Role of Efflux Pumps on Antimicrobial Resistance in Pseudomonas aeruginosa
Abstract
:1. The Silent Pandemic of Antimicrobial-Resistant Bacteria
2. Pseudomonas Aeruginosa
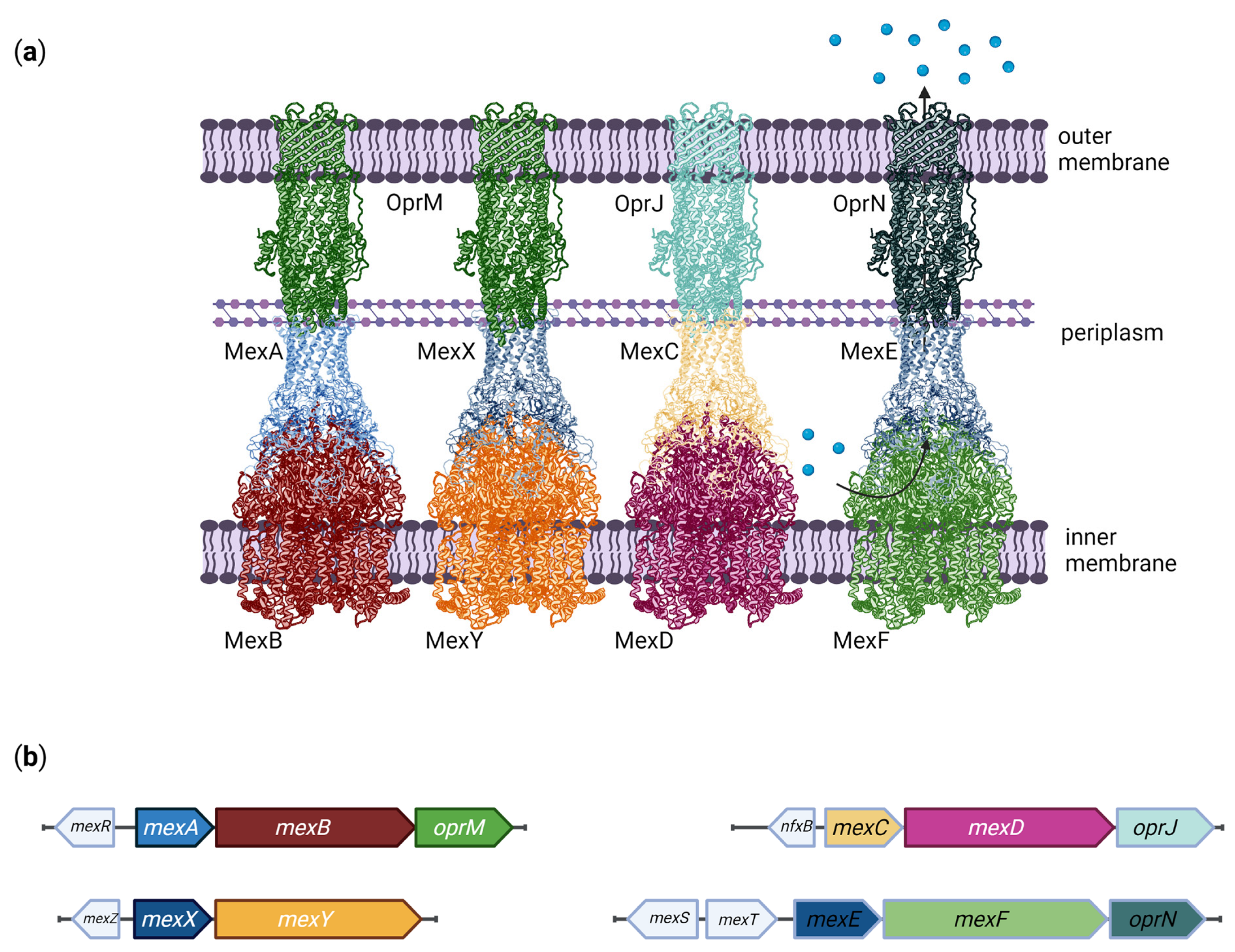
3. Function and Structure of Efflux Pumps
| Family 1 | Efflux Pump | Gene Ids 2 | Substrates 3 | References |
|---|---|---|---|---|
| ABC | Ttg2 (Mla) | PA4456-PA4455-PA4454-PA4453-PA4452 | CHL, CIP, COL, DMF, DOX, LVX, MIN, OFX, TET, TGC, TOB, TMP | [63,64,65] |
| PA1874-77 | PA1874-PA1875-PA1876-PA1877 | CIP, GEN, NOR, TOB | [66,67] | |
| PA3228 | PA3228 | CAR, LVX, NOR | [68] | |
| MFS | Mfs1 | PA1262 | PQT | [69] |
| Mfs2/SmvA | PA1282 | OCT, PQT | [69,70] | |
| CmlA1 | GNT62_RS22140 | CHL | [71,72] | |
| SMR | PASmr/EmrEPae | PA4990 | ACR, EtBr, GEN, KAN, NEO | [73,74] |
| SugE subfamily SMR | PA1882 | Further research is needed | [75] | |
| MATE | PmpM | PA1361 | ACR, BZK, CIP, EtBr, NOR, OFX, TPPCL | [76] |
| PACE | PA2880 | PA2880 | CHX | [77] |
| RND | MexAB-OprM | PA0425-PA0426-PA0427 | AMI, AMX, ATM, CAR, CR, MA, FEP, CFP, CFSL, CTX, FOX, CZOP, CPO, CES, CAZ, CZX, CRO, CXM, CHL, CTET, CIN, CIP, CLX, DP, DOR, ENX, ERY, FMOX, GEN, IPM, LVX, CLM, MEM, MOX, NAF, NAL, NOR, NOV, OFX, OMC, OTC, PG, PPA, PIP, PTZ, PMA, SPX, SPI, STN, SUL, TZB, TET, TIC, TOS | [78,79,80,81] |
| MexCD-OprJ | PA4599-PA4598-PA4597 | AMX, MA, FEP, CFP, CFSL, CTX, FOX, CZOP, CPO, CES, CZX, CRO, CXM, CHL, CHX, CTET, CIN, CIP, CLX, DOR, ENX, ERY, FMOX, LVX, CLM, MEM, NAF, NAL, NOR, NOV, OFX, OMC, OTC, PG, PPA, PIP, PMA, SPX, SPI, TET, TOS | [79,80,82,83] | |
| TMexCD-TOprJ | LSG45_RS29735-LSG45_RS29740-LSG45_RS29745 | FEP, CEQ, CAZ, CTET, CIP, DOX, ERV, FLO, GEN, MIN, NAL, OTC, STR, TET, TGC | [84,85] | |
| MexEF-OprN | PA2493-PA2494-PA2495 | CHL, QN, TET, TMP | [86] | |
| MexGHI-OpmD | PA4205-PA4206-PA4207-PA4208 | 5-Me-PCA, ACR, EtBr, NOR, R6G, TET, V | [87,88,89] | |
| MexJK-OprM | PA3677-PA3676-PA0427 | ERY, TET, TCS | [90] | |
| MexMN-OprM | PA1435-PA1436-PA0427 | BAL30072, ATM, BIPM, CAR, CMN, CAZ, CFT, CHL, MEM, MET, MOX, NOV, PIP, SUL, TMC, TP, TIC | [91,92] | |
| MexPQ-OpmE | PA3523-PA3522-PA3521 | Hoechst 33342, CHL, CIP, ERY, KIT, NOR, RKM, TET, TPPCL | [91] | |
| MexVW-OprM | PA4374-PA4375-PA0427 | ACR, CPO, CHL, ERY, EtBr, NOR, OFX, TET | [93] | |
| MexXY-OprM (-OprA) | PA2019-PA2018-PA0427 (-PSPA7_3271) | ACR, AMI, AMX, MA, FEP, CFP, CFSL, CTX, FOX, CZOP, CPO, CAZ, CZX, CRO, CXM, CHL, CTET, CIN, CIP, CLX, DOR, ENX, ERY, EtBr, FMOX, GEN, IPM, KAN, LVX, CLM, MEM, NAF, NAL, NEO, NOR, OFX, OMC, OTC, PG, PPA, PIP, PTZ, PMA, SPX, SPI, STR, TZB, TET, TIC, TOB, TOS, CAR (only with OprA), SUL (only with OprA) | [79,80,94,95,96] | |
| MuxABC-OpmB | PA2528-PA2527-PA2526-PA2525 | ATM, ERY, KIT, NOV, RKM, TET | [97] | |
| TriABC-OpmH | PA0156-PA0157-PA0158-PA4974 | TCS | [98] |
3.1. General Role of Efflux Pumps in Bacteria in a Natural Environment
3.2. Efflux Pumps Involved in AMR in Bacteria
4. Efflux Pumps as a Mechanism of Antimicrobial Resistance in P. aeruginosa
4.1. MexAB-OprM
4.2. MexXY
4.3. MexCD-OprJ
4.4. MexEF-OprN
5. Efflux Pumps and Biofilm Formation
6. Efflux Pumps as Targets for New Drugs
7. Concluding Remarks
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Williams, K.J. The Introduction of “chemotherapy” Using Arsphenamine—The First Magic Bullet. J. R. Soc. Med. 2009, 102, 343–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleming, A. On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to Their Use in the Isolation of B. Influenzæ. Br. J. Exp. Pathol. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- Sengupta, S.; Chattopadhyay, M.K.; Grossart, H.-P. The Multifaceted Roles of Antibiotics and Antibiotic Resistance in Nature. Front. Microbiol. 2013, 4, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2021; World Health Organization: Geneva, Switzerland, 2021; p. 180. [Google Scholar]
- Bassetti, M.; Kanj, S.S.; Kiratisin, P.; Rodrigues, C.; Van Duin, D.; Villegas, M.V.; Yu, Y. Early Appropriate Diagnostics and Treatment of MDR Gram-Negative Infections. JAC-Antimicrob. Resist. 2022, 4, dlac089. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-H.; Buttery, J. Antimicrobial Resistance: A Global One-Health Problem for All Ages. World J. Pediatr. 2018, 14, 521–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plackett, B. Why Big Pharma Has Abandoned Antibiotics. Nature 2020, 586, S50–S52. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. The Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- World Health Organization. The True Death Toll of COVID-19: Estimating Global Excess Mortality; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- WHO. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 12 September 2022).
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Van Camp, P.-J.; Haslam, D.B.; Porollo, A. Bioinformatics Approaches to the Understanding of Molecular Mechanisms in Antimicrobial Resistance. Int. J. Mol. Sci. 2020, 21, 1363. [Google Scholar] [CrossRef]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular Mechanisms of Antibiotic Resistance Revisited. Nat. Rev. Microbiol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Crone, S.; Vives-Flórez, M.; Kvich, L.; Saunders, A.M.; Malone, M.; Nicolaisen, M.H.; Martínez-García, E.; Rojas-Acosta, C.; Catalina Gomez-Puerto, M.; Calum, H.; et al. The Environmental Occurrence of Pseudomonas Aeruginosa. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2020, 128, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Tuon, F.F.; Dantas, L.R.; Suss, P.H.; Tasca Ribeiro, V.S. Pathogenesis of the Pseudomonas Aeruginosa Biofilm: A Review. Pathogens 2022, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.-L.; Suetens, C.; Savey, A.; Palomar, M.; Hiesmayr, M.; Morales, I.; Agodi, A.; Frank, U.; Mertens, K.; Schumacher, M.; et al. Clinical Outcomes of Health-Care-Associated Infections and Antimicrobial Resistance in Patients Admitted to European Intensive-Care Units: A Cohort Study. Lancet Infect. Dis. 2011, 11, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, D.; Kollef, M. The Epidemiology and Pathogenesis and Treatment of Pseudomonas Aeruginosa Infections: An Update. Drugs 2021, 81, 2117–2131. [Google Scholar] [CrossRef] [PubMed]
- Turkina, M.V.; Vikström, E. Bacteria-Host Crosstalk: Sensing of the Quorum in the Context of Pseudomonas Aeruginosa Infections. J. Innate Immun. 2019, 11, 263–279. [Google Scholar] [CrossRef]
- Pier, G.B.; Grout, M.; Zaidi, T.S. Cystic Fibrosis Transmembrane Conductance Regulator Is an Epithelial Cell Receptor for Clearance of Pseudomonas Aeruginosa from the Lung. Proc. Natl. Acad. Sci. USA 1997, 94, 12088–12093. [Google Scholar] [CrossRef] [Green Version]
- Jolly, A.L.; Takawira, D.; Oke, O.O.; Whiteside, S.A.; Chang, S.W.; Wen, E.R.; Quach, K.; Evans, D.J.; Fleiszig, S.M.J. Pseudomonas Aeruginosa-Induced Bleb-Niche Formation in Epithelial Cells Is Independent of Actinomyosin Contraction and Enhanced by Loss of Cystic Fibrosis Transmembrane-Conductance Regulator Osmoregulatory Function. mBio 2015, 6, e02533. [Google Scholar] [CrossRef] [Green Version]
- Del Barrio-Tofiño, E.; Zamorano, L.; Cortes-Lara, S.; López-Causapé, C.; Sánchez-Diener, I.; Cabot, G.; Bou, G.; Martínez-Martínez, L.; Oliver, A.; GEMARA-SEIMC/REIPI Pseudomonas study Group. Spanish Nationwide Survey on Pseudomonas Aeruginosa Antimicrobial Resistance Mechanisms and Epidemiology. J. Antimicrob. Chemother. 2019, 74, 1825–1835. [Google Scholar] [CrossRef]
- Recio, R.; Mancheño, M.; Viedma, E.; Villa, J.; Orellana, M.Á.; Lora-Tamayo, J.; Chaves, F. Predictors of Mortality in Bloodstream Infections Caused by Pseudomonas Aeruginosa and Impact of Antimicrobial Resistance and Bacterial Virulence. Antimicrob. Agents Chemother. 2020, 64, e01759-19. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CDC. Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019. [CrossRef] [Green Version]
- Diggle, S.P.; Whiteley, M. Microbe Profile: Pseudomonas Aeruginosa: Opportunistic Pathogen and Lab Rat: This Article Is Part of the Microbe Profiles Collection. Microbiology 2020, 166, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, T.; Vasconcelos, U. Colour Me Blue: The History and the Biotechnological Potential of Pyocyanin. Molecules 2021, 26, 927. [Google Scholar] [CrossRef]
- Michel-Briand, Y.; Baysse, C. The Pyocins of Pseudomonas Aeruginosa. Biochimie 2002, 84, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Noto, M.J.; Burns, W.J.; Beavers, W.N.; Skaar, E.P. Mechanisms of Pyocyanin Toxicity and Genetic Determinants of Resistance in Staphylococcus Aureus. J. Bacteriol. 2017, 199, e00221-17. [Google Scholar] [CrossRef] [Green Version]
- Emmerich, R.; Löw, O. Bakteriolytische Enzyme als Ursache der erworbenen Immunität und die Heilung von Infectionskrankheiten durch dieselben. Z. Für Hyg. Infect. 1899, 31, 1–65. [Google Scholar] [CrossRef]
- Holloway, B.W. Genetic Recombination in Pseudomonas Aeruginosa. J. Gen. Microbiol. 1955, 13, 572–581. [Google Scholar] [CrossRef] [Green Version]
- Holloway, B.W.; Morgan, A.F. Genome Organization in Pseudomonas. Annu. Rev. Microbiol. 1986, 40, 79–105. [Google Scholar] [CrossRef]
- Holloway, B.W.; Krishnapillai, V.; Morgan, A.F. Chromosomal Genetics of Pseudomonas. Microbiol. Rev. 1979, 43, 73–102. [Google Scholar] [CrossRef]
- Holloway, B.W.; Römling, U.; Tümmler, B. Genomic Mapping of Pseudomonas Aeruginosa PAO. Microbiol. Read. Engl. 1994, 140 Pt 11, 2907–2929. [Google Scholar] [CrossRef] [Green Version]
- Römling, U.; Grothues, D.; Bautsch, W.; Tümmler, B. A Physical Genome Map of Pseudomonas Aeruginosa PAO. EMBO J. 1989, 8, 4081–4089. [Google Scholar] [CrossRef] [PubMed]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete Genome Sequence of Pseudomonas Aeruginosa PAO1, an Opportunistic Pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, W.K.; Ferrarini, M.; Morello, L.G.; Faoro, H. Resistome Analysis of Bloodstream Infection Bacterial Genomes Reveals a Specific Set of Proteins Involved in Antibiotic Resistance and Drug Efflux. NAR Genom. Bioinforma 2020, 2, lqaa055. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J.V. Multidrug-Resistance Efflux Pumps ? Not Just for Resistance. Nat. Rev. Microbiol. 2006, 4, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Deng, Z.; Yan, A. Bacterial Multidrug Efflux Pumps: Mechanisms, Physiology and Pharmacological Exploitations. Biochem. Biophys. Res. Commun. 2014, 453, 254–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, K.A.; Liu, Q.; Elbourne, L.D.H.; Ahmad, I.; Sharples, D.; Naidu, V.; Chan, C.L.; Li, L.; Harborne, S.P.D.; Pokhrel, A.; et al. Pacing across the Membrane: The Novel PACE Family of Efflux Pumps Is Widespread in Gram-Negative Pathogens. Res. Microbiol. 2018, 169, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B. Active Efflux Mechanisms for Antimicrobial Resistance. Antimicrob. Agents Chemother. 1992, 36, 695–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikaido, H. Prevention of Drug Access to Bacterial Targets: Permeability Barriers and Active Efflux. Science 1994, 264, 382–388. [Google Scholar] [CrossRef] [Green Version]
- Sanz-García, F.; Gil-Gil, T.; Laborda, P.; Ochoa-Sánchez, L.E.; Martínez, J.L.; Hernando-Amado, S. Coming from the Wild: Multidrug Resistant Opportunistic Pathogens Presenting a Primary, Not Human-Linked, Environmental Habitat. Int. J. Mol. Sci. 2021, 22, 8080. [Google Scholar] [CrossRef]
- Saier, M.H., Jr.; Paulsen, I.T.; Marek, K.S.; Pao, S.S.; Ronald, A.S.; Nikaido, H. Evolutionary Origins of Multidrug and Drug-Specific Efflux Pumps in Bacteria. FASEB J. 1998, 12, 265–274. [Google Scholar] [CrossRef] [Green Version]
- Shahini Shams Abadi, M.; Gholipour, A.; Hadi, N. The Highly Conserved Domain of RND Multidrug Efflux Pumps in Pathogenic Gram-Negative Bacteria. Cell. Mol. Biol. Noisy 2018, 64, 79–83. [Google Scholar] [CrossRef]
- McMurry, L.; Petrucci, R.E.; Levy, S.B. Active Efflux of Tetracycline Encoded by Four Genetically Different Tetracycline Resistance Determinants in Escherichia Coli. Proc. Natl. Acad. Sci. USA 1980, 77, 3974–3977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kodama, Y.; Inouye, S. Correlation of Susceptibility and Resistance of Twenty-Five Bacterial Strains by Analysis of MIC Database of Cephalosporins and Oxacephalosporins. J. Antibiot. 1997, 50, 246–255. [Google Scholar] [CrossRef]
- Yuan, L.; Zhai, Y.-J.; Wu, H.; Sun, H.-R.; He, Z.-P.; Wang, Y.-B.; Pan, Y.-S.; Kuang, N.-N.; Hu, G.-Z. Identification and Prevalence of RND Family Multidrug Efflux Pump OqxAB Genes in Enterococci Isolates from Swine Manure in China. J. Med. Microbiol. 2018, 67, 733–739. [Google Scholar] [CrossRef]
- Saier, M.H., Jr.; Tam, R.; Reizer, A.; Reizer, J. Two Novel Families of Bacterial Membrane Proteins Concerned with Nodulation, Cell Division and Transport. Mol. Microbiol. 1994, 11, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H. Phylogenetic Approaches to the Identification and Characterization of Protein Families and Superfamilies. Microb. Comp. Genomics 1996, 1, 129–150. [Google Scholar] [CrossRef]
- Johnson, J.M.; Church, G.M. Alignment and Structure Prediction of Divergent Protein Families: Periplasmic and Outer Membrane Proteins of Bacterial Efflux Pumps11Edited by G. von Heijne. J. Mol. Biol. 1999, 287, 695–715. [Google Scholar] [CrossRef] [Green Version]
- Ma, D.; Cook, D.N.; Hearst, J.E.; Nikaido, H. Efflux Pumps and Drug Resistance in Gram-Negative Bacteria. Trends Microbiol. 1994, 2, 489–493. [Google Scholar] [CrossRef]
- Jamshidi, S.; Sutton, J.M.; Rahman, K.M. Mapping the Dynamic Functions and Structural Features of AcrB Efflux Pump Transporter Using Accelerated Molecular Dynamics Simulations. Sci. Rep. 2018, 8, 10470. [Google Scholar] [CrossRef]
- Wang, Z.; Fan, G.; Hryc, C.F.; Blaza, J.N.; Serysheva, I.I.; Schmid, M.F.; Chiu, W.; Luisi, B.F.; Du, D. An Allosteric Transport Mechanism for the AcrAB-TolC Multidrug Efflux Pump. eLife 2017, 6, e24905. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Nakashima, R.; Sakurai, K. Structural Basis of RND-Type Multidrug Exporters. Front. Microbiol. 2015, 6, 327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zomot, E.; Yardeni, E.H.; Vargiu, A.V.; Tam, H.-K.; Malloci, G.; Ramaswamy, V.K.; Perach, M.; Ruggerone, P.; Pos, K.M.; Bibi, E. A New Critical Conformational Determinant of Multidrug Efflux by an MFS Transporter. J. Mol. Biol. 2018, 430, 1368–1385. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Szewczyk, P.; Karyakin, A.; Evin, M.; Hong, W.-X.; Zhang, Q.; Chang, G. Structure of a Cation-Bound Multidrug and Toxic Compound Extrusion Transporter. Nature 2010, 467, 991–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolla, J.R.; Howes, A.C.; Fiorentino, F.; Robinson, C.V. Assembly and Regulation of the Chlorhexidine-Specific Efflux Pump AceI. Proc. Natl. Acad. Sci. USA 2020, 117, 17011–17018. [Google Scholar] [CrossRef] [PubMed]
- Al-Hamad, A.; Upton, M.; Burnie, J. Molecular Cloning and Characterization of SmrA, a Novel ABC Multidrug Efflux Pump from Stenotrophomonas Maltophilia. J. Antimicrob. Chemother. 2009, 64, 731–734. [Google Scholar] [CrossRef] [Green Version]
- Shcherbakov, A.A.; Hisao, G.; Mandala, V.S.; Thomas, N.E.; Soltani, M.; Salter, E.A.; Davis, J.H.; Henzler-Wildman, K.A.; Hong, M. Structure and Dynamics of the Drug-Bound Bacterial Transporter EmrE in Lipid Bilayers. Nat. Commun. 2021, 12, 172. [Google Scholar] [CrossRef]
- Marshall, N.J.; Piddock, L.J. Antibacterial Efflux Systems. Microbiol. Madr. Spain 1997, 13, 285–300. [Google Scholar]
- Sulavik, M.C.; Houseweart, C.; Cramer, C.; Jiwani, N.; Murgolo, N.; Greene, J.; DiDomenico, B.; Shaw, K.J.; Miller, G.H.; Hare, R.; et al. Antibiotic Susceptibility Profiles of Escherichia Coli Strains Lacking Multidrug Efflux Pump Genes. Antimicrob. Agents Chemother. 2001, 45, 1126–1136. [Google Scholar] [CrossRef] [Green Version]
- McDaniel, C.; Su, S.; Panmanee, W.; Lau, G.W.; Browne, T.; Cox, K.; Paul, A.T.; Ko, S.-H.B.; Mortensen, J.E.; Lam, J.S.; et al. A Putative ABC Transporter Permease Is Necessary for Resistance to Acidified Nitrite and EDTA in Pseudomonas Aeruginosa under Aerobic and Anaerobic Planktonic and Biofilm Conditions. Front. Microbiol. 2016, 7, 291. [Google Scholar] [CrossRef]
- Chen, L.; Duan, K. A PhoPQ-Regulated ABC Transporter System Exports Tetracycline in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 3016–3024. [Google Scholar] [CrossRef]
- Yero, D.; Díaz-Lobo, M.; Costenaro, L.; Conchillo-Solé, O.; Mayo, A.; Ferrer-Navarro, M.; Vilaseca, M.; Gibert, I.; Daura, X. The Pseudomonas Aeruginosa Substrate-Binding Protein Ttg2D Functions as a General Glycerophospholipid Transporter across the Periplasm. Commun. Biol. 2021, 4, 448. [Google Scholar] [CrossRef]
- Zhang, L.; Mah, T.-F. Involvement of a Novel Efflux System in Biofilm-Specific Resistance to Antibiotics. J. Bacteriol. 2008, 190, 4447–4452. [Google Scholar] [CrossRef] [Green Version]
- Poudyal, B.; Sauer, K. The ABC of Biofilm Drug Tolerance: The MerR-Like Regulator BrlR Is an Activator of ABC Transport Systems, with PA1874-77 Contributing to the Tolerance of Pseudomonas Aeruginosa Biofilms to Tobramycin. Antimicrob. Agents Chemother. 2018, 62, e01981-17. [Google Scholar] [CrossRef]
- Hall, C.W.; Zhang, L.; Mah, T.-F. PA3225 Is a Transcriptional Repressor of Antibiotic Resistance Mechanisms in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2017, 61, e02114-16. [Google Scholar] [CrossRef] [Green Version]
- Dulyayangkul, P.; Satapoomin, N.; Avison, M.B.; Charoenlap, N.; Vattanaviboon, P.; Mongkolsuk, S. Over-Expression of Hypochlorite Inducible Major Facilitator Superfamily (MFS) Pumps Reduces Antimicrobial Drug Susceptibility by Increasing the Production of MexXY Mediated by ArmZ in Pseudomonas Aeruginosa. Front. Microbiol. 2021, 11, 592153. [Google Scholar] [CrossRef]
- Bock, L.J.; Ferguson, P.M.; Clarke, M.; Pumpitakkul, V.; Wand, M.E.; Fady, P.-E.; Allison, L.; Fleck, R.A.; Shepherd, M.J.; Mason, A.J.; et al. Pseudomonas Aeruginosa Adapts to Octenidine via a Combination of Efflux and Membrane Remodelling. Commun. Biol. 2021, 4, 1058. [Google Scholar] [CrossRef]
- Bissonnette, L.; Champetier, S.; Buisson, J.P.; Roy, P.H. Characterization of the Nonenzymatic Chloramphenicol Resistance (CmlA) Gene of the In4 Integron of Tn1696: Similarity of the Product to Transmembrane Transport Proteins. J. Bacteriol. 1991, 173, 4493–4502. [Google Scholar] [CrossRef] [Green Version]
- Stokes, H.W.; Hall, R.M. Sequence Analysis of the Inducible Chloramphenicol Resistance Determinant in the Tn1696 Integron Suggests Regulation by Translational Attenuation. Plasmid 1991, 26, 10–19. [Google Scholar] [CrossRef]
- Li, X.-Z.; Poole, K.; Nikaido, H. Contributions of MexAB-OprM and an EmrE Homolog to Intrinsic Resistance of Pseudomonas Aeruginosa to Aminoglycosides and Dyes. Antimicrob. Agents Chemother. 2003, 47, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Ninio, S.; Rotem, D.; Schuldiner, S. Functional Analysis of Novel Multidrug Transporters from Human Pathogens. J. Biol. Chem. 2001, 276, 48250–48256. [Google Scholar] [CrossRef]
- Duan, K.; Dammel, C.; Stein, J.; Rabin, H.; Surette, M.G. Modulation of Pseudomonas Aeruginosa Gene Expression by Host Microflora through Interspecies Communication. Mol. Microbiol. 2003, 50, 1477–1491. [Google Scholar] [CrossRef] [Green Version]
- He, G.-X.; Kuroda, T.; Mima, T.; Morita, Y.; Mizushima, T.; Tsuchiya, T. An H(+)-Coupled Multidrug Efflux Pump, PmpM, a Member of the MATE Family of Transporters, from Pseudomonas Aeruginosa. J. Bacteriol. 2004, 186, 262–265. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Hellwig, N.; Djahanschiri, B.; Khera, R.; Morgner, N.; Ebersberger, I.; Wang, J.; Michel, H. Assembly and Functional Role of PACE Transporter PA2880 from Pseudomonas Aeruginosa. Microbiol. Spectr. 2022, 10, e0145321. [Google Scholar] [CrossRef]
- Poole, K.; Krebes, K.; McNally, C.; Neshat, S. Multiple Antibiotic Resistance in Pseudomonas Aeruginosa: Evidence for Involvement of an Efflux Operon. J. Bacteriol. 1993, 175, 7363–7372. [Google Scholar] [CrossRef] [Green Version]
- Masuda, N.; Sakagawa, E.; Ohya, S.; Gotoh, N.; Tsujimoto, H.; Nishino, T. Substrate Specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM Efflux Pumps in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2000, 44, 3322–3327. [Google Scholar] [CrossRef] [Green Version]
- Bialvaei, A.Z.; Rahbar, M.; Hamidi-Farahani, R.; Asgari, A.; Esmailkhani, A.; Mardani Dashti, Y.; Soleiman-Meigooni, S. Expression of RND Efflux Pumps Mediated Antibiotic Resistance in Pseudomonas Aeruginosa Clinical Strains. Microb. Pathog. 2021, 153, 104789. [Google Scholar] [CrossRef]
- Pesingi, P.V.; Singh, B.R.; Pesingi, P.K.; Bhardwaj, M.; Singh, S.V.; Kumawat, M.; Sinha, D.K.; Gandham, R.K. MexAB-OprM Efflux Pump of Pseudomonas Aeruginosa Offers Resistance to Carvacrol: A Herbal Antimicrobial Agent. Front. Microbiol. 2019, 10, 2664. [Google Scholar] [CrossRef]
- Poole, K.; Gotoh, N.; Tsujimoto, H.; Zhao, Q.; Wada, A.; Yamasaki, T.; Neshat, S.; Yamagishi, J.; Li, X.Z.; Nishino, T. Overexpression of the MexC-MexD-OprJ Efflux Operon in NfxB-Type Multidrug-Resistant Strains of Pseudomonas Aeruginosa. Mol. Microbiol. 1996, 21, 713–724. [Google Scholar] [CrossRef]
- Fraud, S.; Campigotto, A.J.; Chen, Z.; Poole, K. MexCD-OprJ Multidrug Efflux System of Pseudomonas Aeruginosa: Involvement in Chlorhexidine Resistance and Induction by Membrane-Damaging Agents Dependent upon the AlgU Stress Response Sigma Factor. Antimicrob. Agents Chemother. 2008, 52, 4478–4482. [Google Scholar] [CrossRef] [Green Version]
- Dong, N.; Zeng, Y.; Wang, Y.; Liu, C.; Lu, J.; Cai, C.; Liu, X.; Chen, Y.; Wu, Y.; Fang, Y.; et al. Distribution and Spread of the Mobilised RND Efflux Pump Gene Cluster TmexCD-ToprJ in Clinical Gram-Negative Bacteria: A Molecular Epidemiological Study. Lancet Microbe 2022, 3, e846–e856. [Google Scholar] [CrossRef]
- Lv, L.; Wan, M.; Wang, C.; Gao, X.; Yang, Q.; Partridge, S.R.; Wang, Y.; Zong, Z.; Doi, Y.; Shen, J.; et al. Emergence of a Plasmid-Encoded Resistance-Nodulation-Division Efflux Pump Conferring Resistance to Multiple Drugs, Including Tigecycline, in Klebsiella Pneumoniae. mBio 2020, 11, e02930-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köhler, T.; Michéa-Hamzehpour, M.; Henze, U.; Gotoh, N.; Curty, L.K.; Pechère, J.C. Characterization of MexE-MexF-OprN, a Positively Regulated Multidrug Efflux System of Pseudomonas Aeruginosa. Mol. Microbiol. 1997, 23, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Aendekerk, S.; Ghysels, B.; Cornelis, P.; Baysse, C. Characterization of a New Efflux Pump, MexGHI-OpmD, from Pseudomonas Aeruginosa That Confers Resistance to Vanadium. Microbiol. Read. Engl. 2002, 148, 2371–2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakhtah, H.; Koyama, L.; Zhang, Y.; Morales, D.K.; Fields, B.L.; Price-Whelan, A.; Hogan, D.A.; Shepard, K.; Dietrich, L.E.P. The Pseudomonas Aeruginosa Efflux Pump MexGHI-OpmD Transports a Natural Phenazine That Controls Gene Expression and Biofilm Development. Proc. Natl. Acad. Sci. USA 2016, 113, E3538–E3547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekiya, H.; Mima, T.; Morita, Y.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. Functional Cloning and Characterization of a Multidrug Efflux Pump, MexHI-OpmD, from a Pseudomonas Aeruginosa Mutant. Antimicrob. Agents Chemother. 2003, 47, 2990–2992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuanchuen, R.; Narasaki, C.T.; Schweizer, H.P. The MexJK Efflux Pump of Pseudomonas Aeruginosa Requires OprM for Antibiotic Efflux but Not for Efflux of Triclosan. J. Bacteriol. 2002, 184, 5036–5044. [Google Scholar] [CrossRef] [Green Version]
- Mima, T.; Sekiya, H.; Mizushima, T.; Kuroda, T.; Tsuchiya, T. Gene Cloning and Properties of the RND-Type Multidrug Efflux Pumps MexPQ-OpmE and MexMN-OprM from Pseudomonas Aeruginosa. Microbiol. Immunol. 2005, 49, 999–1002. [Google Scholar] [CrossRef]
- Ranjitkar, S.; Jones, A.K.; Mostafavi, M.; Zwirko, Z.; Iartchouk, O.; Barnes, S.W.; Walker, J.R.; Willis, T.W.; Lee, P.S.; Dean, C.R. Target (MexB)- and Efflux-Based Mechanisms Decreasing the Effectiveness of the Efflux Pump Inhibitor D13-9001 in Pseudomonas Aeruginosa PAO1: Uncovering a New Role for MexMN-OprM in Efflux of β-Lactams and a Novel Regulatory Circuit (MmnRS) Controlling MexMN Expression. Antimicrob. Agents Chemother. 2019, 63, e01718-18. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Mima, T.; Komori, Y.; Morita, Y.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. A New Member of the Tripartite Multidrug Efflux Pumps, MexVW–OprM, in Pseudomonas Aeruginosa. J. Antimicrob. Chemother. 2003, 52, 572–575. [Google Scholar] [CrossRef] [Green Version]
- Mine, T.; Morita, Y.; Kataoka, A.; Mizushima, T.; Tsuchiya, T. Expression in Escherichia Coli of a New Multidrug Efflux Pump, MexXY, from Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 1999, 43, 415–417. [Google Scholar] [CrossRef]
- Singh, M.; Sykes, E.M.E.; Li, Y.; Kumar, A. MexXY RND Pump of Pseudomonas Aeruginosa PA7 Effluxes Bi-Anionic β-Lactams Carbenicillin and Sulbenicillin When It Partners with the Outer Membrane Factor OprA but Not with OprM. Microbiol. Read. Engl. 2020, 166, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Masuda, N.; Sakagawa, E.; Ohya, S.; Gotoh, N.; Tsujimoto, H.; Nishino, T. Contribution of the MexX-MexY-OprM Efflux System to Intrinsic Resistance in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2000, 44, 2242–2246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mima, T.; Kohira, N.; Li, Y.; Sekiya, H.; Ogawa, W.; Kuroda, T.; Tsuchiya, T. Gene Cloning and Characteristics of the RND-Type Multidrug Efflux Pump MuxABC-OpmB Possessing Two RND Components in Pseudomonas Aeruginosa. Microbiol. Read. Engl. 2009, 155, 3509–3517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mima, T.; Joshi, S.; Gomez-Escalada, M.; Schweizer, H.P. Identification and Characterization of TriABC-OpmH, a Triclosan Efflux Pump of Pseudomonas Aeruginosa Requiring Two Membrane Fusion Proteins. J. Bacteriol. 2007, 189, 7600–7609. [Google Scholar] [CrossRef] [Green Version]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S.L. Enhanced Annotations and Features for Comparing Thousands of Pseudomonas Genomes in the Pseudomonas Genome Database. Nucleic Acids Res. 2016, 44, D646–D653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanz-García, F.; Hernando-Amado, S.; López-Causapé, C.; Oliver, A.; Martínez, J.L. Low Ciprofloxacin Concentrations Select Multidrug-Resistant Mutants Overproducing Efflux Pumps in Clinical Isolates of Pseudomonas Aeruginosa. Microbiol. Spectr. 2022, 10, e0072322. [Google Scholar] [CrossRef]
- Langevin, A.M.; Dunlop, M.J. Stress Introduction Rate Alters the Benefit of AcrAB-TolC Efflux Pumps. J. Bacteriol. 2018, 200, e00525-17. [Google Scholar] [CrossRef] [Green Version]
- Adamiak, J.W.; Jhawar, V.; Bonifay, V.; Chandler, C.E.; Leus, I.V.; Ernst, R.K.; Schweizer, H.P.; Zgurskaya, H.I. Loss of RND-Type Multidrug Efflux Pumps Triggers Iron Starvation and Lipid A Modifications in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2021, 65, e0059221. [Google Scholar] [CrossRef]
- Nag, A.; Mehra, S. Involvement of the SCO3366 Efflux Pump from S. Coelicolor in Rifampicin Resistance and Its Regulation by a TetR Regulator. Appl. Microbiol. Biotechnol. 2022, 106, 2175–2190. [Google Scholar] [CrossRef]
- Yao, X.; Tao, F.; Zhang, K.; Tang, H.; Xu, P. Multiple Roles for Two Efflux Pumps in the Polycyclic Aromatic Hydrocarbon-Degrading Pseudomonas Putida Strain B6-2 (DSM 28064). Appl. Environ. Microbiol. 2017, 83, e01882-17. [Google Scholar] [CrossRef]
- Zwama, M.; Yamaguchi, A.; Nishino, K. Phylogenetic and Functional Characterisation of the Haemophilus Influenzae Multidrug Efflux Pump AcrB. Commun. Biol. 2019, 2, 340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehla, J.; Malloci, G.; Mansbach, R.; López, C.A.; Tsivkovski, R.; Haynes, K.; Leus, I.V.; Grindstaff, S.B.; Cascella, R.H.; D’Cunha, N.; et al. Predictive Rules of Efflux Inhibition and Avoidance in Pseudomonas Aeruginosa. mBio 2021, 12, e02785-20. [Google Scholar] [CrossRef] [PubMed]
- Elkins, C.A.; Nikaido, H. Substrate Specificity of the RND-Type Multidrug Efflux Pumps AcrB and AcrD of Escherichia Coli Is Determined Predominantly by Two Large Periplasmic Loops. J. Bacteriol. 2002, 184, 6490–6498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishino, K.; Yamaguchi, A. Analysis of a Complete Library of Putative Drug Transporter Genes in Escherichia Coli. J. Bacteriol. 2001, 183, 5803–5812. [Google Scholar] [CrossRef] [Green Version]
- Laudy, A.E. Non-Antibiotics, Efflux Pumps and Drug Resistance of Gram-Negative Rods. Pol. J. Microbiol. 2018, 67, 129–135. [Google Scholar] [CrossRef] [Green Version]
- Ramos, J.L.; Duque, E.; Gallegos, M.-T.; Godoy, P.; Ramos-Gonzalez, M.I.; Rojas, A.; Teran, W.; Segura, A. Mechanisms of Solvent Tolerance in Gram-Negative Bacteria. Annu. Rev. Microbiol. 2002, 56, 743–768. [Google Scholar] [CrossRef]
- Garratt, I.; Aranega-Bou, P.; Sutton, J.M.; Moore, G.; Wand, M.E. Long-Term Exposure to Octenidine in a Simulated Sink Trap Environment Results in Selection of Pseudomonas Aeruginosa, Citrobacter, and Enterobacter Isolates with Mutations in Efflux Pump Regulators. Appl. Environ. Microbiol. 2021, 87, e00210-21. [Google Scholar] [CrossRef]
- Lennen, R.M.; Politz, M.G.; Kruziki, M.A.; Pfleger, B.F. Identification of Transport Proteins Involved in Free Fatty Acid Efflux in Escherichia Coli. J. Bacteriol. 2013, 195, 135–144. [Google Scholar] [CrossRef] [Green Version]
- Sabrin, A.; Gioe, B.W.; Gupta, A.; Grove, A. An EmrB Multidrug Efflux Pump in Burkholderia Thailandensis with Unexpected Roles in Antibiotic Resistance. J. Biol. Chem. 2019, 294, 1891–1903. [Google Scholar] [CrossRef] [Green Version]
- Ramos, J.L.; Martínez-Bueno, M.; Molina-Henares, A.J.; Terán, W.; Watanabe, K.; Zhang, X.; Gallegos, M.T.; Brennan, R.; Tobes, R. The TetR Family of Transcriptional Repressors. Microbiol. Mol. Biol. Rev. MMBR 2005, 69, 326–356. [Google Scholar] [CrossRef]
- Liu, Q.; Hassan, K.A.; Ashwood, H.E.; Gamage, H.K.A.H.; Li, L.; Mabbutt, B.C.; Paulsen, I.T. Regulation of the AceI Multidrug Efflux Pump Gene in Acinetobacter Baumannii. J. Antimicrob. Chemother. 2018, 73, 1492–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stubenrauch, C.J.; Bamert, R.S.; Wang, J.; Lithgow, T. A Noncanonical Chaperone Interacts with Drug Efflux Pumps during Their Assembly into Bacterial Outer Membranes. PLoS Biol. 2022, 20, e3001523. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Cook, D.N.; Alberti, M.; Pon, N.G.; Nikaido, H.; Hearst, J.E. Molecular Cloning and Characterization of AcrA and AcrE Genes of Escherichia Coli. J. Bacteriol. 1993, 175, 6299–6313. [Google Scholar] [CrossRef] [Green Version]
- Schmalstieg, A.M.; Srivastava, S.; Belkaya, S.; Deshpande, D.; Meek, C.; Leff, R.; van Oers, N.S.C.; Gumbo, T. The Antibiotic Resistance Arrow of Time: Efflux Pump Induction Is a General First Step in the Evolution of Mycobacterial Drug Resistance. Antimicrob. Agents Chemother. 2012, 56, 4806–4815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, J.L.; Kwon, T.; Marcotte, E.M.; Whiteley, M. Intrinsic Antimicrobial Resistance Determinants in the Superbug Pseudomonas Aeruginosa. mBio 2015, 6, e01603-01615. [Google Scholar] [CrossRef] [Green Version]
- Chevalier, S.; Bouffartigues, E.; Bodilis, J.; Maillot, O.; Lesouhaitier, O.; Feuilloley, M.G.J.; Orange, N.; Dufour, A.; Cornelis, P. Structure, Function and Regulation of Pseudomonas Aeruginosa Porins. FEMS Microbiol. Rev. 2017, 41, 698–722. [Google Scholar] [CrossRef] [Green Version]
- Valot, B.; Guyeux, C.; Rolland, J.Y.; Mazouzi, K.; Bertrand, X.; Hocquet, D. What It Takes to Be a Pseudomonas Aeruginosa? The Core Genome of the Opportunistic Pathogen Updated. PLoS ONE 2015, 10, e0126468. [Google Scholar] [CrossRef]
- Venter, H.; Mowla, R.; Ohene-Agyei, T.; Ma, S. RND-Type Drug Efflux Pumps from Gram-Negative Bacteria: Molecular Mechanism and Inhibition. Front. Microbiol. 2015, 6, 377. [Google Scholar] [CrossRef]
- Daury, L.; Orange, F.; Taveau, J.-C.; Verchère, A.; Monlezun, L.; Gounou, C.; Marreddy, R.K.R.; Picard, M.; Broutin, I.; Pos, K.M.; et al. Tripartite Assembly of RND Multidrug Efflux Pumps. Nat. Commun. 2016, 7, 10731. [Google Scholar] [CrossRef] [Green Version]
- Du, D.; Wang-Kan, X.; Neuberger, A.; van Veen, H.W.; Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Multidrug Efflux Pumps: Structure, Function and Regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef]
- López-Causapé, C.; Sommer, L.M.; Cabot, G.; Rubio, R.; Ocampo-Sosa, A.A.; Johansen, H.K.; Figuerola, J.; Cantón, R.; Kidd, T.J.; Molin, S.; et al. Evolution of the Pseudomonas Aeruginosa Mutational Resistome in an International Cystic Fibrosis Clone. Sci. Rep. 2017, 7, 5555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colclough, A.L.; Alav, I.; Whittle, E.E.; Pugh, H.L.; Darby, E.M.; Legood, S.W.; McNeil, H.E.; Blair, J.M. RND Efflux Pumps in Gram-Negative Bacteria; Regulation, Structure and Role in Antibiotic Resistance. Future Microbiol. 2020, 15, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Cabot, G.; Ocampo-Sosa, A.A.; Tubau, F.; Macia, M.D.; Rodríguez, C.; Moya, B.; Zamorano, L.; Suárez, C.; Peña, C.; Martínez-Martínez, L.; et al. Overexpression of AmpC and Efflux Pumps in Pseudomonas Aeruginosa Isolates from Bloodstream Infections: Prevalence and Impact on Resistance in a Spanish Multicenter Study. Antimicrob. Agents Chemother. 2011, 55, 1906–1911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, A.K.; Caughlan, R.E.; Woods, A.L.; Uehara, K.; Xie, L.; Barnes, S.W.; Walker, J.R.; Thompson, K.V.; Ranjitkar, S.; Lee, P.S.; et al. Mutations Reducing In Vitro Susceptibility to Novel LpxC Inhibitors in Pseudomonas Aeruginosa and Interplay of Efflux and Nonefflux Mechanisms. Antimicrob. Agents Chemother. 2019, 64, e01490-19. [Google Scholar] [CrossRef]
- Krause, K.M.; Haglund, C.M.; Hebner, C.; Serio, A.W.; Lee, G.; Nieto, V.; Cohen, F.; Kane, T.R.; Machajewski, T.D.; Hildebrandt, D.; et al. Potent LpxC Inhibitors with In Vitro Activity against Multidrug-Resistant Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e00977-19. [Google Scholar] [CrossRef]
- Ito, A.; Sato, T.; Ota, M.; Takemura, M.; Nishikawa, T.; Toba, S.; Kohira, N.; Miyagawa, S.; Ishibashi, N.; Matsumoto, S.; et al. In Vitro Antibacterial Properties of Cefiderocol, a Novel Siderophore Cephalosporin, against Gram-Negative Bacteria. Antimicrob. Agents Chemother. 2018, 62, e01454-17. [Google Scholar] [CrossRef] [Green Version]
- Amieva, R.; Gil-Gil, T.; Martínez, J.L.; Alcalde-Rico, M. The MexJK Multidrug Efflux Pump Is Not Involved in Acquired or Intrinsic Antibiotic Resistance in Pseudomonas Aeruginosa, but Modulates the Bacterial Quorum Sensing Response. Int. J. Mol. Sci. 2022, 23, 7492. [Google Scholar] [CrossRef]
- López, C.A.; Travers, T.; Pos, K.M.; Zgurskaya, H.I.; Gnanakaran, S. Dynamics of Intact MexAB-OprM Efflux Pump: Focusing on the MexA-OprM Interface. Sci. Rep. 2017, 7, 16521. [Google Scholar] [CrossRef] [Green Version]
- Tsutsumi, K.; Yonehara, R.; Ishizaka-Ikeda, E.; Miyazaki, N.; Maeda, S.; Iwasaki, K.; Nakagawa, A.; Yamashita, E. Structures of the Wild-Type MexAB-OprM Tripartite Pump Reveal Its Complex Formation and Drug Efflux Mechanism. Nat. Commun. 2019, 10, 1520. [Google Scholar] [CrossRef] [Green Version]
- Poole, K.; Tetro, K.; Zhao, Q.; Neshat, S.; Heinrichs, D.E.; Bianco, N. Expression of the Multidrug Resistance Operon MexA-MexB-OprM in Pseudomonas Aeruginosa: MexR Encodes a Regulator of Operon Expression. Antimicrob. Agents Chemother. 1996, 40, 2021–2028. [Google Scholar] [CrossRef]
- Cao, L.; Srikumar, R.; Poole, K. MexAB-OprM Hyperexpression in NalC-Type Multidrug-Resistant Pseudomonas Aeruginosa: Identification and Characterization of the NalC Gene Encoding a Repressor of PA3720-PA3719. Mol. Microbiol. 2004, 53, 1423–1436. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Cao, L.; Gould, V.C.; Avison, M.B.; Poole, K. NalD Encodes a Second Repressor of the MexAB-OprM Multidrug Efflux Operon of Pseudomonas Aeruginosa. J. Bacteriol. 2006, 188, 8649–8654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tafti, F.A.; Eslami, G.; Zandi, H.; Barzegar, K. Mutations in Nalc Gene of Mex AB-OprM Efflux Pump in Carbapenem Resistant Pseudomonas Aeruginosa Isolated from Burn Wounds in Yazd, Iran. Iran. J. Microbiol. 2020, 12, 32–36. [Google Scholar] [PubMed]
- Ziha-Zarifi, I.; Llanes, C.; Köhler, T.; Pechere, J.-C.; Plesiat, P. In Vivo Emergence of Multidrug-Resistant Mutants of Pseudomonas Aeruginosa Overexpressing the Active Efflux System MexA-MexB-OprM. Antimicrob. Agents Chemother. 1999, 43, 287–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horna, G.; López, M.; Guerra, H.; Saénz, Y.; Ruiz, J. Interplay between MexAB-OprM and MexEF-OprN in Clinical Isolates of Pseudomonas Aeruginosa. Sci. Rep. 2018, 8, 16463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choudhury, D.; Ghose, A.; Ghosh, A.; Dhar Chanda, D.; Das Talukdar, A.; Dutta Choudhury, M.; Paul, D.; Maurya, A.P.; Chakravarty, A.; Chakravorty, A.; et al. Premature Termination of MexR Leads to Overexpression of MexAB-OprM Efflux Pump in Pseudomonas Aeruginosa in a Tertiary Referral Hospital in India. PLoS ONE 2016, 11, e0149156. [Google Scholar] [CrossRef]
- Boutoille, D.; Corvec, S.; Caroff, N.; Giraudeau, C.; Espaze, E.; Caillon, J.; Plésiat, P.; Reynaud, A. Detection of an IS21 Insertion Sequence in the MexR Gene of Pseudomonas Aeruginosa Increasing Beta-Lactam Resistance. FEMS Microbiol. Lett. 2004, 230, 143–146. [Google Scholar] [CrossRef]
- Ma, Z.; Xu, C.; Zhang, X.; Wang, D.; Pan, X.; Liu, H.; Zhu, G.; Bai, F.; Cheng, Z.; Wu, W.; et al. A MexR Mutation Which Confers Aztreonam Resistance to Pseudomonas Aeruginosa. Front. Microbiol. 2021, 12, 659808. [Google Scholar] [CrossRef]
- Aguilar-Rodea, P.; Zúñiga, G.; Cerritos, R.; Rodríguez-Espino, B.A.; Gomez-Ramirez, U.; Nolasco-Romero, C.G.; López-Marceliano, B.; Rodea, G.E.; Mendoza-Elizalde, S.; Reyes-López, A.; et al. Nucleotide Substitutions in the MexR, NalC and NalD Regulator Genes of the MexAB-OprM Efflux Pump Are Maintained in Pseudomonas Aeruginosa Genetic Lineages. PLoS ONE 2022, 17, e0266742. [Google Scholar] [CrossRef]
- Suresh, M.; Nithya, N.; Jayasree, P.R.; Vimal, K.P.; Manish Kumar, P.R. Mutational Analyses of Regulatory Genes, MexR, NalC, NalD and MexZ of MexAB-OprM and MexXY Operons, in Efflux Pump Hyperexpressing Multidrug-Resistant Clinical Isolates of Pseudomonas Aeruginosa. World J. Microbiol. Biotechnol. 2018, 34, 83. [Google Scholar] [CrossRef]
- Beig, M.; Taheri, M.; Arabestani, M.R. Expression of MexAB-OprM Efflux Pump and OprD Porin in Carbapenemase Producing Pseudomonas Aeruginosa Clinical Isolates. Gene Rep. 2020, 20, 100744. [Google Scholar] [CrossRef]
- Glen, K.A.; Lamont, I.L. β-Lactam Resistance in Pseudomonas Aeruginosa: Current Status, Future Prospects. Pathogens 2021, 10, 1638. [Google Scholar] [CrossRef] [PubMed]
- Lodge, J.M.; Minchin, S.D.; Piddock, L.J.; Busby, J.W. Cloning, Sequencing and Analysis of the Structural Gene and Regulatory Region of the Pseudomonas Aeruginosa Chromosomal AmpC Beta-Lactamase. Biochem. J. 1990, 272, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, M.; Tazrout, S.; Bour, M.; Triponey, P.; Muller, C.; Jeannot, K.; Plésiat, P. Reassessment of the Cooperativity between Efflux System MexAB-OprM and Cephalosporinase AmpC in the Resistance of Pseudomonas Aeruginosa to β-Lactams. J. Antimicrob. Chemother. 2021, 76, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.H.; Tetu, S.G.; Larouche, A.; Elbourne, L.; Tremblay, S.; Ren, Q.; Dodson, R.; Harkins, D.; Shay, R.; Watkins, K.; et al. Complete Genome Sequence of the Multiresistant Taxonomic Outlier Pseudomonas Aeruginosa PA7. PLoS ONE 2010, 5, e8842. [Google Scholar] [CrossRef]
- Morita, Y.; Tomida, J.; Kawamura, Y. Primary Mechanisms Mediating Aminoglycoside Resistance in the Multidrug-Resistant Pseudomonas Aeruginosa Clinical Isolate PA7. Microbiol. Read. Engl. 2012, 158, 1071–1083. [Google Scholar] [CrossRef]
- Issa, K.H.B.; Phan, G.; Broutin, I. Functional Mechanism of the Efflux Pumps Transcription Regulators From Pseudomonas Aeruginosa Based on 3D Structures. Front. Mol. Biosci. 2018, 5, 57. [Google Scholar] [CrossRef] [Green Version]
- Muller, C.; Plésiat, P.; Jeannot, K. A Two-Component Regulatory System Interconnects Resistance to Polymyxins, Aminoglycosides, Fluoroquinolones, and β-Lactams in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2011, 55, 1211–1221. [Google Scholar] [CrossRef] [Green Version]
- Hay, T.; Fraud, S.; Lau, C.H.-F.; Gilmour, C.; Poole, K. Antibiotic Inducibility of the MexXY Multidrug Efflux Operon of Pseudomonas Aeruginosa: Involvement of the MexZ Anti-Repressor ArmZ. PLoS ONE 2013, 8, e56858. [Google Scholar] [CrossRef]
- Kawalek, A.; Modrzejewska, M.; Zieniuk, B.; Bartosik, A.A.; Jagura-Burdzy, G. Interaction of ArmZ with the DNA-Binding Domain of MexZ Induces Expression of MexXY Multidrug Efflux Pump Genes and Antimicrobial Resistance in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e01199-19. [Google Scholar] [CrossRef]
- Guénard, S.; Muller, C.; Monlezun, L.; Benas, P.; Broutin, I.; Jeannot, K.; Plésiat, P. Multiple Mutations Lead to MexXY-OprM-Dependent Aminoglycoside Resistance in Clinical Strains of Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2014, 58, 221–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seupt, A.; Schniederjans, M.; Tomasch, J.; Häussler, S. Expression of the MexXY Aminoglycoside Efflux Pump and Presence of an Aminoglycoside-Modifying Enzyme in Clinical Pseudomonas Aeruginosa Isolates Are Highly Correlated. Antimicrob. Agents Chemother. 2020, 65, e01166-20. [Google Scholar] [CrossRef] [PubMed]
- López-Causapé, C.; Rubio, R.; Cabot, G.; Oliver, A. Evolution of the Pseudomonas Aeruginosa Aminoglycoside Mutational Resistome In Vitro and in the Cystic Fibrosis Setting. Antimicrob. Agents Chemother. 2018, 62, e02583-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, M.; Yau, Y.C.W.; Wang, S.; Waters, V.; Kumar, A. MexXY Efflux Pump Overexpression and Aminoglycoside Resistance in Cystic Fibrosis Isolates of Pseudomonas Aeruginosa from Chronic Infections. Can. J. Microbiol. 2017, 63, 929–938. [Google Scholar] [CrossRef] [Green Version]
- Westbrock-Wadman, S.; Sherman, D.R.; Hickey, M.J.; Coulter, S.N.; Zhu, Y.Q.; Warrener, P.; Nguyen, L.Y.; Shawar, R.M.; Folger, K.R.; Stover, C.K. Characterization of a Pseudomonas Aeruginosa Efflux Pump Contributing to Aminoglycoside Impermeability. Antimicrob. Agents Chemother. 1999, 43, 2975–2983. [Google Scholar] [CrossRef] [Green Version]
- Vettoretti, L.; Plésiat, P.; Muller, C.; El Garch, F.; Phan, G.; Attrée, I.; Ducruix, A.; Llanes, C. Efflux Unbalance in Pseudomonas Aeruginosa Isolates from Cystic Fibrosis Patients. Antimicrob. Agents Chemother. 2009, 53, 1987–1997. [Google Scholar] [CrossRef] [Green Version]
- Shigemura, K.; Osawa, K.; Kato, A.; Tokimatsu, I.; Arakawa, S.; Shirakawa, T.; Fujisawa, M. Association of Overexpression of Efflux Pump Genes with Antibiotic Resistance in Pseudomonas Aeruginosa Strains Clinically Isolated from Urinary Tract Infection Patients. J. Antibiot. (Tokyo) 2015, 68, 568–572. [Google Scholar] [CrossRef]
- Jeannot, K.; Elsen, S.; Köhler, T.; Attree, I.; van Delden, C.; Plésiat, P. Resistance and Virulence of Pseudomonas Aeruginosa Clinical Strains Overproducing the MexCD-OprJ Efflux Pump. Antimicrob. Agents Chemother. 2008, 52, 2455–2462. [Google Scholar] [CrossRef] [Green Version]
- Gomis-Font, M.A.; Pitart, C.; Del Barrio-Tofiño, E.; Zboromyrska, Y.; Cortes-Lara, S.; Mulet, X.; Marco, F.; Vila, J.; López-Causapé, C.; Oliver, A. Emergence of Resistance to Novel Cephalosporin-β-Lactamase Inhibitor Combinations through the Modification of the Pseudomonas Aeruginosa MexCD-OprJ Efflux Pump. Antimicrob. Agents Chemother. 2021, 65, e0008921. [Google Scholar] [CrossRef]
- Morita, Y.; Komori, Y.; Mima, T.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. Construction of a Series of Mutants Lacking All of the Four Major Mex Operons for Multidrug Efflux Pumps or Possessing Each One of the Operons from Pseudomonas Aeruginosa PAO1: MexCD-OprJ Is an Inducible Pump. FEMS Microbiol. Lett. 2001, 202, 139–143. [Google Scholar] [CrossRef]
- Srikumar, R.; Kon, T.; Gotoh, N.; Poole, K. Expression of Pseudomonas Aeruginosa Multidrug Efflux Pumps MexA-MexB-OprM and MexC-MexD-OprJ in a Multidrug-Sensitive Escherichia Coli Strain. Antimicrob. Agents Chemother. 1998, 42, 65–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuda, N.; Gotoh, N.; Ohya, S.; Nishino, T. Quantitative Correlation between Susceptibility and OprJ Production in NfxB Mutants of Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 1996, 40, 909–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, W.; Warren, M.S.; Black, D.S.; Satou, T.; Murata, T.; Nishino, T.; Gotoh, N.; Lomovskaya, O. On the Mechanism of Substrate Specificity by Resistance Nodulation Division (RND)-Type Multidrug Resistance Pumps: The Large Periplasmic Loops of MexD from Pseudomonas Aeruginosa Are Involved in Substrate Recognition. Mol. Microbiol. 2002, 46, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, N.; Tsujimoto, H.; Tsuda, M.; Okamoto, K.; Nomura, A.; Wada, T.; Nakahashi, M.; Nishino, T. Characterization of the MexC-MexD-OprJ Multidrug Efflux System in DeltamexA-MexB-OprM Mutants of Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 1998, 42, 1938–1943. [Google Scholar] [CrossRef] [Green Version]
- Mulet, X.; Moyá, B.; Juan, C.; Macià, M.D.; Pérez, J.L.; Blázquez, J.; Oliver, A. Antagonistic Interactions of Pseudomonas Aeruginosa Antibiotic Resistance Mechanisms in Planktonic but Not Biofilm Growth▿. Antimicrob. Agents Chemother. 2011, 55, 4560–4568. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Ramos, I.; Mulet, X.; Moyá, B.; Barbier, M.; Oliver, A.; Albertí, S. Overexpression of MexCD-OprJ Reduces Pseudomonas Aeruginosa Virulence by Increasing Its Susceptibility to Complement-Mediated Killing. Antimicrob. Agents Chemother. 2014, 58, 2426–2429. [Google Scholar] [CrossRef] [Green Version]
- Cazares, A.; Moore, M.P.; Hall, J.P.J.; Wright, L.L.; Grimes, M.; Emond-Rhéault, J.-G.; Pongchaikul, P.; Santanirand, P.; Levesque, R.C.; Fothergill, J.L.; et al. A Megaplasmid Family Driving Dissemination of Multidrug Resistance in Pseudomonas. Nat. Commun. 2020, 11, 1370. [Google Scholar] [CrossRef] [Green Version]
- Köhler, T.; Epp, S.F.; Curty, L.K.; Pechère, J.-C. Characterization of MexT, the Regulator of the MexE-MexF-OprN Multidrug Efflux System of Pseudomonas Aeruginosa. J. Bacteriol. 1999, 181, 6300–6305. [Google Scholar] [CrossRef] [Green Version]
- Sobel, M.L.; Neshat, S.; Poole, K. Mutations in PA2491 (MexS) Promote MexT-Dependent MexEF-OprN Expression and Multidrug Resistance in a Clinical Strain of Pseudomonas Aeruginosa. J. Bacteriol. 2005, 187, 1246–1253. [Google Scholar] [CrossRef] [Green Version]
- Maseda, H.; Saito, K.; Nakajima, A.; Nakae, T. Variation of the MexT Gene, a Regulator of the MexEF-OprN Efflux Pump Expression in Wild-Type Strains of Pseudomonas Aeruginosa. FEMS Microbiol. Lett. 2000, 192, 107–112. [Google Scholar] [CrossRef]
- Fetar, H.; Gilmour, C.; Klinoski, R.; Daigle, D.M.; Dean, C.R.; Poole, K. MexEF-OprN Multidrug Efflux Operon of Pseudomonas Aeruginosa: Regulation by the MexT Activator in Response to Nitrosative Stress and Chloramphenicol. Antimicrob. Agents Chemother. 2011, 55, 508–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morita, Y.; Tomida, J.; Kawamura, Y. Efflux-Mediated Fluoroquinolone Resistance in the Multidrug-Resistant Pseudomonas Aeruginosa Clinical Isolate PA7: Identification of a Novel MexS Variant Involved in Upregulation of the MexEF-OprN Multidrug Efflux Operon. Front. Microbiol. 2015, 6, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardot, C.; Juarez, P.; Jeannot, K.; Patry, I.; Plésiat, P.; Llanes, C. Amino Acid Substitutions Account for Most MexS Alterations in Clinical NfxC Mutants of Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 2302–2310. [Google Scholar] [CrossRef] [Green Version]
- Iftikhar, A.; Asif, A.; Manzoor, A.; Azeem, M.; Sarwar, G.; Rashid, N.; Qaisar, U. Mutation in PvcABCD Operon of Pseudomonas Aeruginosa Modulates MexEF-OprN Efflux System and Hence Resistance to Chloramphenicol and Ciprofloxacin. Microb. Pathog. 2020, 149, 104491. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, D.; Schneper, L.; Merighi, M.; Smith, R.; Narasimhan, G.; Lory, S.; Mathee, K. The Regulatory Repertoire of Pseudomonas Aeruginosa AmpC SS-Lactamase Regulator AmpR Includes Virulence Genes. PLoS ONE 2012, 7, e34067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westfall, L.W.; Carty, N.L.; Layland, N.; Kuan, P.; Colmer-Hamood, J.A.; Hamood, A.N. MvaT Mutation Modifies the Expression of the Pseudomonas Aeruginosa Multidrug Efflux Operon MexEF-OprN. FEMS Microbiol. Lett. 2006, 255, 247–254. [Google Scholar] [CrossRef] [Green Version]
- Ochs, M.M.; McCusker, M.P.; Bains, M.; Hancock, R.E.W. Negative Regulation of the Pseudomonas Aeruginosa Outer Membrane Porin OprD Selective for Imipenem and Basic Amino Acids. Antimicrob. Agents Chemother. 1999, 43, 1085–1090. [Google Scholar] [CrossRef] [Green Version]
- Sherrard, L.J.; Wee, B.A.; Duplancic, C.; Ramsay, K.A.; Dave, K.A.; Ballard, E.; Wainwright, C.E.; Grimwood, K.; Sidjabat, H.E.; Whiley, D.M.; et al. Emergence and Impact of OprD Mutations in Pseudomonas Aeruginosa Strains in Cystic Fibrosis. J. Cyst. Fibros. Off. J. Eur. Cyst. Fibros. Soc. 2022, 21, e35–e43. [Google Scholar] [CrossRef]
- Mlynarcik, P.; Kolar, M. Molecular Mechanisms of Polymyxin Resistance and Detection of Mcr Genes. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czechoslov. 2019, 163, 28–38. [Google Scholar] [CrossRef] [Green Version]
- Yin, R.; Cheng, J.; Wang, J.; Li, P.; Lin, J. Treatment of Pseudomonas Aeruginosa Infectious Biofilms: Challenges and Strategies. Front. Microbiol. 2022, 13, 955286. [Google Scholar] [CrossRef]
- Nikaido, H.; Pagès, J.-M. Broad-Specificity Efflux Pumps and Their Role in Multidrug Resistance of Gram-Negative Bacteria. FEMS Microbiol. Rev. 2012, 36, 340–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reza, A.; Sutton, J.M.; Rahman, K.M. Effectiveness of Efflux Pump Inhibitors as Biofilm Disruptors and Resistance Breakers in Gram-Negative (ESKAPEE) Bacteria. Antibiotics 2019, 8, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of Bacterial Efflux Pumps in Biofilm Formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawan, J.; Li, Y.; Lu, F.; He, X.; Ahn, J. Role of Efflux Pump-Mediated Antibiotic Resistance in Quorum Sensing-Regulated Biofilm Formation by Salmonella Typhimurium. Pathogens 2022, 11, 147. [Google Scholar] [CrossRef] [PubMed]
- Kvist, M.; Hancock, V.; Klemm, P. Inactivation of Efflux Pumps Abolishes Bacterial Biofilm Formation. Appl. Environ. Microbiol. 2008, 74, 7376–7382. [Google Scholar] [CrossRef] [Green Version]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The Involvement of Cell-to-Cell Signals in the Development of a Bacterial Biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef] [Green Version]
- Pearson, J.P.; Van Delden, C.; Iglewski, B.H. Active Efflux and Diffusion Are Involved in Transport of Pseudomonas Aeruginosa Cell-to-Cell Signals. J. Bacteriol. 1999, 181, 1203–1210. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, P.; Linares, J.F.; Ruiz-Díez, B.; Campanario, E.; Navas, A.; Baquero, F.; Martínez, J.L. Fitness of in Vitro Selected Pseudomonas Aeruginosa NalB and NfxB Multidrug Resistant Mutants. J. Antimicrob. Chemother. 2002, 50, 657–664. [Google Scholar] [CrossRef] [Green Version]
- Lamarche, M.G.; Déziel, E. MexEF-OprN Efflux Pump Exports the Pseudomonas Quinolone Signal (PQS) Precursor HHQ (4-Hydroxy-2-Heptylquinoline). PLoS ONE 2011, 6, e24310. [Google Scholar] [CrossRef]
- De Kievit, T.R.; Parkins, M.D.; Gillis, R.J.; Srikumar, R.; Ceri, H.; Poole, K.; Iglewski, B.H.; Storey, D.G. Multidrug Efflux Pumps: Expression Patterns and Contribution to Antibiotic Resistance in Pseudomonas Aeruginosa Biofilms. Antimicrob. Agents Chemother. 2001, 45, 1761–1770. [Google Scholar] [CrossRef]
- Waite, R.D.; Papakonstantinopoulou, A.; Littler, E.; Curtis, M.A. Transcriptome Analysis of Pseudomonas Aeruginosa Growth: Comparison of Gene Expression in Planktonic Cultures and Developing and Mature Biofilms. J. Bacteriol. 2005, 187, 6571–6576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrington, N.E.; Littler, J.L.; Harrison, F. Transcriptome Analysis of Pseudomonas Aeruginosa Biofilm Infection in an Ex Vivo Pig Model of the Cystic Fibrosis Lung. Appl. Environ. Microbiol. 2022, 88, e0178921. [Google Scholar] [CrossRef] [PubMed]
- Kaatz, G.W. Inhibition of Bacterial Efflux Pumps: A New Strategy to Combat Increasing Antimicrobial Agent Resistance. Expert Opin. Emerg. Drugs 2002, 7, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Aínsa, J.A.; Blokpoel, M.C.J.; Otal, I.; Young, D.B.; De Smet, K.A.L.; Martín, C. Molecular Cloning and Characterization of Tap, a Putative Multidrug Efflux Pump Present in Mycobacterium Fortuitum and Mycobacterium Tuberculosis. J. Bacteriol. 1998, 180, 5836–5843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulen, C.; Racine, P.-J.; Feuilloley, M.; Elomri, A.; Lomri, N.-E. Effects of Verapamil and Two Bisbenzylisoquinolines, Curine and Guattegaumerine Extracted from Isolona Hexaloba, on the Inhibition of ABC Transporters from Pseudomonas Aeruginosa. Antibiotics 2022, 11, 700. [Google Scholar] [CrossRef] [PubMed]
- Lomovskaya, O.; Warren, M.S.; Lee, A.; Galazzo, J.; Fronko, R.; Lee, M.; Blais, J.; Cho, D.; Chamberland, S.; Renau, T.; et al. Identification and Characterization of Inhibitors of Multidrug Resistance Efflux Pumps in Pseudomonas Aeruginosa: Novel Agents for Combination Therapy. Antimicrob. Agents Chemother. 2001, 45, 105–116. [Google Scholar] [CrossRef] [Green Version]
- Piddock, L.J.; Williams, K.J.; Ricci, V. Accumulation of Rifampicin by Mycobacterium Aurum, Mycobacterium Smegmatis and Mycobacterium Tuberculosis. J. Antimicrob. Chemother. 2000, 45, 159–165. [Google Scholar] [CrossRef] [Green Version]
- Silva, P.E.; Bigi, F.; Santangelo, M.P.; Romano, M.I.; Martín, C.; Cataldi, A.; Aínsa, J.A. Characterization of P55, a Multidrug Efflux Pump in Mycobacterium Bovis and Mycobacterium Tuberculosis. Antimicrob. Agents Chemother. 2001, 45, 800–804. [Google Scholar] [CrossRef] [Green Version]
- Coban, A.Y.; Ekinci, B.; Durupinar, B. A Multidrug Efflux Pump Inhibitor Reduces Fluoroquinolone Resistance in Pseudomonas Aeruginosa Isolates. Chemotherapy 2004, 50, 22–26. [Google Scholar] [CrossRef]
- Pagès, J.-M.; Amaral, L. Mechanisms of Drug Efflux and Strategies to Combat Them: Challenging the Efflux Pump of Gram-Negative Bacteria. Biochim. Biophys. Acta 2009, 1794, 826–833. [Google Scholar] [CrossRef]
- Hirakata, Y.; Kondo, A.; Hoshino, K.; Yano, H.; Arai, K.; Hirotani, A.; Kunishima, H.; Yamamoto, N.; Hatta, M.; Kitagawa, M.; et al. Efflux Pump Inhibitors Reduce the Invasiveness of Pseudomonas Aeruginosa. Int. J. Antimicrob. Agents 2009, 34, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Kriengkauykiat, J.; Porter, E.; Lomovskaya, O.; Wong-Beringer, A. Use of an Efflux Pump Inhibitor to Determine the Prevalence of Efflux Pump-Mediated Fluoroquinolone Resistance and Multidrug Resistance in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2005, 49, 565–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, C.J.; Stone, T.A.; Deber, C.M. Peptide-Based Efflux Pump Inhibitors of the Small Multidrug Resistance Protein from Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e00730-19. [Google Scholar] [CrossRef] [Green Version]
- Stavri, M.; Piddock, L.J.V.; Gibbons, S. Bacterial Efflux Pump Inhibitors from Natural Sources. J. Antimicrob. Chemother. 2007, 59, 1247–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueroa-Romero, A.; Pons-Duran, C.; Gonzalez, R. Drugs for Intermittent Preventive Treatment of Malaria in Pregnancy: Current Knowledge and Way Forward. Trop. Med. Infect. Dis. 2022, 7, 152. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Aroca, F.; Meng, A.; Minz, T.; Page, M.G.P.; Dreier, J. Use of Resazurin to Detect Mefloquine as an Efflux-Pump Inhibitor in Pseudomonas Aeruginosa and Escherichia Coli. J. Microbiol. Methods 2009, 79, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J.V.; Garvey, M.I.; Rahman, M.M.; Gibbons, S. Natural and Synthetic Compounds Such as Trimethoprim Behave as Inhibitors of Efflux in Gram-Negative Bacteria. J. Antimicrob. Chemother. 2010, 65, 1215–1223. [Google Scholar] [CrossRef] [Green Version]
- Adamson, D.H.; Krikstopaityte, V.; Coote, P.J. Enhanced Efficacy of Putative Efflux Pump Inhibitor/Antibiotic Combination Treatments versus MDR Strains of Pseudomonas Aeruginosa in a Galleria Mellonella in Vivo Infection Model. J. Antimicrob. Chemother. 2015, 70, 2271–2278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laborda, P.; Alcalde-Rico, M.; Chini, A.; Martínez, J.L.; Hernando-Amado, S. Discovery of Inhibitors of Pseudomonas aeruginosa Virulence through the Search for Natural-like Compounds with a Dual Role as Inducers and Substrates of Efflux Pumps. Environ. Microbiol. 2021, 23, 7396–7411. [Google Scholar] [CrossRef]
- Tambat, R.; Mahey, N.; Chandal, N.; Verma, D.K.; Jangra, M.; Thakur, K.G.; Nandanwar, H. A Microbe-Derived Efflux Pump Inhibitor of the Resistance-Nodulation-Cell Division Protein Restores Antibiotic Susceptibility in Escherichia Coli and Pseudomonas Aeruginosa. ACS Infect. Dis. 2022, 8, 255–270. [Google Scholar] [CrossRef]
- Yamasaki, S.; Koga, N.; Zwama, M.; Sakurai, K.; Nakashima, R.; Yamaguchi, A.; Nishino, K. Spatial Characteristics of the Efflux Pump MexB Determine Inhibitor Binding. Antimicrob. Agents Chemother. 2022, 66, e0067222. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, L.; Molin, S. Synergistic Activities of an Efflux Pump Inhibitor and Iron Chelators against Pseudomonas Aeruginosa Growth and Biofilm Formation. Antimicrob. Agents Chemother. 2010, 54, 3960–3963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rampioni, G.; Pillai, C.R.; Longo, F.; Bondì, R.; Baldelli, V.; Messina, M.; Imperi, F.; Visca, P.; Leoni, L. Effect of Efflux Pump Inhibition on Pseudomonas Aeruginosa Transcriptome and Virulence. Sci. Rep. 2017, 7, 11392. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lorusso, A.B.; Carrara, J.A.; Barroso, C.D.N.; Tuon, F.F.; Faoro, H. Role of Efflux Pumps on Antimicrobial Resistance in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2022, 23, 15779. https://doi.org/10.3390/ijms232415779
Lorusso AB, Carrara JA, Barroso CDN, Tuon FF, Faoro H. Role of Efflux Pumps on Antimicrobial Resistance in Pseudomonas aeruginosa. International Journal of Molecular Sciences. 2022; 23(24):15779. https://doi.org/10.3390/ijms232415779
Chicago/Turabian StyleLorusso, Andre Bittencourt, João Antônio Carrara, Carolina Deuttner Neumann Barroso, Felipe Francisco Tuon, and Helisson Faoro. 2022. "Role of Efflux Pumps on Antimicrobial Resistance in Pseudomonas aeruginosa" International Journal of Molecular Sciences 23, no. 24: 15779. https://doi.org/10.3390/ijms232415779
APA StyleLorusso, A. B., Carrara, J. A., Barroso, C. D. N., Tuon, F. F., & Faoro, H. (2022). Role of Efflux Pumps on Antimicrobial Resistance in Pseudomonas aeruginosa. International Journal of Molecular Sciences, 23(24), 15779. https://doi.org/10.3390/ijms232415779






