 and 9 that synergistically promote cell invasion in vitro. PLoS ONE 2013, 8, e56121. [Google Scholar] [CrossRef] [Green Version]
- Chatziantoniou, A.; Zaravinos, A. Signatures of Co-Deregulated Genes and Their Transcriptional Regulators in Lung Cancer. Int. J. Mol. Sci. 2022, 23, 10933. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Miao, B.; Wang, S.; Dong, W.; Xu, H.; Si, C.; Wang, W.; Duan, S.; Lou, J.; Bao, Z.; et al. Hiplot: A comprehensive and easy-to-use web service for boosting publication-ready biomedical data visualization. Brief. Bioinform. 2022, 23, bbac261. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids. Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids. Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tai, J.; Park, J.; Han, M.; Kim, T.H. Screening Key Genes and Biological Pathways in Nasopharyngeal Carcinoma by Integrated Bioinformatics Analysis. Int. J. Mol. Sci. 2022, 23, 15701. https://doi.org/10.3390/ijms232415701
Tai J, Park J, Han M, Kim TH. Screening Key Genes and Biological Pathways in Nasopharyngeal Carcinoma by Integrated Bioinformatics Analysis. International Journal of Molecular Sciences. 2022; 23(24):15701. https://doi.org/10.3390/ijms232415701
Chicago/Turabian StyleTai, Junhu, Jaehyung Park, Munsoo Han, and Tae Hoon Kim. 2022. "Screening Key Genes and Biological Pathways in Nasopharyngeal Carcinoma by Integrated Bioinformatics Analysis" International Journal of Molecular Sciences 23, no. 24: 15701. https://doi.org/10.3390/ijms232415701




