Temporal Analysis Reveals the Transient Differential Expression of Transcription Factors That Underlie the Trans-Differentiation of Human Monocytes to Macrophages
Abstract
:1. Introduction
2. Results
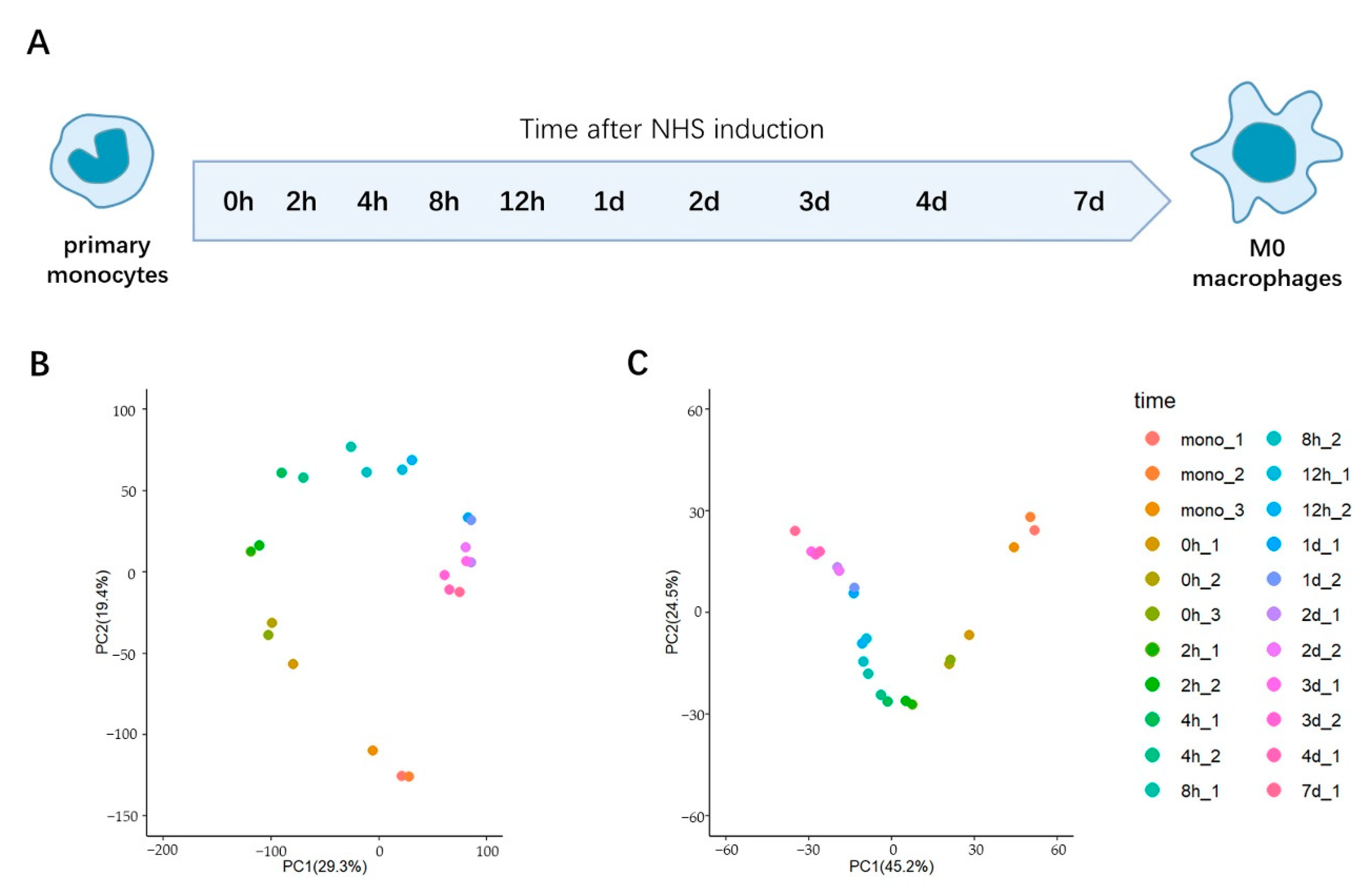
2.1. Temporal Profiling of the Transcriptomic Changes during the Trans-Differentiation of Primary Monocytes to M0 Macrophages In Vitro
2.2. PCA Analysis Indicates That the Transcriptional Changes during This Transition Are Largely Completed in One Day
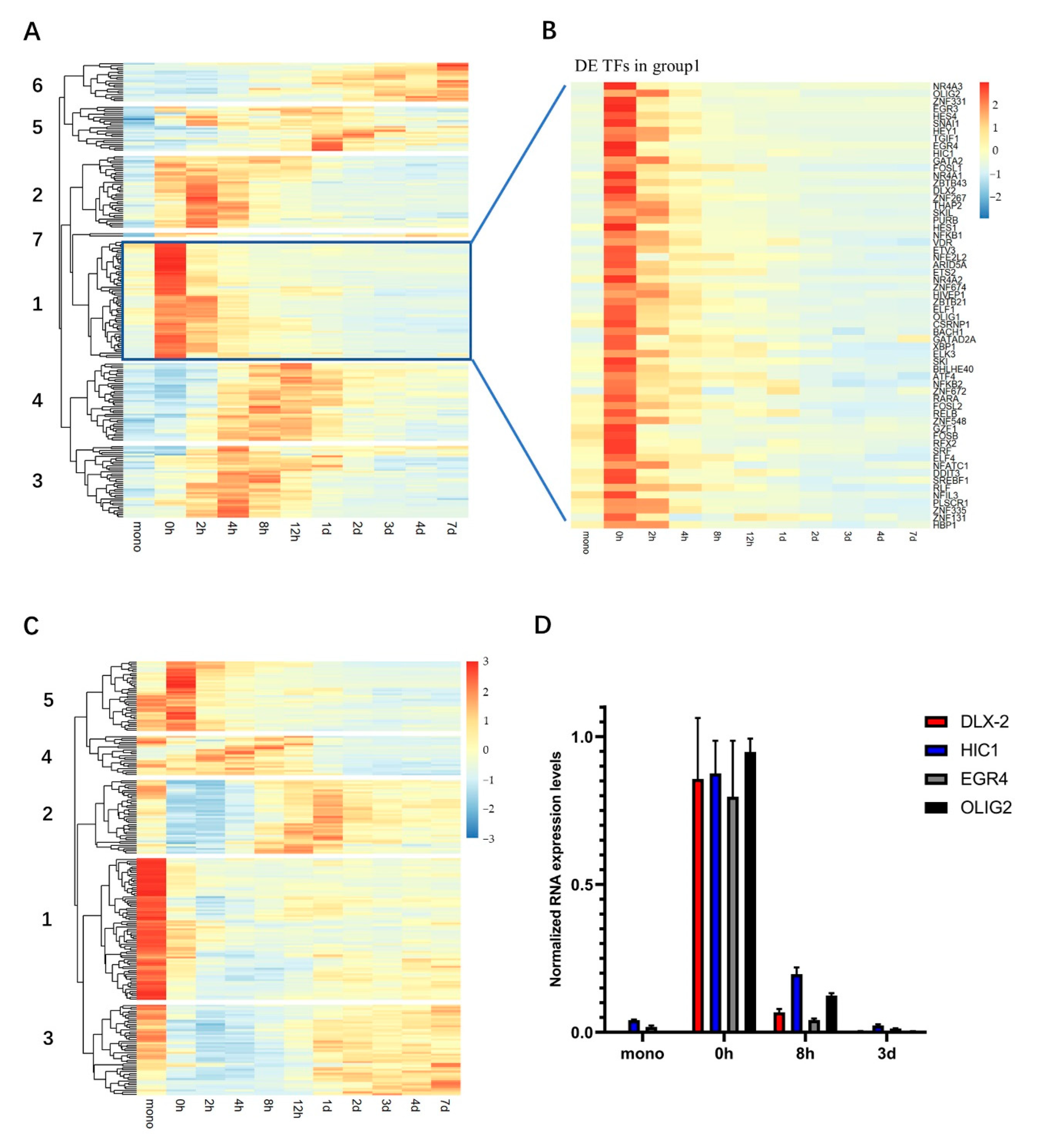
2.3. Hierarchical Clustering Shows “Wave-Like” Changes in the Transcription of TFs during the Trans-Differentiation
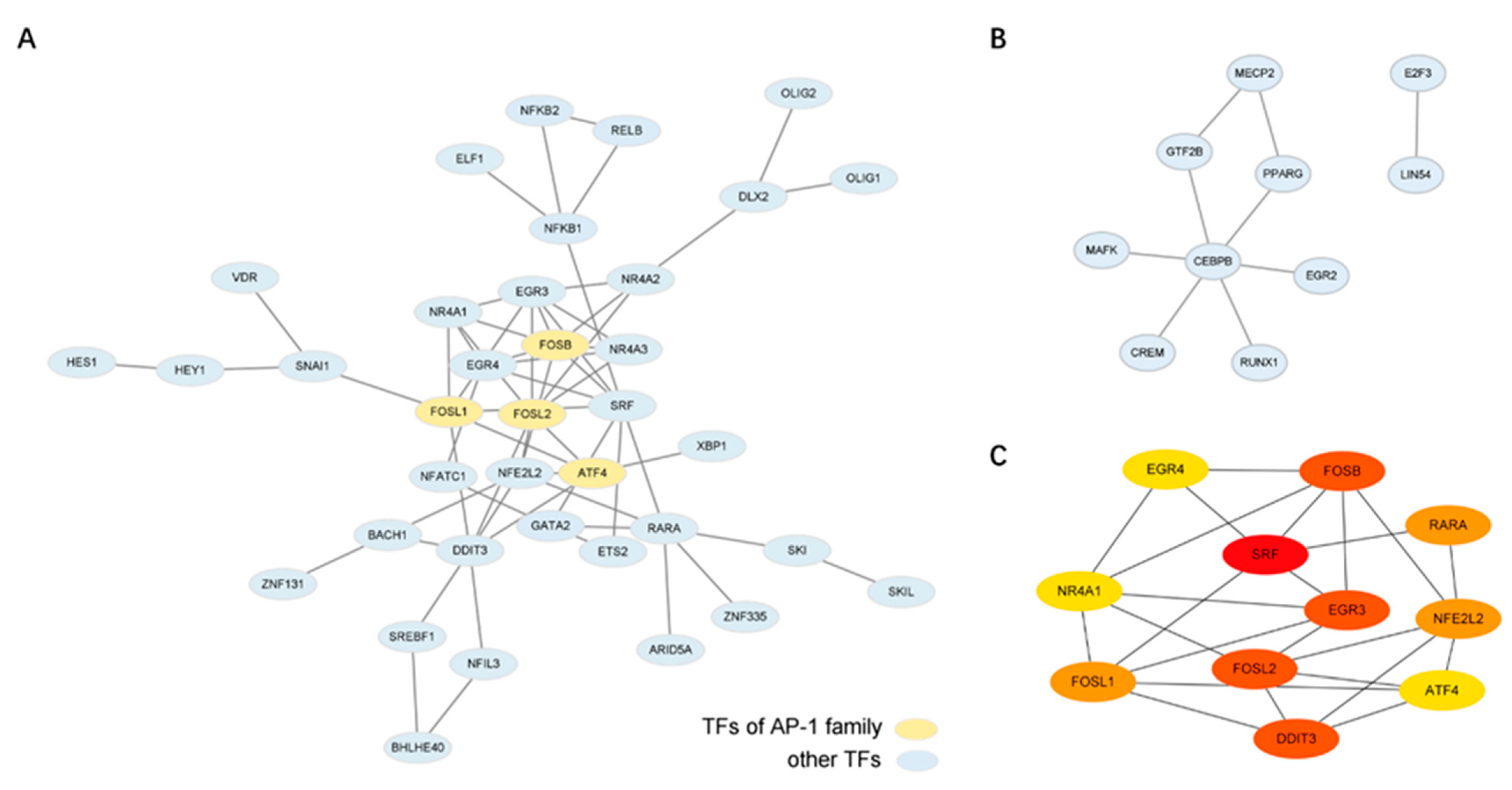
2.4. Bioinformatics Analysis Identifies the Potential Key Regulator TFs of This Transition
3. Discussion
4. Materials and Methods
4.1. Human Primary Monocyte Isolation and the M0 Macrophage Differentiation
4.2. Flow Cytometry
4.3. Immunofluorescence Microscopy
4.4. RNA-Seq Library Preparation and Sequencing
4.5. RNA-Seq Quality Control and Alignment to the Human Genome
4.6. Differentially Expressed Genes (DEGs) and the PCA Analysis
4.7. Identifying and Clustering the Transiently Differentially Expressed Transcription Factors
4.8. Protein-Protein Interaction Analysis
4.9. Transcription Factor (TF) Binding Site Motif Analysis
4.10. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.-A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage Plasticity, Polarization, and Function in Health and Disease. J. Cell Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef]
- Ginhoux, F.; Jung, S. Monocytes and Macrophages: Developmental Pathways and Tissue Homeostasis. Nat. Rev. Immunol. 2014, 14, 392–404. [Google Scholar] [CrossRef]
- Coillard, A.; Segura, E. In Vivo Differentiation of Human Monocytes. Front. Immunol. 2019, 10, 1907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epelman, S.; Lavine, K.J.; Randolph, G.J. Origin and Functions of Tissue Macrophages. Immunity 2014, 41, 21–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gessain, G.; Blériot, C.; Ginhoux, F. Non-Genetic Heterogeneity of Macrophages in Diseases-A Medical Perspective. Front. Cell Dev. Biol. 2020, 8, 613116. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lorenzini, P.A.; Zhang, F.; Xu, S.; Wong, M.S.M.; Zheng, J.; Roca, X. Alternative Splicing Analysis in Human Monocytes and Macrophages Reveals MBNL1 as Major Regulator. Nucleic Acids Res. 2018, 46, 6069–6086. [Google Scholar] [CrossRef]
- Phanstiel, D.H.; Van Bortle, K.; Spacek, D.; Hess, G.T.; Shamim, M.S.; Machol, I.; Love, M.I.; Aiden, E.L.; Bassik, M.C.; Snyder, M.P. Static and Dynamic DNA Loops Form AP-1-Bound Activation Hubs during Macrophage Development. Mol. Cell 2017, 67, 1037–1048.e6. [Google Scholar] [CrossRef] [Green Version]
- Baek, Y.-S.; Haas, S.; Hackstein, H.; Bein, G.; Hernandez-Santana, M.; Lehrach, H.; Sauer, S.; Seitz, H. Identification of Novel Transcriptional Regulators Involved in Macrophage Differentiation and Activation in U937 Cells. BMC Immunol. 2009, 10, 18. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Li, H.; Czajkowsky, D.M.; Shao, Z. Monocytic THP-1 Cells Diverge Significantly from Their Primary Counterparts: A Comparative Examination of the Chromosomal Conformations and Transcriptomes. Hereditas 2021, 158, 43. [Google Scholar] [CrossRef]
- Shiratori, H.; Feinweber, C.; Luckhardt, S.; Linke, B.; Resch, E.; Geisslinger, G.; Weigert, A.; Parnham, M.J. THP-1 and Human Peripheral Blood Mononuclear Cell-Derived Macrophages Differ in Their Capacity to Polarize in Vitro. Mol. Immunol. 2017, 88, 58–68. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, Q.; Liu, Y.; Sun, Q.; Li, H.; Czajkowsky, D.M.; Shao, Z. Massive Reorganization of the Genome during Primary Monocyte Differentiation into Macrophage. Acta Biochim. Biophys. Sin. (Shanghai) 2020, 52, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Pamer, E.G. Monocyte Recruitment during Infection and Inflammation. Nat. Rev. Immunol. 2011, 11, 762–774. [Google Scholar] [CrossRef] [Green Version]
- Martinez, F.O.; Gordon, S.; Locati, M.; Mantovani, A. Transcriptional Profiling of the Human Monocyte-to-Macrophage Differentiation and Polarization: New Molecules and Patterns of Gene Expression. J. Immunol. 2006, 177, 7303–7311. [Google Scholar] [CrossRef] [Green Version]
- Perera, L.P.; Waldmann, T.A. Activation of Human Monocytes Induces Differential Resistance to Apoptosis with Rapid down Regulation of Caspase-8/FLICE. Proc. Natl. Acad. Sci. USA 1998, 95, 14308–14313. [Google Scholar] [CrossRef] [Green Version]
- Klinder, A.; Markhoff, J.; Jonitz-Heincke, A.; Sterna, P.; Salamon, A.; Bader, R. Comparison of Different Cell Culture Plates for the Enrichment of Non-Adherent Human Mononuclear Cells. Exp. Ther. Med. 2019, 17, 2004–2012. [Google Scholar] [CrossRef] [Green Version]
- Shen, M.; Horbett, T.A. The Effects of Surface Chemistry and Adsorbed Proteins on Monocyte/Macrophage Adhesion to Chemically Modified Polystyrene Surfaces. J. Biomed. Mater. Res. 2001, 57, 336–345. [Google Scholar] [CrossRef]
- Musson, R.A. Human Serum Induces Maturation of Human Monocytes in Vitro. Changes in Cytolytic Activity, Intracellular Lysosomal Enzymes, and Nonspecific Esterase Activity. Am. J. Pathol. 1983, 111, 331–340. [Google Scholar] [PubMed]
- Vogel, D.Y.S.; Glim, J.E.; Stavenuiter, A.W.D.; Breur, M.; Heijnen, P.; Amor, S.; Dijkstra, C.D.; Beelen, R.H.J. Human Macrophage Polarization in Vitro: Maturation and Activation Methods Compared. Immunobiology 2014, 219, 695–703. [Google Scholar] [CrossRef]
- Herate, C.; Ramdani, G.; Grant, N.J.; Marion, S.; Gasman, S.; Niedergang, F.; Benichou, S.; Bouchet, J. Phospholipid Scramblase 1 Modulates FcR-Mediated Phagocytosis in Differentiated Macrophages. PLoS ONE 2016, 11, e0145617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, C.; Zhao, G.; Zhong, M.; Yue, Y.; Wu, L.; Xiong, S. RNA Sequencing and Transcriptomal Analysis of Human Monocyte to Macrophage Differentiation. Gene 2013, 519, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Wallner, S.; Schröder, C.; Leitão, E.; Berulava, T.; Haak, C.; Beißer, D.; Rahmann, S.; Richter, A.S.; Manke, T.; Bönisch, U.; et al. Epigenetic Dynamics of Monocyte-to-Macrophage Differentiation. Epigenet. Chromatin 2016, 9, 33. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, T.; Natoli, G. Transcriptional Regulation of Macrophage Polarization: Enabling Diversity with Identity. Nat. Rev. Immunol. 2011, 11, 750–761. [Google Scholar] [CrossRef]
- Lavin, Y.; Mortha, A.; Rahman, A.; Merad, M. Regulation of Macrophage Development and Function in Peripheral Tissues. Nat. Rev. Immunol. 2015, 15, 731–744. [Google Scholar] [CrossRef] [Green Version]
- Kelly, L.M.; Englmeier, U.; Lafon, I.; Sieweke, M.H.; Graf, T. MafB Is an Inducer of Monocytic Differentiation. EMBO J. 2000, 19, 1987–1997. [Google Scholar] [CrossRef] [PubMed]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple Combinations of Lineage-Determining Transcription Factors Prime Cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.; Wang, M.; Zhao, Y.; Wang, W. Taiji: System-Level Identification of Key Transcription Factors Reveals Transcriptional Waves in Mouse Embryonic Development. Sci. Adv. 2019, 5, eaav3262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuda, M.; Yamanaka, Y.; Uemura, M.; Osawa, M.; Saito, M.K.; Nagahashi, A.; Nishio, M.; Guo, L.; Ikegawa, S.; Sakurai, S.; et al. Recapitulating the Human Segmentation Clock with Pluripotent Stem Cells. Nature 2020, 580, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Telley, L.; Govindan, S.; Prados, J.; Stevant, I.; Nef, S.; Dermitzakis, E.; Dayer, A.; Jabaudon, D. Sequential Transcriptional Waves Direct the Differentiation of Newborn Neurons in the Mouse Neocortex. Science 2016, 351, 1443–1446. [Google Scholar] [CrossRef] [PubMed]
- Italiani, P.; Boraschi, D. From Monocytes to M1/M2 Macrophages: Phenotypical vs. Functional Differentiation. Front. Immunol. 2014, 5, 514. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Shi, B.; Huang, C.-C.; Eksarko, P.; Pope, R.M. Transcriptional Diversity during Monocyte to Macrophage Differentiation. Immunol. Lett. 2008, 117, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Skinner, B.M.; Johnson, E.E.P. Nuclear Morphologies: Their Diversity and Functional Relevance. Chromosoma 2017, 126, 195–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, R.; Gao, Y.; Dozmorov, I.; Malladi, V.; Saha, I.; McDaniel, M.M.; Parameswaran, S.; Liang, C.; Arana, C.; Zhang, B.; et al. IRF1 Governs the Differential Interferon-Stimulated Gene Responses in Human Monocytes and Macrophages by Regulating Chromatin Accessibility. Cell Rep. 2021, 34, 108891. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef] [Green Version]
- Sonnen, K.F.; Lauschke, V.M.; Uraji, J.; Falk, H.J.; Petersen, Y.; Funk, M.C.; Beaupeux, M.; François, P.; Merten, C.A.; Aulehla, A. Modulation of Phase Shift between Wnt and Notch Signaling Oscillations Controls Mesoderm Segmentation. Cell 2018, 172, 1079–1090.e12. [Google Scholar] [CrossRef] [Green Version]
- Göös, H.; Kinnunen, M.; Salokas, K.; Tan, Z.; Liu, X.; Yadav, L.; Zhang, Q.; Wei, G.-H.; Varjosalo, M. Human Transcription Factor Protein Interaction Networks. Nat. Commun. 2022, 13, 766. [Google Scholar] [CrossRef]
- Vierbuchen, T.; Ling, E.; Cowley, C.J.; Couch, C.H.; Wang, X.; Harmin, D.A.; Roberts, C.W.M.; Greenberg, M.E. AP-1 Transcription Factors and the BAF Complex Mediate Signal-Dependent Enhancer Selection. Mol. Cell 2017, 68, 1067–1082.e12. [Google Scholar] [CrossRef] [Green Version]
- Mufson, R.A. Induction of Immediate Early Response Genes by Macrophage Colony-Stimulating Factor in Normal Human Monocytes. J. Immunol. 1990, 145, 2333–2339. [Google Scholar] [PubMed]
- Kharbanda, S.; Nakamura, T.; Stone, R.; Hass, R.; Bernstein, S.; Datta, R.; Sukhatme, V.P.; Kufe, D. Expression of the Early Growth Response 1 and 2 Zinc Finger Genes during Induction of Monocytic Differentiation. J. Clin. Investig. 1991, 88, 571–577. [Google Scholar] [CrossRef]
- Hie, B.; Bryson, B.; Berger, B. Efficient Integration of Heterogeneous Single-Cell Transcriptomes Using Scanorama. Nat. Biotechnol. 2019, 37, 685–691. [Google Scholar] [CrossRef]
- Kurotaki, D.; Sasaki, H.; Tamura, T. Transcriptional Control of Monocyte and Macrophage Development. Int. Immunol. 2017, 29, 97–107. [Google Scholar] [CrossRef]
- Valledor, A.F.; Borràs, F.E.; Cullell-Young, M.; Celada, A. Transcription Factors That Regulate Monocyte/Macrophage Differentiation. J. Leukoc. Biol. 1998, 63, 405–417. [Google Scholar] [CrossRef]
- Liebermann, D.A.; Gregory, B.; Hoffman, B. AP-1 (Fos/Jun) Transcription Factors in Hematopoietic Differentiation and Apoptosis. Int. J. Oncol. 1998, 12, 685–700. [Google Scholar] [CrossRef]
- Wang, Y.-Z.; Fan, H.; Ji, Y.; Reynolds, K.; Gu, R.; Gan, Q.; Yamagami, T.; Zhao, T.; Hamad, S.; Bizen, N.; et al. Olig2 Regulates Terminal Differentiation and Maturation of Peripheral Olfactory Sensory Neurons. Cell Mol. Life Sci. 2020, 77, 3597–3609. [Google Scholar] [CrossRef]
- De Cesare, D.; Fimia, G.M.; Sassone-Corsi, P. CREM, a Master-Switch of the Transcriptional Cascade in Male Germ Cells. J. Endocrinol. Investig. 2000, 23, 592–596. [Google Scholar] [CrossRef]
- Mancinelli, S.; Vitiello, M.; Donnini, M.; Mantile, F.; Palma, G.; Luciano, A.; Arra, C.; Cerchia, L.; Liguori, G.L.; Fedele, M. The Transcription Regulator Patz1 Is Essential for Neural Stem Cell Maintenance and Proliferation. Front. Cell Dev. Biol. 2021, 9, 657149. [Google Scholar] [CrossRef]
- Oaks, A.W.; Zamarbide, M.; Tambunan, D.E.; Santini, E.; Di Costanzo, S.; Pond, H.L.; Johnson, M.W.; Lin, J.; Gonzalez, D.M.; Boehler, J.F.; et al. Cc2d1a Loss of Function Disrupts Functional and Morphological Development in Forebrain Neurons Leading to Cognitive and Social Deficits. Cereb. Cortex 2017, 27, 1670–1685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiyama, Y.; Griffith, R.; Miller, P.; Stevenson, G.W.; Lund, S.; Kanapa, D.J.; Stevenson, H.C. Effects of Adherence, Activation and Distinct Serum Proteins on the in Vitro Human Monocyte Maturation Process. J. Leukoc. Biol. 1988, 43, 224–231. [Google Scholar] [CrossRef]
- Nielsen, M.C.; Andersen, M.N.; Møller, H.J. Monocyte Isolation Techniques Significantly Impact the Phenotype of Both Isolated Monocytes and Derived Macrophages in Vitro. Immunology 2020, 159, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Zhang, X.; Chen, Z.; Robinson, M.K.; Simon, D.I. Leukocyte Integrin Mac-1 Recruits Toll/Interleukin-1 Receptor Superfamily Signaling Intermediates to Modulate NF-KappaB Activity. Circ. Res. 2001, 89, 859–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianchi, E.; Denti, S.; Granata, A.; Bossi, G.; Geginat, J.; Villa, A.; Rogge, L.; Pardi, R. Integrin LFA-1 Interacts with the Transcriptional Co-Activator JAB1 to Modulate AP-1 Activity. Nature 2000, 404, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Agarwal, M.; Mukherjee, R.; Sen, P.; Sinha, D.K. 3D Micro-Environment Regulates NF-Κβ Dependent Adhesion to Induce Monocyte Differentiation. Cell Death Dis. 2018, 9, 914. [Google Scholar] [CrossRef]
- Ivashkiv, L.B. Inflammatory Signaling in Macrophages: Transitions from Acute to Tolerant and Alternative Activation States. Eur. J. Immunol. 2011, 41, 2477–2481. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, W.; Chen, M.; Tang, Y.; Zhang, L.; Xu, Z.; Li, X.; Czajkowsky, D.M.; Shao, Z. Temporal Analysis Reveals the Transient Differential Expression of Transcription Factors That Underlie the Trans-Differentiation of Human Monocytes to Macrophages. Int. J. Mol. Sci. 2022, 23, 15830. https://doi.org/10.3390/ijms232415830
Deng W, Chen M, Tang Y, Zhang L, Xu Z, Li X, Czajkowsky DM, Shao Z. Temporal Analysis Reveals the Transient Differential Expression of Transcription Factors That Underlie the Trans-Differentiation of Human Monocytes to Macrophages. International Journal of Molecular Sciences. 2022; 23(24):15830. https://doi.org/10.3390/ijms232415830
Chicago/Turabian StyleDeng, Weihang, Min Chen, Ying Tang, Le Zhang, Zeqian Xu, Xinhui Li, Daniel M. Czajkowsky, and Zhifeng Shao. 2022. "Temporal Analysis Reveals the Transient Differential Expression of Transcription Factors That Underlie the Trans-Differentiation of Human Monocytes to Macrophages" International Journal of Molecular Sciences 23, no. 24: 15830. https://doi.org/10.3390/ijms232415830





