The NF-Y Transcription Factor Family in Watermelon: Re-Characterization, Assembly of ClNF-Y Complexes, Hormone- and Pathogen-Inducible Expression and Putative Functions in Disease Resistance
Abstract
1. Introduction
2. Results
2.1. Identification of the Watermelon ClNF-Y Family
2.2. Structural Features of ClNF-Y Genes and Proteins
2.3. Evolution of the ClNF-Y Family
2.4. Cis-Elements in Promoters of the ClNF-Y Genes
2.5. Subcellular Localization of the ClNF-Y Proteins
2.6. Interactions between the ClNF-Y Subunits and Assembly of the ClNF-Y Complexes
2.7. Expression Changes of ClNF-Y Genes in Response to Defense Hormones and a Fungal Pathogen
2.8. Generation of ClNF-Y-Overexpressing Arabidopsis Lines and the Involvement of ClNF-Y in Growth and Development
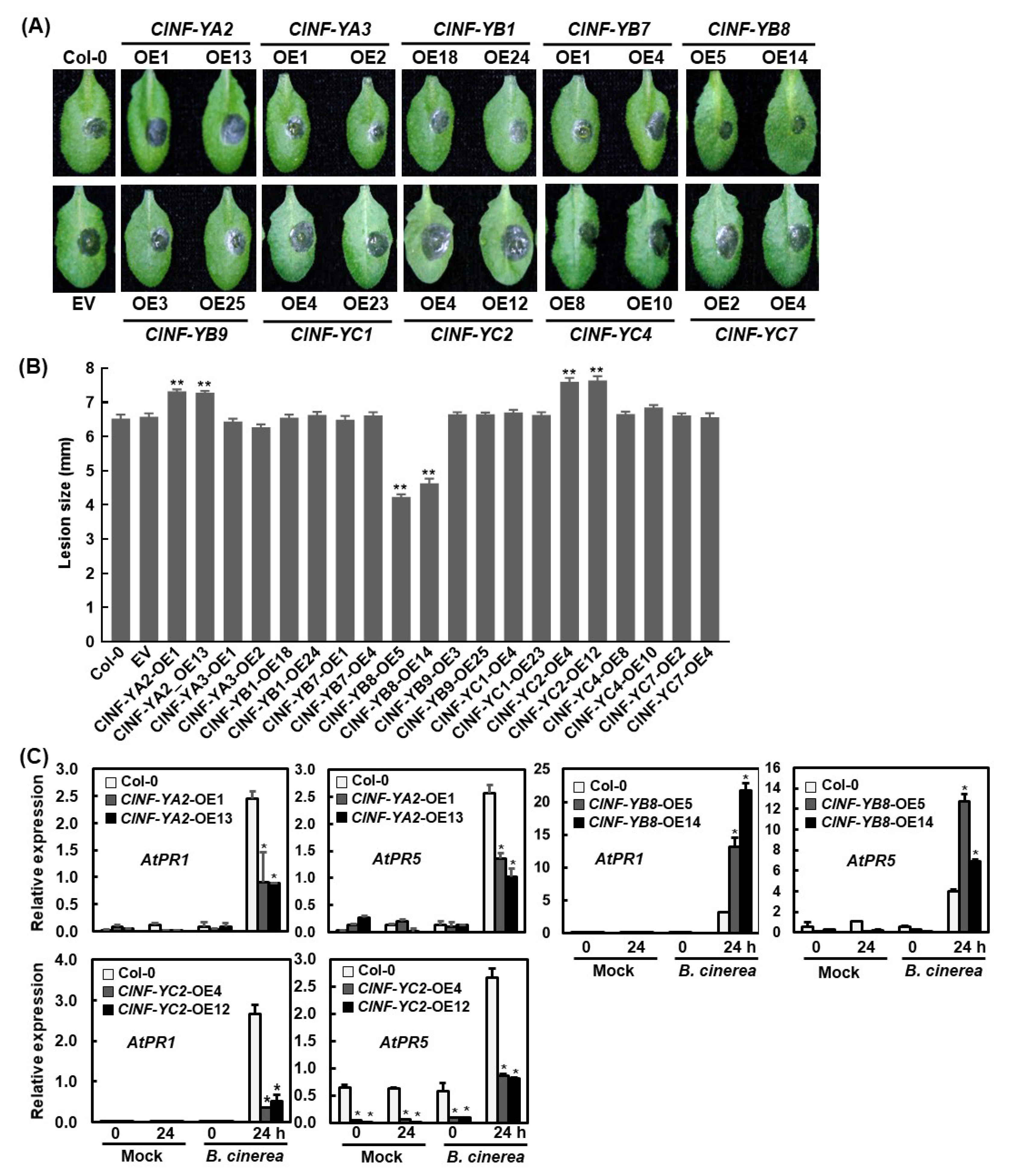
2.9. Functions of the ClNF-Y Genes in Disease Resistance
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Treatments
4.2. Identification and Bioinformatics Analysis of the Watermelon ClNF-Y Family
4.3. Cloning of ClNF-Y Genes
4.4. Subcellular Localization Assays
4.5. BiFC Assays
4.6. Y3H Assays
4.7. RT-qPCR Assays
4.8. Generation and Characterization of ClNF-Y-Overexpressing Transgenic Lines
4.9. Disease Assays
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Birkenbihl, R.P.; Liu, S.; Somssich, I.E. Transcriptional events defining plant immune responses. Curr. Opin. Plant Biol. 2017, 38, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H. Multifaceted chromatin structure and transcription changes in plant stress response. Int. J. Mol. Sci. 2021, 22, 2013. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chang, C. Concerto on chromatin: Interplays of different epigenetic mechanisms in plant development and environmental adaptation. Plants 2021, 10, 2766. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Prado, J.S.; Abulfaraj, A.A.; Rayapuram, N.; Benhamed, M.; Hirt, H. Plant immunity: From signaling to epigenetic control of defense. Trends Plant Sci. 2018, 23, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, M.E.; Rípodas, C.; Niebel, A. Plant NF-Y transcription factors: Key players in plant-microbe interactions, root development and adaptation to stress. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 645–654. [Google Scholar] [CrossRef]
- Myers, Z.A.; Holt, B.F. NUCLEAR FACTOR-Y: Still complex after all these years? Curr. Opin. Plant Biol. 2018, 45, 96–102. [Google Scholar] [CrossRef]
- Laloum, T.; De Mita, S.; Gamas, P.; Baudin, M.; Niebel, A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013, 18, 157–166. [Google Scholar] [CrossRef]
- Nardini, M.; Gnesutta, N.; Donati, G.; Gatta, R.; Forni, C.; Fossati, A.; Vonrhein, C.; Moras, D.; Romier, C.; Bolognesi, M.; et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 2013, 152, 132–143. [Google Scholar] [CrossRef]
- Li, X.Y.; Hooft Van Huijsduijnen, R.; Mantovani, R.; Benoist, C.; Mathis, D. Intron-exon organization of the NF-Y genes. Tissue-specific splicing modifies an activation domain. J. Biol. Chem. 1992, 267, 8984–8990. [Google Scholar] [CrossRef]
- Siefers, N.; Dang, K.K.; Kumimoto, R.W.; Bynum, W.E.; Tayrose, G.; Holt, B.F. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009, 149, 625–641. [Google Scholar] [CrossRef]
- Yang, W.; Lu, Z.; Xiong, Y.; Yao, J. Genome-wide identification and co-expression network analysis of the OsNF-Y gene family in rice. Crop J. 2017, 5, 21–31. [Google Scholar] [CrossRef]
- Chen, L.; Zhou, Y.; Lai, W.; Hu, L.; Jiang, L.; Liu, S. In silico identification and expression analysis of Nuclear Factor Y (NF-Y) transcription factors in cucumber. Agronomy 2020, 10, 236. [Google Scholar] [CrossRef]
- Quach, T.N.; Nguyen, H.T.M.; Valliyodan, B.; Joshi, T.; Xu, D.; Nguyen, H.T. Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. Mol. Genet. Genom. 2015, 290, 1095–1115. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X.; Zhang, C.; Zou, H.; Wu, Z. Isolation, structural analysis, and expression characteristics of the maize nuclear factor Y gene families. Biochem. Biophys. Res. Commun. 2016, 478, 752–758. [Google Scholar] [CrossRef]
- Panahi, B.; Mohammadi, S.A.; Ruzicka, K.; Holaso, H.A.; Mehrjerdi, M.Z. Genome-wide identification and co-expression network analysis of nuclear factor-Y in barley revealed potential functions in salt stress. Physiol. Mol. Biol. Plants 2019, 25, 485–495. [Google Scholar] [CrossRef]
- Li, S.; Li, K.; Ju, Z.; Cao, D.; Fu, D.; Zhu, H.; Zhu, B.; Luo, Y. Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genom. 2016, 17, 36. [Google Scholar] [CrossRef]
- Xuanyuan, G.; Lian, Q.; Jia, R.; Du, M.; Kang, L.; Pu, Y.; Zhang, Z.; Qi, J.; Zhao, J. Genome-wide screening and identification of nuclear factor-Y family genes and exploration their function on regulating abiotic and biotic stress in potato (Solanum tuberosum L.). Gene 2022, 812, 146089. [Google Scholar] [CrossRef]
- Zheng, X.; Zhang, H.; Zhang, L.; Xu, F.; Shi, L.; Wang, S.; Hong, J.; Ding, G. Identification and comprehensive analysis of the Nuclear Factor-Y family genes reveal their multiple roles in response to nutrient deficiencies in Brassica napus. Int. J. Mol. Sci. 2021, 22, 10354. [Google Scholar] [CrossRef]
- Romier, C.; Cocchiarella, F.; Mantovani, R.; Moras, D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003, 278, 1336–1345. [Google Scholar] [CrossRef]
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt, B.F.; Mantovani, R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar] [CrossRef]
- Chaves-Sanjuan, A.; Gnesutta, N.; Gobbini, A.; Martignago, D.; Bernardini, A.; Fornara, F.; Mantovani, R.; Nardini, M. Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants. Plant J. 2021, 105, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Frontini, M.; Imbriano, C.; Manni, I.; Mantovani, R. Cell-cycle regulation of NF-YC nuclear localization. Cell Cycle 2004, 3, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Calvenzani, V.; Testoni, B.; Gusmaroli, G.; Lorenzo, M.; Gnesutta, N.; Petroni, K.; Mantovani, R.; Tonelli, C. Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS ONE 2012, 7, e42902. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, R. The molecular biology of the CCAAT-binding factor NF-Y. Gene 1999, 239, 15–27. [Google Scholar] [CrossRef]
- Hackenberg, D.; Wu, Y.; Voigt, A.; Adams, R.; Schramm, P.; Grimm, B. Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol. Plant 2012, 5, 876–888. [Google Scholar] [CrossRef]
- Swain, S.; Myers, Z.; Chamindika, S.; Holt, B. The multifaceted roles of NUCLEAR FACTOR-Y in Arabidopsis thaliana development and stress responses. Biochim. Biophys. Acta-Gene Regul. Mech. 2016, 1860, 636–644. [Google Scholar] [CrossRef]
- Mantovani, R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998, 26, 1135–1143. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, D.; Kong, F.; Lin, K.; Zhang, H.; Li, G. The Arabidopsis thaliana Nuclear Factor Y transcription factors. Front. Plant Sci. 2017, 7, 2045. [Google Scholar] [CrossRef]
- Liu, X.; Hu, P.; Huang, M.; Tang, Y.; Li, Y.; Li, L.; Hou, X. The NF-YC–RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 2016, 7, 12768. [Google Scholar] [CrossRef]
- Mu, J.; Tan, H.; Hong, S.; Liang, Y.; Zuo, J. Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol. Plant. 2013, 6, 188–201. [Google Scholar] [CrossRef]
- Sorin, C.; Declerck, M.; Christ, A.; Blein, T.; Ma, L.; Lelandais-Brière, C.; Njo, M.F.; Beeckman, T.; Crespi, M.; Hartmann, C. A miR 169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 2014, 202, 1197–1211. [Google Scholar] [CrossRef]
- Soyano, T.; Shimoda, Y.; Kawaguchi, M.; Hayashi, M. A shared gene drives lateral root development and root nodule symbiosis pathways in Lotus. Science 2019, 366, 1021–1023. [Google Scholar] [CrossRef]
- Wenkel, S.; Turck, F.; Singer, K.; Gissot, L.; Le Gourrierec, J.; Samach, A.; Coupland, G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 2006, 18, 2971–2984. [Google Scholar] [CrossRef]
- Cai, X.; Ballif, J.; Endo, S.; Davis, E.; Liang, M.; Chen, D.; DeWald, D.; Kreps, J.; Zhu, T.; Wu, Y. A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol. 2007, 145, 98–105. [Google Scholar] [CrossRef]
- Kumimoto, R.W.; Zhang, Y.; Siefers, N.; Holt, B.F. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 2010, 63, 379–391. [Google Scholar] [CrossRef]
- Cao, S.; Kumimoto, R.W.; Gnesutta, N.; Calogero, A.M.; Mantovani, R.; Holt, B.F. A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 2014, 26, 1009–1017. [Google Scholar] [CrossRef]
- Hou, X.; Zhou, J.; Liu, C.; Liu, L.; Shen, L.; Yu, H. Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 2014, 5, 4601. [Google Scholar] [CrossRef]
- Siriwardana, C.L.; Gnesutta, N.; Kumimoto, R.W.; Jones, D.S.; Myers, Z.A.; Mantovani, R.; Holt, B.F. NUCLEAR FACTOR Y, subunit A (NF-YA) proteins positively regulate flowering and act through FLOWERING LOCUS T. PLoS Genet. 2016, 12, e1006496. [Google Scholar] [CrossRef]
- Shen, C.; Liu, H.; Guan, Z.; Yan, J.; Zheng, T.; Yan, W.; Wu, C.; Zhang, Q.; Yin, P.; Xing, Y. Structural insight into DNA recognition by CCT/NF-YB/YC complexes in plant photoperiodic flowering. Plant Cell 2020, 32, 3469–3484. [Google Scholar] [CrossRef]
- Wang, J.; Li, G.; Li, C.; Zhang, C.; Cui, L.; Ai, G.; Wang, X.; Zheng, F.; Zhang, D.; Larkin, R.M.; et al. NF-Y plays essential roles in flavonoid biosynthesis by modulating histone modifications in tomato. New Phytol. 2021, 229, 3237–3252. [Google Scholar] [CrossRef]
- Nelson, D.E.; Repetti, P.P.; Adams, T.R.; Creelman, R.A.; Wu, J.; Warner, D.C.; Anstrom, D.C.; Bensen, R.J.; Castiglioni, P.P.; Donnarummo, M.G.; et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA 2007, 104, 16450–16455. [Google Scholar] [CrossRef] [PubMed]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.K.; Kim, H.I.; Jang, G.; Chung, P.J.; Jeong, J.S.; Kim, Y.S.; Bang, S.W.; Jung, H.; Choi, Y.D.; Kim, J.K. The NF-YA transcription factor OsNF-YA7 confers drought stress tolerance of rice in an abscisic acid independent manner. Plant Sci. 2015, 241, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.M.; Tanaka, T.; Nakamura, H.; Ichikawa, H.; Kobayashi, K.; Yaeno, T.; Yamaoka, N.; Shimomoto, K.; Takayama, K.; Nishina, H.; et al. Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant Biotechnol. J. 2015, 13, 85–96. [Google Scholar] [CrossRef]
- Su, H.; Cao, Y.; Ku, L.; Yao, W.; Cao, Y.; Ren, Z.; Dou, D.; Wang, H.; Ren, Z.; Liu, H.; et al. Dual functions of ZmNF-YA3 in photoperiod-dependent flowering and abiotic stress responses in maize. J. Exp. Bot. 2018, 69, 5177–5189. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.; Wang, X.; Han, X.; An, Y.; Lin, S.; Shen, C.; Wen, J.; Liu, C.; Yin, W.; et al. Root-specific NF-Y family transcription factor, PdNF-YB21, positively regulates root growth and drought resistance by abscisic acid-mediated indoylacetic acid transport in Populus. New Phytol. 2020, 227, 407–426. [Google Scholar] [CrossRef]
- Yu, T.F.; Liu, Y.; Fu, J.D.; Ma, J.; Fang, Z.W.; Chen, J.; Zheng, L.; Lu, Z.W.; Zhou, Y.B.; Chen, M.; et al. The NF-Y-PYR module integrates the abscisic acid signal pathway to regulate plant stress tolerance. Plant Biotechnol. J. 2021, 19, 2589–2605. [Google Scholar] [CrossRef]
- Zanetti, M.E.; Blanco, F.A.; Beker, M.P.; Battaglia, M.; Aguilar, O.M. A C subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean-Rhizobium etli symbiosis. Plant Cell 2010, 22, 4142–4157. [Google Scholar] [CrossRef]
- Mazziotta, L.; Reynoso, M.A.; Aguilar, O.M.; Blanco, F.A.; Zanetti, M.E. Transcriptional and functional variation of NF-YC1 in genetically diverse accessions of Phaseolus vulgaris during the symbiotic association with Rhizobium etli. Plant Biol. 2013, 15, 808–818. [Google Scholar] [CrossRef]
- Soyano, T.; Kouchi, H.; Hirota, A.; Hayashi, M. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 2013, 9, e1003352. [Google Scholar] [CrossRef]
- Baudin, M.; Laloum, T.; Lepage, A.; Rípodas, C.; Ariel, F.; Frances, L.; Crespi, M.; Gamas, P.; Blanco, F.A.; Zanetti, M.E.; et al. A phylogenetically conserved group of Nuclear Factor-Y transcription factors interact to control nodulation in Legumes. Plant Physiol. 2015, 169, 2761–2773. [Google Scholar]
- Hossain, M.S.; Shrestha, A.; Zhong, S.; Miri, M.; Austin, R.S.; Sato, S.; Ross, L.; Huebert, T.; Tromas, A.; Torres-Jerez, I.; et al. Lotus japonicus NF-YA1 plays an essential role during nodule differentiation and targets members of the SHI/STY gene family. Mol. Plant-Microbe Interact. 2016, 29, 950–964. [Google Scholar] [CrossRef]
- Rípodas, C.; Castaingts, M.; Clúa, J.; Villafañe, J.; Blanco, F.A.; Zanetti, M.E. The PvNF-YA1 and PvNF-YB7 subunits of the heterotrimeric NF-Y transcription factor influence strain preference in the Phaseolus vulgaris-Rhizobium etli symbiosis. Front. Plant Sci. 2019, 10, 221. [Google Scholar] [CrossRef]
- Bu, F.; Rutten, L.; Roswanjaya, Y.P.; Kulikova, O.; Rodriguez-Franco, M.; Ott, T.; Bisseling, T.; van Zeijl, A.; Geurts, R. Mutant analysis in the nonlegume Parasponia andersonii identifies NIN and NF-YA1 transcription factors as a core genetic network in nitrogen-fixing nodule symbioses. New Phytol. 2020, 226, 541–554. [Google Scholar] [CrossRef]
- Tan, X.; Zhang, H.; Yang, Z.; Wei, Z.; Li, Y.; Chen, J.; Sun, Z. NF-YA transcription factors suppress jasmonic acid-mediated antiviral defense and facilitate viral infection in rice. PLoS Pathog. 2022, 18, e1010548. [Google Scholar] [CrossRef]
- O′Conner, S.; Zheng, W.; Qi, M.; Kandel, Y.; Fuller, R.; Whitham, S.A.; Li, L. GmNF-YC4-2 increases protein, exhibits broad disease resistance and expedites maturity in soybean. Int. J. Mol. Sci. 2021, 22, 3586. [Google Scholar] [CrossRef]
- Rey, T.; Laporte, P.; Bonhomme, M.; Jardinaud, M.F.; Huguet, S.; Balzergue, S.; Dumas, B.; Niebel, A.; Jacquet, C. MtNF-YA1, a central transcriptional regulator of symbiotic nodule development, is also a determinant of Medicago truncatula susceptibility toward a root pathogen. Front. Plant Sci. 2016, 7, 1837. [Google Scholar] [CrossRef]
- He, X.; Liu, G.; Li, B.; Xie, Y.; Wei, Y.; Shang, S.; Tian, L.; Shi, H. Functional analysis of the heterotrimeric NF-Y transcription factor complex in cassava disease resistance. Ann. Bot. 2020, 124, 1185–1198. [Google Scholar] [CrossRef]
- Martyn, R.D. Fusarium wilt of watermelon: 120 years of research. Hort. Rev. 2014, 42, 349–442. [Google Scholar]
- Yang, J.; Zhu, J.; Yang, Y. Genome-wide identification and expression analysis of NF-Y transcription factor families in watermelon (Citrullus lanatus). J. Plant Growth Reg. 2017, 36, 590–607. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.J.; Zhang, H.; Zheng, Y.; Mao, L.; Ren, Y.; Wang, Z.; et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, X.; Reddy, U.; Sun, H.; Bao, K.; Gao, L.; Mao, L.; Patel, T.; Ortiz, C.; Abburi, V.L.; et al. Genome of ‘Charleston Gray’, the principal American watermelon cultivar, and genetic characterization of 1,365 accessions in the U.S. National Plant Germplasm System watermelon collection. Plant Biotechnol. J. 2019, 17, 2246–2258. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, R.; Banerjee, R.; Chung, S.M.; Farman, M.; Citovsky, V.; Hogenhout, S.A.; Tzfira, T.; Goodin, M. PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: Probing Nicotiana benthamiana-virus interactions. Mol. Plant-Microbe Interact. 2007, 20, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Howell, S. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 2010, 22, 782–796. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, D.; Liu, Y.; Luo, C.; Zhou, Y.; Zhang, L. Overexpression of a NF-YB3 transcription factor from Picea wilsonii confers tolerance to salinity and drought stress in transformed Arabidopsis thaliana. Plant Physiol. Biochem. 2015, 94, 153–164. [Google Scholar] [CrossRef]
- Kumimoto, R.W.; Adam, L.; Hymus, G.J.; Repetti, P.P.; Reuber, T.L.; Marion, C.M.; Hempel, F.D.; Ratcliffe, O.J. The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 2008, 228, 709–723. [Google Scholar] [CrossRef]
- Hackenberg, D.; Keetman, U.; Grimm, B. Homologous NF-YC2 subunit from Arabidopsis and tobacco is activated by photooxidative stress and induces flowering. Int. J. Mol. Sci. 2012, 13, 3458–3477. [Google Scholar] [CrossRef]
- Kirsch, C.; Takamiya-Wik, M.; Schmelzer, E.; Hahlbrock, K.; Somssich, I.E. A novel regulatory element involved in rapid activation of parsley ELI7 gene family members by fungal elicitor or pathogen infection. Mol. Plant Pathol. 2000, 1, 243–251. [Google Scholar] [CrossRef]
- Rushton, P.J.; Reinstadler, A.; Lipka, V.; Lippok, B.; Somssich, I.E. Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen-and wound-induced signaling. Plant Cell 2002, 14, 749–762. [Google Scholar] [CrossRef]
- Persad-Russell, R.; Mazarei, M.; Schimel, T.M.; Howe, L.; Schmid, M.J.; Kakeshpour, T.; Barnes, C.N.; Brabazon, H.; Seaberry, E.M.; Reuter, D.N.; et al. Specific bacterial pathogen phytosensing is enabled by a synthetic promoter-transcription factor system in potato. Front. Plant Sci. 2022, 13, 873480. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Grant, M.R.; Jones, J.D. Hormone (dis)harmony moulds plant health and disease. Science 2009, 324, 750–752. [Google Scholar] [CrossRef]
- Koornneef, A.; Pieterse, C.M. Cross talk in defense signaling. Plant Physiol. 2008, 146, 839–844. [Google Scholar] [CrossRef]
- Verhage, A.; van Wees, S.C.; Pieterse, C.M. Plant immunity: It’s the hormones talking, but what do they say? Plant Physiol. 2010, 154, 536–540. [Google Scholar] [CrossRef]
- Song, Q.; Li, D.; Dai, Y.; Liu, S.; Huang, L.; Hong, Y.; Zhang, H.; Song, F. Characterization, expression patterns and functional analysis of the MAPK and MAPKK genes in watermelon (Citrullus lanatus). BMC Plant Biol. 2015, 15, 298. [Google Scholar] [CrossRef]
- Liang, J.; Li, X.; Wen, Y.; Wu, X.; Wang, H.; Li, D.; Song, F. Genome-wide characterization of the methyl CpG binding domain-containing proteins in watermelon and functional analysis of their roles in disease resistance through ectopic overexpression in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 886965. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; Kissinger, J.C.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- AbuQamar, S.; Chai, M.F.; Luo, H.L.; Song, F.M.; Mengiste, T. Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 2008, 20, 1964–1983. [Google Scholar] [CrossRef]
- Zhang, H.; Hong, Y.; Huang, L.; Li, D.; Song, F. Arabidopsis AtERF014 acts as a dual regulator that differentially modulates immunity against Pseudomonas syringae pv. tomato and Botrytis cinerea. Sci. Rep. 2016, 6, 30251. [Google Scholar] [CrossRef]









| Subunits | Genes | Locus ID (97103 v2) | Chromosomes | ORF (bp) | Protein (aa) | MW (Da) | pI |
|---|---|---|---|---|---|---|---|
| ClNF-YA | ClNF-YA1 | Cla97C01G025170 | 1 | 666 | 221 | 24,444.14 | 8.27 |
| ClNF-YA2 | Cla97C02G049270 | 2 | 1230 | 409 | 44,283.62 | 6.49 | |
| ClNF-YA3 | Cla97C03G063440 | 3 | 810 | 269 | 29,487.43 | 6.97 | |
| ClNF-YA4 | Cla97C04G068610 | 4 | 816 | 271 | 30,768.39 | 9.08 | |
| ClNF-YA5 | Cla97C09G166390 | 9 | 954 | 317 | 35,487.74 | 8.47 | |
| ClNF-YA6 | Cla97C10G201180 | 10 | 972 | 323 | 35,228.14 | 9.14 | |
| ClNF-YA7 | Cla97C11G207940 | 11 | 1044 | 347 | 38,318.83 | 7.73 | |
| ClNF-YB | ClNF-YB1 | Cla97C02G035590 | 2 | 618 | 205 | 20,920.07 | 6.61 |
| ClNF-YB2 | Cla97C02G039490 | 2 | 525 | 174 | 18,999.25 | 5.23 | |
| ClNF-YB3 | Cla97C02G048360 | 2 | 486 | 161 | 17,972.03 | 5.0 | |
| ClNF-YB4 | Cla97C03G067370 | 3 | 405 | 134 | 15,544.21 | 4.67 | |
| ClNF-YB5 | Cla97C06G124610 | 6 | 576 | 191 | 21,345.08 | 4.57 | |
| ClNF-YB6 | Cla97C07G136140 | 7 | 678 | 225 | 24,739.47 | 7.98 | |
| ClNF-YB7 | Cla97C09G168740 | 9 | 480 | 159 | 17,290.24 | 5.08 | |
| ClNF-YB8 | Cla97C10G188720 | 10 | 528 | 175 | 18,844.55 | 5.73 | |
| ClNF-YB9 | Cla97C10G188900 | 10 | 672 | 223 | 24,150.34 | 6.01 | |
| ClNF-YB10 | Cla97C10G203800 | 10 | 522 | 173 | 18,705.82 | 6.35 | |
| ClNF-YC | ClNF-YC1 | Cla97C02G047110 | 2 | 603 | 280 | 22,498.46 | 9.17 |
| ClNF-YC2 | Cla97C02G048620 | 2 | 843 | 260 | 31,251.64 | 4.77 | |
| ClNF-YC3 | Cla97C03G067230 | 3 | 783 | 220 | 28,842.77 | 6.16 | |
| ClNF-YC4 | Cla97C06G127670 | 6 | 663 | 117 | 24,223.41 | 4.97 | |
| ClNF-YC5 | Cla97C07G135150 | 7 | 354 | 140 | 12,986.36 | 7.96 | |
| ClNF-YC6 | Cla97C08G152900 | 8 | 423 | 266 | 15,794.05 | 8.99 | |
| ClNF-YC7 | Cla97C09G176240 | 9 | 801 | 283 | 29,969.79 | 6.33 | |
| ClNF-YC8 | Cla97C11G218950 | 11 | 852 | 280 | 31,695.19 | 5.09 |
| ClNF-YBs | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| ClNF-YCs | 1 | + | + | − | − | − | − | − | − |
| 2 | − | − | + | + | + | + | + | + | |
| 3 | − | − | + | + | + | + | + | + | |
| 4 | − | − | + | + | + | + | + | + | |
| 5 | − | − | + | + | + | + | + | + | |
| 6 | − | − | + | + | + | + | + | + | |
| 7 | − | − | + | + | + | + | + | + | |
| 8 | − | − | + | + | + | + | + | + | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, S.; Wang, H.; Wen, Y.; Liang, J.; Li, D.; Song, F. The NF-Y Transcription Factor Family in Watermelon: Re-Characterization, Assembly of ClNF-Y Complexes, Hormone- and Pathogen-Inducible Expression and Putative Functions in Disease Resistance. Int. J. Mol. Sci. 2022, 23, 15778. https://doi.org/10.3390/ijms232415778
Jiang S, Wang H, Wen Y, Liang J, Li D, Song F. The NF-Y Transcription Factor Family in Watermelon: Re-Characterization, Assembly of ClNF-Y Complexes, Hormone- and Pathogen-Inducible Expression and Putative Functions in Disease Resistance. International Journal of Molecular Sciences. 2022; 23(24):15778. https://doi.org/10.3390/ijms232415778
Chicago/Turabian StyleJiang, Siyu, Hui Wang, Ya Wen, Jiayu Liang, Dayong Li, and Fengming Song. 2022. "The NF-Y Transcription Factor Family in Watermelon: Re-Characterization, Assembly of ClNF-Y Complexes, Hormone- and Pathogen-Inducible Expression and Putative Functions in Disease Resistance" International Journal of Molecular Sciences 23, no. 24: 15778. https://doi.org/10.3390/ijms232415778
APA StyleJiang, S., Wang, H., Wen, Y., Liang, J., Li, D., & Song, F. (2022). The NF-Y Transcription Factor Family in Watermelon: Re-Characterization, Assembly of ClNF-Y Complexes, Hormone- and Pathogen-Inducible Expression and Putative Functions in Disease Resistance. International Journal of Molecular Sciences, 23(24), 15778. https://doi.org/10.3390/ijms232415778






