Genome-Wide Association Studies of Salt Tolerance at the Seed Germination Stage and Yield-Related Traits in Brassica napus L.
Abstract
1. Introduction
2. Results
2.1. Phenotypic Variation
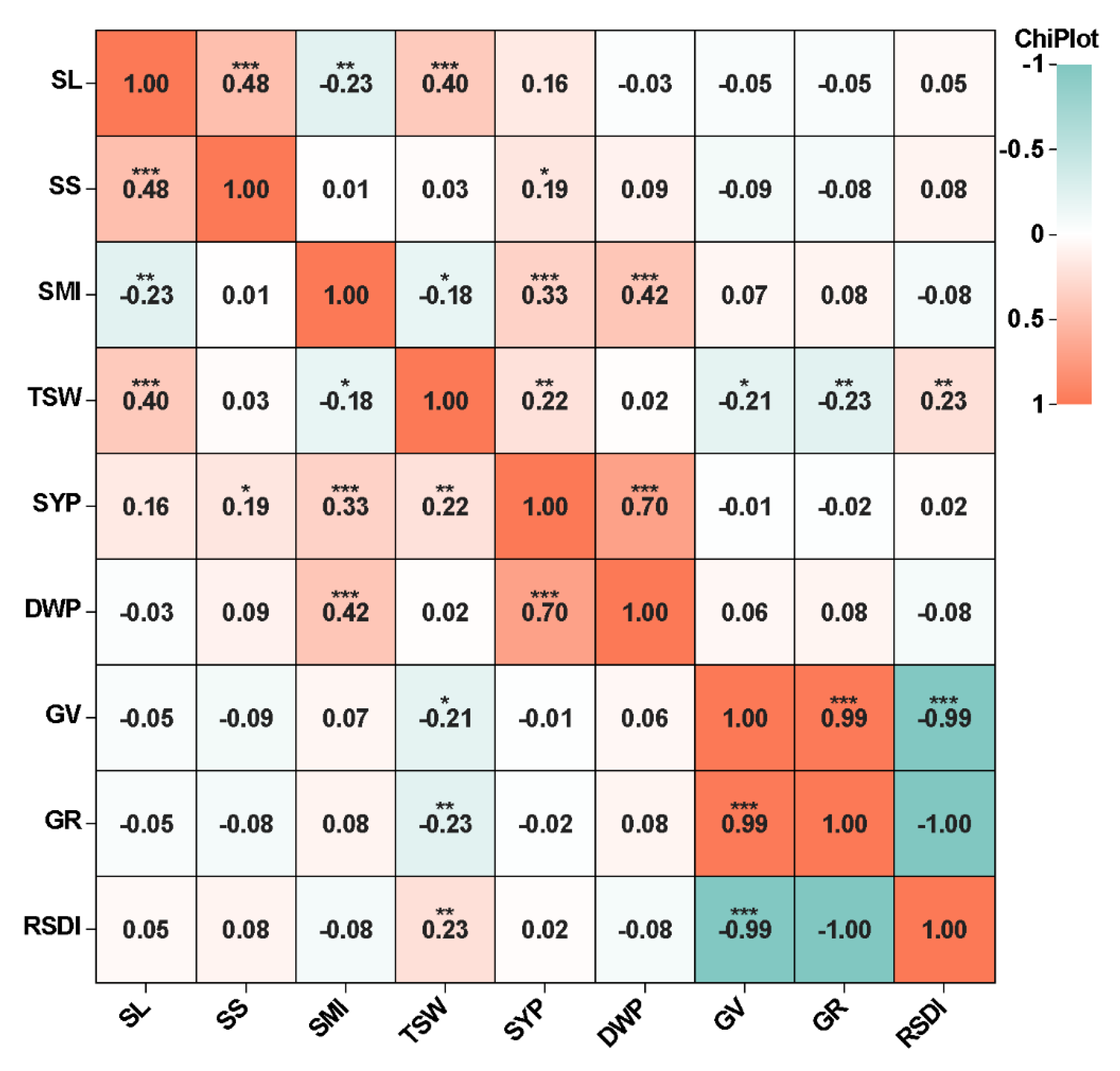
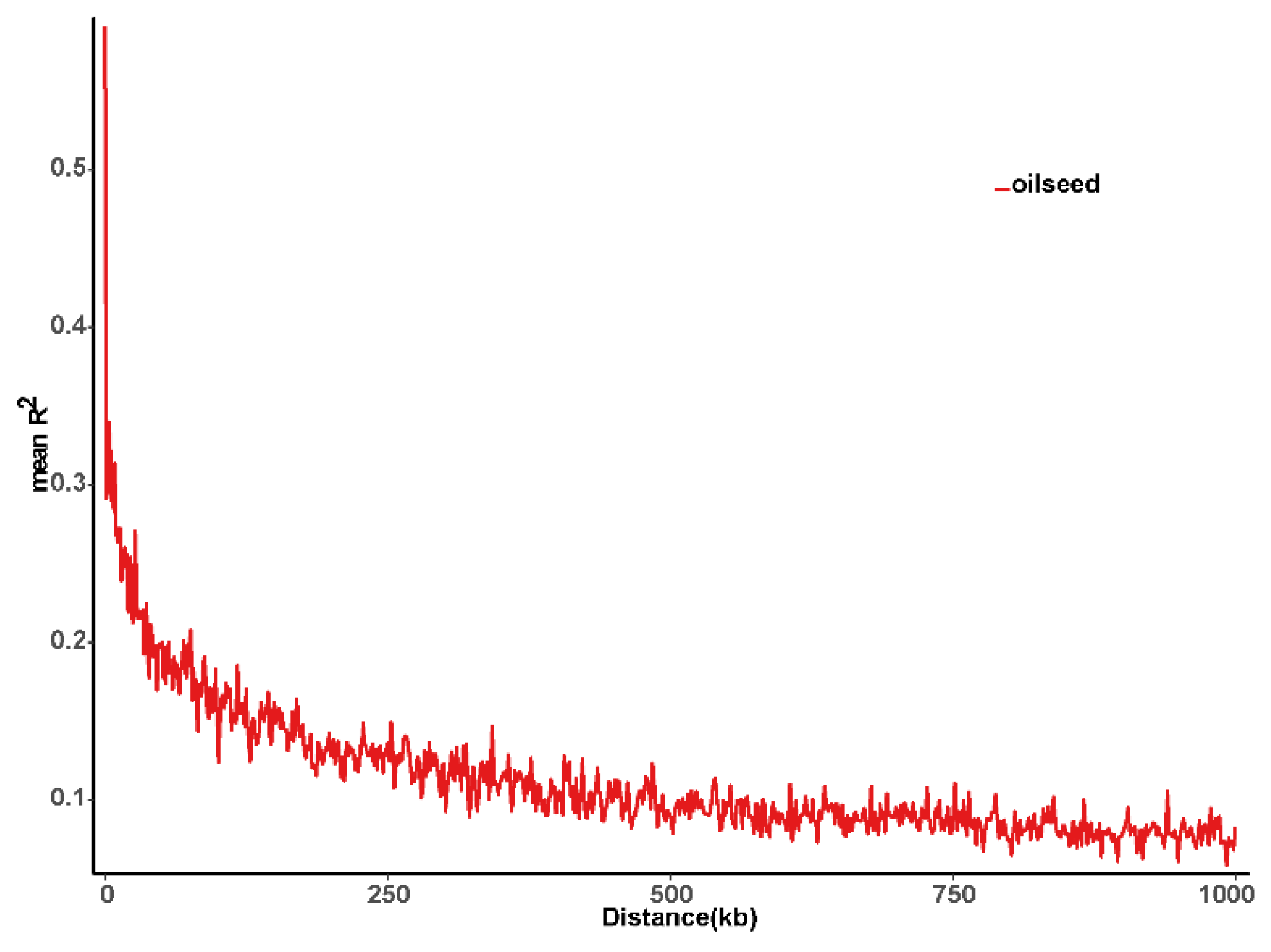
2.2. Genotyping Analysis and Linkage Disequilibrium
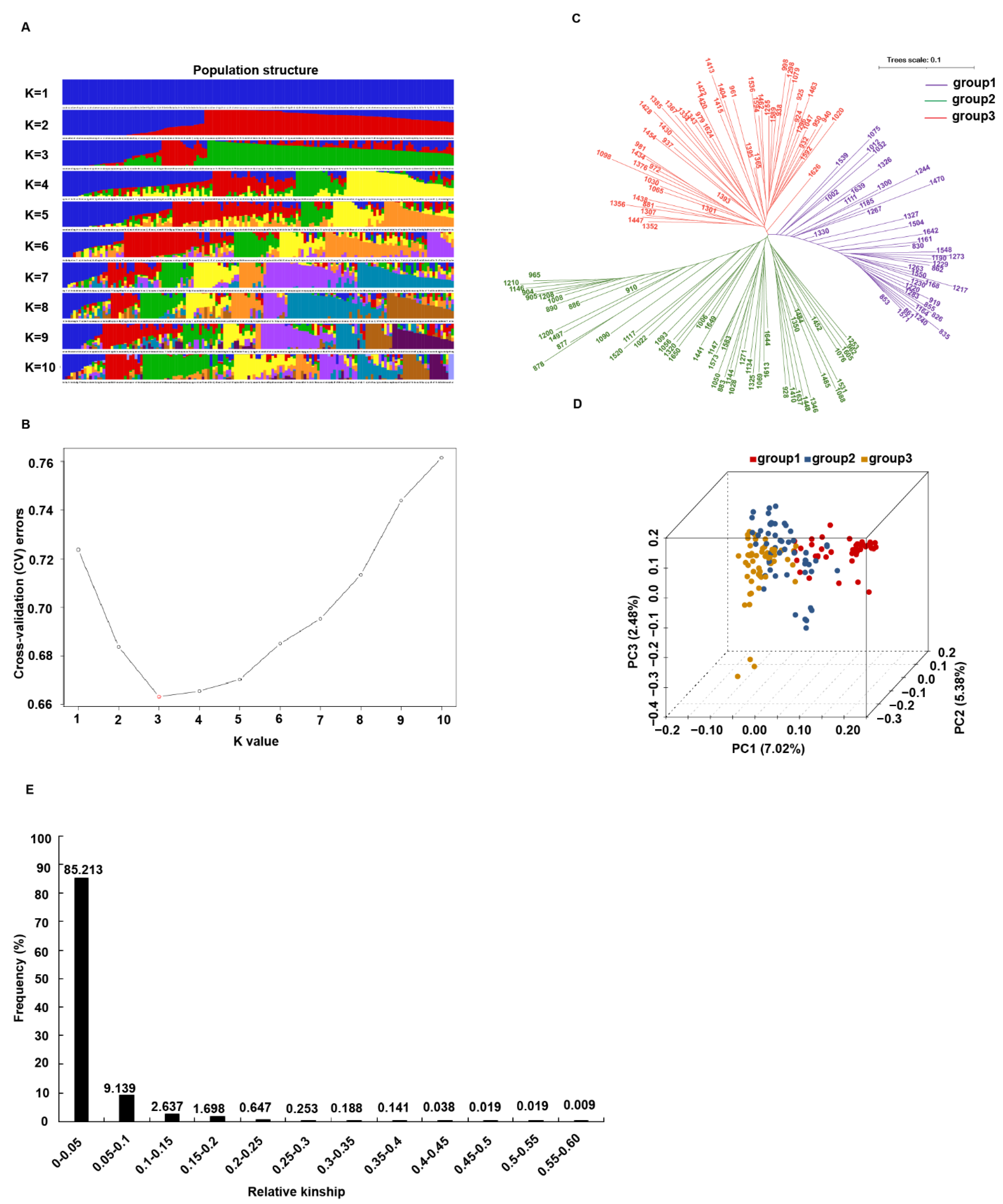
2.3. Population Structure and Kinship Analyses
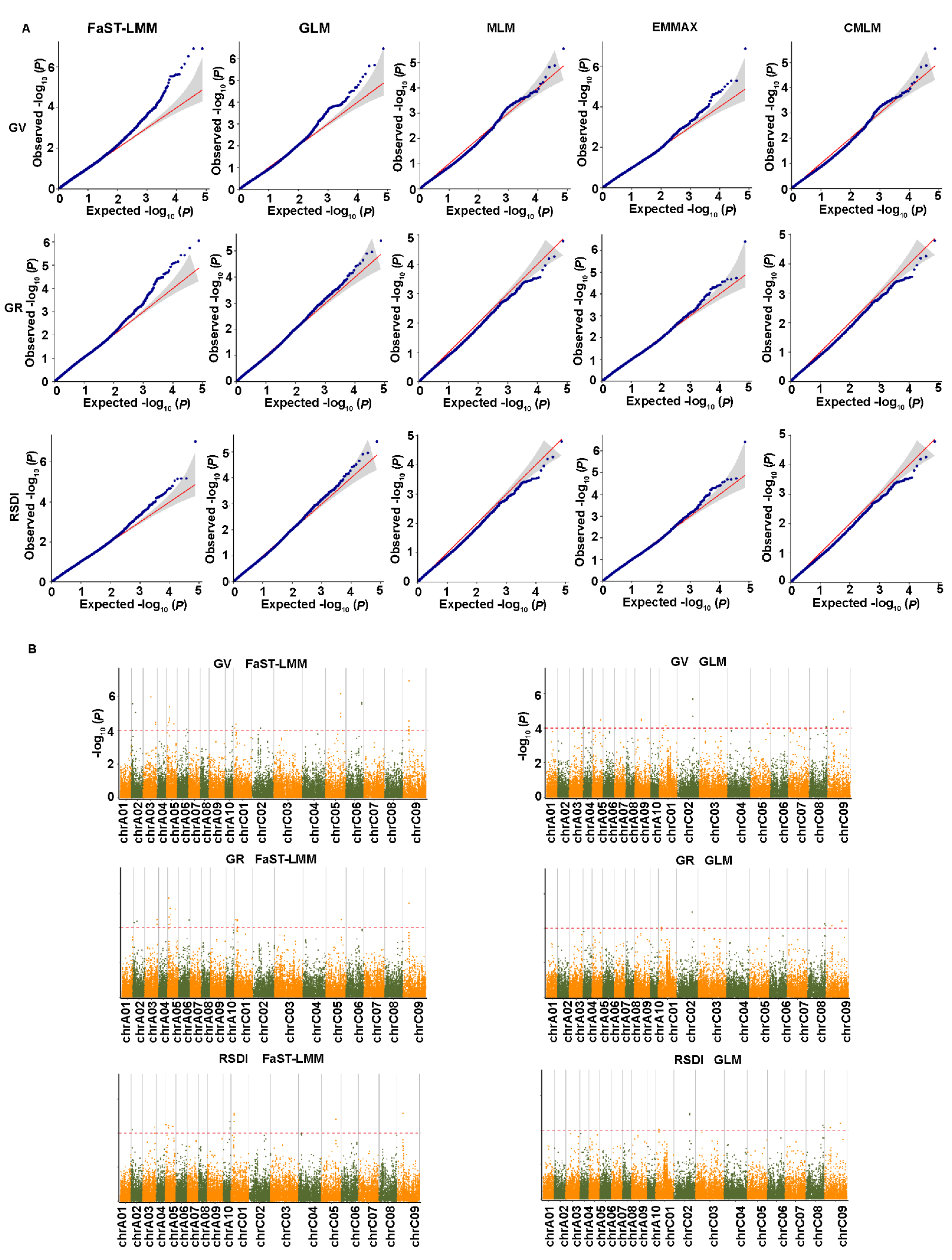
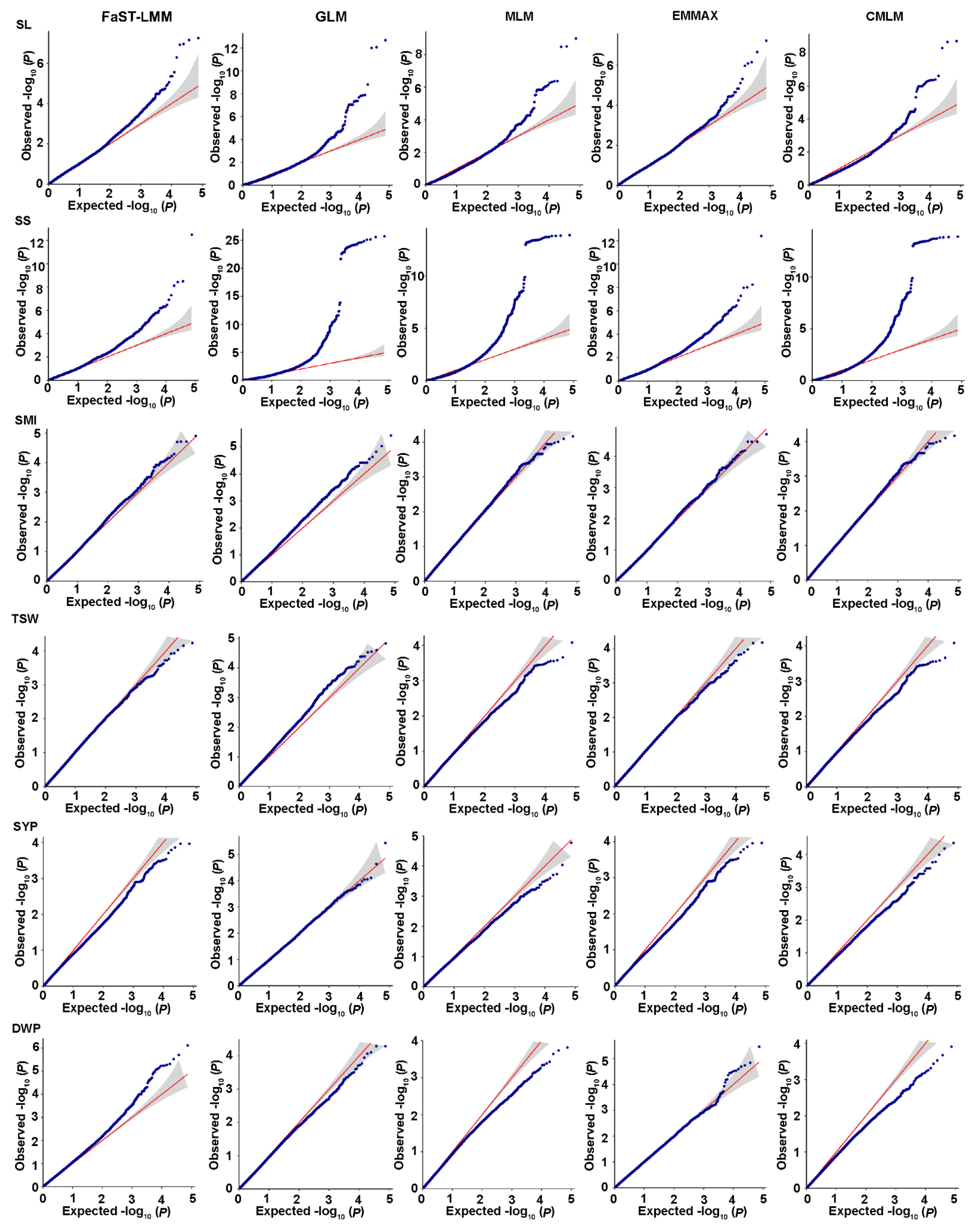
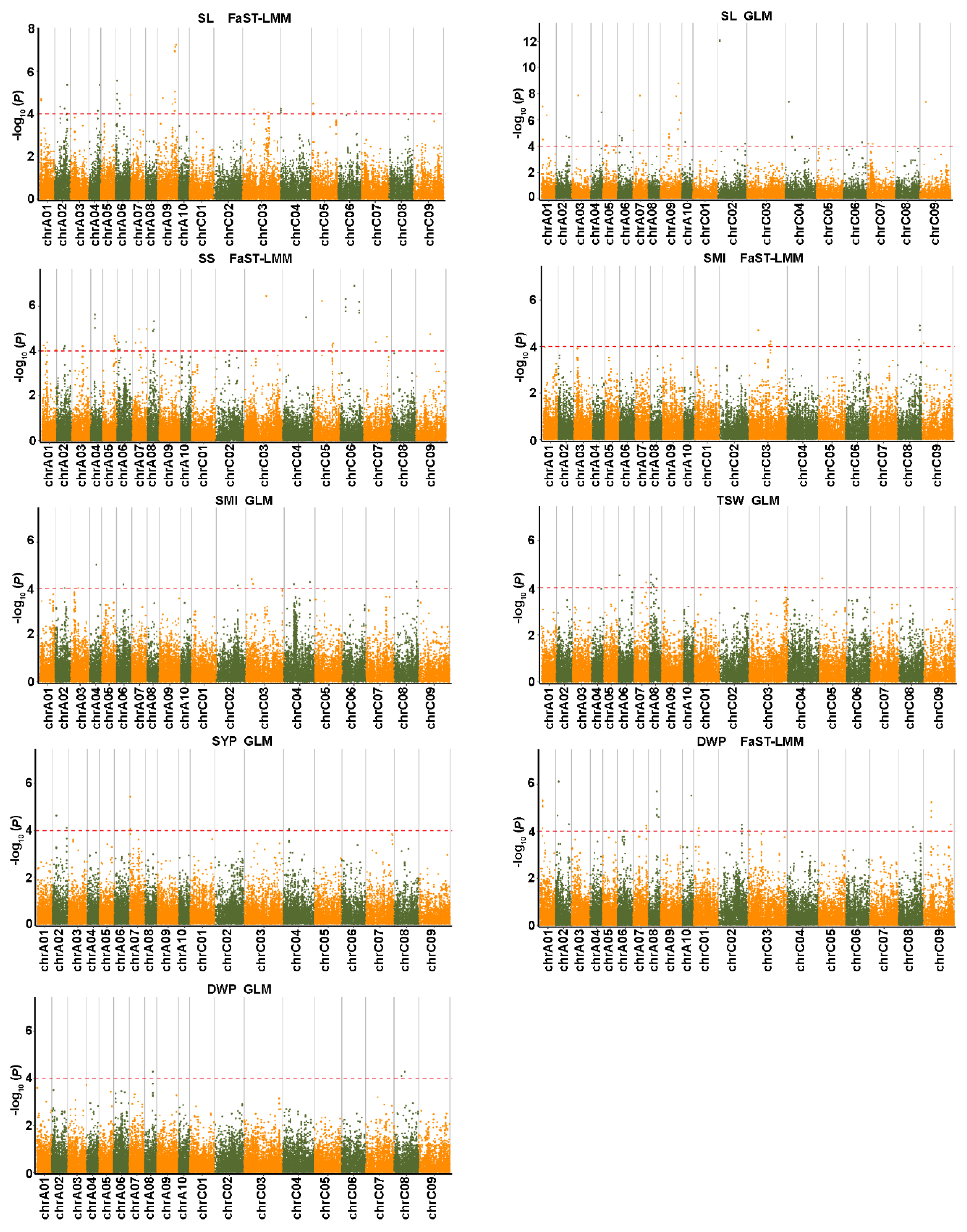
2.4. Genome-Wide Association Analysis for Salt-Related Traits
2.5. Genome-Wide Association Analysis for Yield-Related Traits
2.6. Associated SNPs for Salt Tolerance Co-Localized with QTLs for Yield-Related Traits
2.7. Comparative Genome Analysis for Salt Tolerance and Yield-Related Traits of the Present and Previous QTLs
2.8. Candidate Genes for Controlling Both Salt-Alkali Tolerance and Yield
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Phenotypic Evaluation and Statistical Analysis
4.3. SLAF Library Construction and Sequencing
4.4. SNP Genotyping Analysis
4.5. Population Structure and Linkage Disequilibrium Analysis
4.6. Genome-Wide Association Analysis
4.7. Genomic Comparison of Present and Previous QTLs for All Traits
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saeidnia, S.; Gohari, A.R. Importance of Brassica napus as a medicinal food plant. J. Med. Plant Res. 2012, 6, 2700–2703. [Google Scholar] [CrossRef]
- Wang, H. Analysis and Strategy for Current Domestic Edible Oil Supply. Chin. J. Oil Crop Sci. 2007, 29, 347–349. [Google Scholar]
- Singh, H.; Kumar, P.; Kumar, A.; Kyriacou, M.; Colla, G.; Rouphael, Y. Grafting Tomato as a Tool to Improve Salt Tolerance. Agronomy 2020, 10, 263. [Google Scholar] [CrossRef]
- Machado, R.M.A.; Serralheiro, R.P. Soil Salinity: Effect on Vegetable Crop Growth. Management Practices to Prevent and Mitigate Soil Salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Puppala, N.; Fowler, J.L.; Poindexter, L.; Bhardwaj, H.L. Evaluation of Salinity Tolerance of Canola Germination; Janick, J., Ed.; ASHS: Alexandria, VA, USA, 1999. [Google Scholar]
- Shabani, A.; Sepaskhah, A.R.; Kamgar-Haghighi, A.A. A model to predict the dry matter and yield of rapeseed under salinity and deficit irrigation. Arch. Agron. Soil Sci. 2014, 61, 525–542. [Google Scholar] [CrossRef]
- Hamdy, A.; Abdel-Dayem, S.; Abu-Zeid, M. Saline water management for optimum crop production. Agric. Water Manag. 1993, 24, 189–203. [Google Scholar] [CrossRef]
- Paterson, A.H.; Lander, E.S.; Hewitt, J.D.; Peterson, S.; Lincoln, S.E.; Tanksley, S.D. Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 1988, 335, 721–726. [Google Scholar] [CrossRef]
- Flowers, T.J. Improving crop salt tolerance. J. Exp. Bot. 2004, 55, 307–319. [Google Scholar] [CrossRef]
- Udall, J.A.; Quijada, P.A.; Lambert, B.; Osborn, T.C. Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 2. Identification of alleles from unadapted germplasm. Theor. Appl. Genet. 2006, 113, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Jiaqin, S.; Ruiyuan, L.; Dan, Q.; Congcong, J.; Yan, L.; Morgan, C.; Bancroft, I.; Jianyi, Z.; Jinling, M. Unraveling the Complex Trait of Crop Yield with Quantitative Trait Loci Mapping in Brassica Napus. Genetics 2009, 182, 851–861. [Google Scholar]
- Cai, D.; Xiao, Y.; Yang, W.; Ye, W.; Wang, B.; Younas, M.; Wu, J.; Liu, K. Association mapping of six yield-related traits in rapeseed (Brassica napus L.). Theor. Appl. Genet. 2013, 127, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Lang, L.N.; Xu, A.X.; Ding, J.; Zhang, Y.; Zhao, N.; Tian, Z.S.; Liu, Y.P.; Wang, Y.; Liu, X.; Liang, F.H.; et al. Quantitative Trait Locus Mapping of Salt Tolerance and Identification of Salt-Tolerant Genes in Brassica Napus L. Front. Plant Sci. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Sun, J.; Gu, S.; Wang, J.; Su, C.; Li, Y.; Ma, D.; Zhao, M.; Chen, W. Mining Beneficial Genes for Salt Tolerance from a Core Collection of Rice Landraces at the Seedling Stage Through Genome-Wide Association Mapping. Front. Plant Sci. 2022, 13, 1320. [Google Scholar] [CrossRef] [PubMed]
- Mandozai, A.; Moussa, A.A.; Zhang, Q.; Qu, J.; Du, Y.; Anwari, G.; Al Amin, N.; Wang, P. Genome-Wide Association Study of Root and Shoot Related Traits in Spring Soybean (Glycine max L.) at Seedling Stages Using SLAF-Seq. Front. Plant Sci. 2021, 12, 1598. [Google Scholar] [CrossRef]
- Wang, J.; Xian, X.; Xu, X.; Qu, C.; Lu, K.; Li, J.; Liu, L. Genome-Wide Association Mapping of Seed Coat Color in Brassica napus. J. Agric. Food Chem. 2017, 65, 5229–5237. [Google Scholar] [CrossRef]
- Yong, H.-Y.; Wang, C.; Bancroft, I.; Li, F.; Wu, X.; Kitashiba, H.; Nishio, T. Identification of a gene controlling variation in the salt tolerance of rapeseed (Brassica napus L.). Planta 2015, 242, 313–326. [Google Scholar] [CrossRef]
- He, Y.; Wu, D.; You, J.; Qian, W. Genome-Wide Association Analysis of Salt Tolerance Related Traits in Brassica Napus and Candidate Gene Prediction. Sci. Agric. Sin. 2017, 50, 1189–1201. [Google Scholar]
- Wan, H.; Chen, L.; Guo, J.; Li, Q.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; Fu, T.; Shen, J. Genome-Wide Association Study Reveals the Genetic Architecture Underlying Salt Tolerance-Related Traits in Rapeseed (Brassica napus L.). Front. Plant Sci. 2017, 8, 593. [Google Scholar] [CrossRef]
- Zhang, R.; Deng, W.; Yang, L.; Wang, Y.; Xiao, F.; He, J.; Lu, K. Genome-Wide Association Study of Root Length and Hypocotyl Length at Germination Stage under Saline Conditions in Brassica Napus. Sci. Agric. Sin. 2017, 50, 15–27. [Google Scholar]
- Wassan, G.M.; Khanzada, H.; Zhou, Q.; Mason, A.S.; Keerio, A.A.; Khanzada, S.; Solangi, A.M.; Faheem, M.; Fu, D.; He, H. Identification of genetic variation for salt tolerance in Brassica napus using genome-wide association mapping. Mol. Genet. Genom. 2021, 296, 391–408. [Google Scholar] [CrossRef]
- Zhang, G.; Zhou, J.; Peng, Y.; Tan, Z.; Li, L.; Yu, L.; Jin, C.; Fang, S.; Lu, S.; Guo, L.; et al. Genome-Wide Association Studies of Salt Tolerance at Seed Germination and Seedling Stages in Brassica napus. Front. Plant Sci. 2022, 12, 3003. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Peng, Y.; Zhou, J.; Tan, Z.; Jin, C.; Fang, S.; Zhong, S.; Jin, C.; Wang, R.; Wen, X.; et al. Genome-Wide Association Studies of Salt-Alkali Tolerance at Seedling and Mature Stages in Brassica napus. Front. Plant Sci. 2022, 13, 857149. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chen, B.; Xu, K.; Wu, J.; Song, W.; Bancroft, I.; Harper, A.L.; Trick, M.; Liu, S.; Gao, G.; et al. Genome-Wide Association Study Dissects the Genetic Architecture of Seed Weight and Seed Quality in Rapeseed (Brassica napus L.). DNA Res. 2014, 21, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Körber, N.; Bus, A.; Li, J.; Parkin, I.A.P.; Wittkop, B.; Snowdon, R.J.; Stich, B. Agronomic and Seed Quality Traits Dissected by Genome-Wide Association Mapping in Brassica napus. Front. Plant Sci. 2016, 7, 386. [Google Scholar] [CrossRef]
- Menendez, Y.C.; Sanchez, D.H.; Snowdon, R.J.; Rondanini, D.P.; Botto, J.F. Unraveling the impact on agronomic traits of the genetic architecture underlying plant-density responses in canola. J. Exp. Bot. 2021, 72, 5426–5441. [Google Scholar] [CrossRef]
- Song, J.; Li, J.; Sun, J.; Hu, T.; Wu, A.; Liu, S.; Wang, W.; Ma, D.; Zhao, M. Genome-Wide Association Mapping for Cold Tolerance in a Core Collection of Rice (Oryza sativa L.) Landraces by Using High-Density Single Nucleotide Polymorphism Markers from Specific-Locus Amplified Fragment Sequencing. Front. Plant Sci. 2018, 9, 875. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Ma, Q.; Cai, Z.; Xia, Q.; Li, S.; Jia, J.; Chu, L.; Lian, T.; Nian, H.; Cheng, Y. Identification and Mapping of Stable QTLs for Seed Oil and Protein Content in Soybean [Glycine max (L.) Merr.]. J. Agric. Food Chem. 2020, 68, 6448–6460. [Google Scholar] [CrossRef]
- Wang, R.J.; Gao, X.F.; Yang, J.; Kong, X.R. Genome-Wide Association Study to Identify Favorable SNP Allelic Variations and Candidate Genes That Control the Timing of Spring Bud Flush of Tea (Camellia sinensis) Using SLAF-seq. J. Agric. Food Chem. 2019, 67, 10380–10391. [Google Scholar] [CrossRef]
- Zheng, X.; Tang, Y.; Ye, J.; Pan, Z.; Tan, M.; Xie, Z.; Chai, L.; Xu, Q.; Fraser, P.D.; Deng, X. SLAF-Based Construction of a High-Density Genetic Map and Its Application in QTL Mapping of Carotenoids Content in Citrus Fruit. J. Agric. Food Chem. 2018, 67, 994–1002. [Google Scholar] [CrossRef] [PubMed]
- Khanzada, H.; Wassan, G.M.; He, H.; Mason, A.S.; Keerio, A.A.; Khanzada, S.; Faheem, M.; Solangi, A.M.; Zhou, Q.; Fu, D.; et al. Differentially evolved drought stress indices determine the genetic variation of Brassica napus at seedling traits by genome-wide association mapping. J. Adv. Res. 2020, 24, 447–461. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Clay, H.A.; Lynn, J.R.; Morris, K. Towards a genetic understanding of seed vigour in small-seeded crops using natural variation in Brassica oleracea. Plant Sci. 2010, 179, 582–589. [Google Scholar] [CrossRef]
- Hatzig, S.V.; Frisch, M.; Breuer, F.; Nesi, N.; Ducournau, S.; Wagner, M.H.; Leckband, G.; Abbadi, A.; Snowdon, R.J. Ge-nome-Wide Association Mapping Unravels the Genetic Control of Seed Germination and Vigor in Brassica Napus. Front. Plant Sci. 2015, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Foolad, M.R.; Lin, G.Y.; Chen, F.Q. Comparison of QTLs for Seed Germination under Non-Stress, Cold Stress and Salt Stress in Tomato. Plant Breed. 1999, 118, 167–173. [Google Scholar] [CrossRef]
- Wu, H.; Guo, J.; Wang, C.; Li, K.; Zhang, X.; Yang, Z.; Li, M.; Wang, B. An Effective Screening Method and a Reliable Screening Trait for Salt Tolerance of Brassica napus at the Germination Stage. Front. Plant Sci. 2019, 10, 530. [Google Scholar] [CrossRef]
- Wang, X.; Chen, L.; Wang, A.; Wang, H.; Tian, J.; Zhao, X.; Chao, H.; Zhao, Y.; Zhao, W.; Xiang, J.; et al. Quantitative trait loci analysis and genome-wide comparison for silique related traits in Brassica napus. BMC Plant Biol. 2016, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Delourme, R.; Falentin, C.; Fomeju, B.F.; Boillot, M.; Lassalle, G.; André, I.; Duarte, J.; Gauthier, V.; Lucante, N.; Marty, A.; et al. High-Density SNP-Based Genetic Map Development and Linkage Disequilibrium Assessment in Brassica Napus L. BMC Genom. 2013, 14, 1–18. [Google Scholar] [CrossRef]
- Qian, W.; Meng, J.; Li, M.; Frauen, M.; Sass, O.; Noack, J.; Jung, C. Introgression of genomic components from Chinese Brassica rapa contributes to widening the genetic diversity in rapeseed (B. napus L.), with emphasis on the evolution of Chinese rapeseed. Theor. Appl. Genet. 2006, 113, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Qian, W.; Snowdon, R.J. Sub-genomic selection patterns as a signature of breeding in the allopolyploid Brassica napus genome. BMC Genom. 2014, 15, 1170. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler, E.S. Structure of Linkage Disequilibrium in Plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhou, C.; Zheng, W.; Mason, A.S.; Fan, S.; Wu, C.; Fu, D.; Huang, Y. Genome-Wide SNP Markers Based on SLAF-Seq Uncover Breeding Traces in Rapeseed (Brassica napus L.). Front. Plant Sci. 2017, 8, 648. [Google Scholar] [CrossRef]
- Shiga, T. Rape Breeding by Interspecific Crossing between Brassica Napus and Brassica Campestris in Japan. Japan Agric. Res. Q. 1970, 5, 5–10. [Google Scholar]
- Zhou, H.; Xiao, X.; Asjad, A.; Han, D.; Zheng, W.; Xiao, G.; Huang, Y.; Zhou, Q. Integration of GWAS and transcriptome analyses to identify SNPs and candidate genes for aluminum tolerance in rapeseed (Brassica napus L.). BMC Plant Biol. 2022, 22, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Xie, M.; Liang, L.; Yang, L.; Han, H.; Qin, X.; Zhao, J.; Hou, Y.; Dai, W.; Du, C.; et al. Ge-nome-Wide Association Analysis Combined With Quantitative Trait Loci Mapping and Dynamic Transcriptome Unveil the Genetic Control of Seed Oil Content in Brassica Napus L. Front. Plant Sci. 2022, 13, 929197. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.-T.; Wang, T.-Y.; Jian, H.-J.; Wang, J.; Li, J.-N.; Liu, L.-Z. QTL Mapping for Seedling Dry Weight and Fresh Weight under Salt Stress and Candidate Genes Analysis in Brassica napus L. Acta Agron. Sin. 2017, 43, 179–189. [Google Scholar] [CrossRef]
- Zorić, M.; Dodig, D.; Kobiljski, B.; Quarrie, S.; Barnes, J. Population structure in a wheat core collection and genomic loci associated with yield under contrasting environments. Genetica 2012, 140, 259–275. [Google Scholar] [CrossRef]
- Ungerer, M.C.; Halldorsdottir, S.S.; Purugganan, M.D.; Mackay, T.F.C. Genotype-Environment Interactions at Quantitative Trait Loci Affecting Inflorescence Development in Arabidopsis thaliana. Genetics 2003, 165, 353–365. [Google Scholar] [CrossRef]
- Hu, J.; Chen, B.; Zhao, J.; Zhang, F.; Xie, T.; Xu, K.; Gao, G.; Yan, G.; Li, H.; Li, L. Genomic Selection and Genetic Architecture of Ag-ronomic Traits during Modern Rapeseed Breeding. Nat. Genet. 2022, 54, 694–704. [Google Scholar] [CrossRef]
- Inoue, S.; Hayashi, M.; Huang, S.; Yokosho, K.; Gotoh, E.; Ikematsu, S.; Okumura, M.; Suzuki, T.; Kamura, T.; Kinoshita, T.; et al. A tonoplast-localized magnesium transporter is crucial for stomatal opening in Arabidopsis under high Mg2+ conditions. New Phytol. 2022, 236, 864–877. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.-Z.; Song, L.-F.; Zou, J.-J.; Su, Z.; Wu, W.-H. Transcriptome Analyses Show Changes in Gene Expression to Accompany Pollen Germination and Tube Growth in Arabidopsis. Plant Physiol. 2008, 148, 1201–1211. [Google Scholar] [CrossRef]
- Jin, G.; Liu, F.; Ma, H.; Hao, S.; Zhao, Q.; Bian, Z.; Jia, Z.; Song, S. Two G-protein-coupled-receptor candidates, Cand2 and Cand7, are involved in Arabidopsis root growth mediated by the bacterial quorum-sensing signals N-acyl-homoserine lactones. Biochem. Biophys. Res. Commun. 2012, 417, 991–995. [Google Scholar] [CrossRef]
- Malamy, J.E. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 2005, 28, 67–77. [Google Scholar] [CrossRef]
- Merlot, S.; Leonhardt, N.; Fenzi, F.; Valon, C.; Costa, J.; Piette, L.; Vavasseur, A.; Genty, B.; Boivin, K.; Müller, A.; et al. Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 2007, 26, 3216–3226. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.-K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Kunz, H.-H.; Schroeder, J.I.; Kemp, G.; Young, H.S.; Neuhaus, H.E. Decreased capacity for sodium export out of Arabidopsis chloroplasts impairs salt tolerance, photosynthesis and plant performance. Plant J. 2014, 78, 646–658. [Google Scholar] [CrossRef] [PubMed]
- Galon, Y.; Finkler, A.; Fromm, H. Calcium-Regulated Transcription in Plants. Mol. Plant 2010, 3, 653–669. [Google Scholar] [CrossRef]
- Pandey, N.; Ranjan, A.; Pant, P.; Tripathi, R.K.; Ateek, F.; Pandey, H.P.; Patre, U.V.; Sawant, S.V. CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genom. 2013, 14, 216. [Google Scholar] [CrossRef]
- Kawaura, K.; Mochida, K.; Ogihara, Y. Genome-wide analysis for identification of salt-responsive genes in common wheat. Funct. Integr. Genom. 2008, 8, 277–286. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Jung, C.; Jun, S.S.; Sang, W.H.; Yeon, J.K.; Chung, H.K.; Sang, I.S.; Baek, H.N.; Yang, D.C.; Cheong, J.J. Overexpression of AtMYB44 Enhances Stomatal Closure to Confer Abiotic Stress Tolerance in Transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.-S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and Functional Analysis of Arabidopsis Stress-Inducible NAC Transcription Factors That Bind to a Drought-Responsive cis-Element in the early responsive to dehydration stress 1 Promoter[W]. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Alshareef, N.O.; Otterbach, S.L.; Allu, A.D.; Woo, Y.H.; de Werk, T.; Kamranfar, I.; Mueller-Roeber, B.; Tester, M.; Balazadeh, S.; Schmöckel, S.M. NAC transcription factors ATAF1 and ANAC055 affect the heat stress response in Arabidopsis. Sci. Rep. 2022, 12, 1–15. [Google Scholar] [CrossRef]
- Qin, F.; Sakuma, Y.; Tran, L.-S.P.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Fujita, M.; Umezawa, T.; Sawano, Y.; Miyazono, K.-I.; et al. Arabidopsis DREB2A-Interacting Proteins Function as RING E3 Ligases and Negatively Regulate Plant Drought Stress–Responsive Gene Expression. Plant Cell 2008, 20, 1693–1707. [Google Scholar] [CrossRef]
- Chen, Z.; Hong, X.; Zhang, H.; Wang, Y.; Li, X.; Zhu, J.-K.; Gong, Z. Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 2005, 43, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Boudsocq, M.; Barbier-Brygoo, H.; Laurière, C. Identification of Nine Sucrose Nonfermenting 1-related Protein Kinases 2 Activated by Hyperosmotic and Saline Stresses in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 41758–41766. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Zhang, J.; Hsu, P.-K.; Ceciliato, P.H.O.; Zhang, L.; Dubeaux, G.; Munemasa, S.; Ge, C.; Zhao, Y.; Hauser, F.; et al. MAP3Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kant, P.; Kant, S.; Gordon, M.; Shaked, R.; Barak, S. STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, Two DEAD-Box RNA Helicases That Attenuate Arabidopsis Responses to Multiple Abiotic Stresses. Plant Physiol. 2007, 145, 814–830. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, Y.; Xu, C.; Ren, J.; Liu, X.; Black, K.; Gai, X.; Wang, Q.; Ren, H. Cucumber ECERIFERUM1 (CsCER1), which influences the cuticle properties and drought tolerance of cucumber, plays a key role in VLC alkanes biosynthesis. Plant Mol. Biol. 2014, 87, 219–233. [Google Scholar] [CrossRef]
- Liang, J.; Ma, Y.; Wu, J.; Cheng, F.; Liu, B.; Wang, X. Map-based cloning of the dominant genic male sterile Ms-cd1 gene in cabbage (Brassica oleracea). Theor. Appl. Genet. 2016, 130, 71–79. [Google Scholar] [CrossRef]
- Lobell, D.B.; Roberts, M.J.; Schlenker, W.; Braun, N.; Little, B.B.; Rejesus, R.M.; Hammer, G.L. Greater Sensitivity to Drought Accompanies Maize Yield Increase in the U.S. Midwest. Science 2014, 344, 516–519. [Google Scholar] [CrossRef]
- Chen, B.; Xu, K.; Li, J.; Li, F.; Qiao, J.; Li, H.; Gao, G.; Yan, G.; Wu, X. Evaluation of yield and agronomic traits and their genetic variation in 488 global collections of Brassica napus L. Genet. Resour. Crop Evol. 2014, 61, 979–999. [Google Scholar] [CrossRef]
- Doyle, J.J. Isolation of Plant DNA from Fresh Tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Sun, X.; Liu, D.; Zhang, X.; Li, W.; Liu, H.; Hong, W.; Jiang, C.; Guan, N.; Ma, C.; Zeng, H.; et al. SLAF-seq: An Efficient Method of Large-Scale De Novo SNP Discovery and Genotyping Using High-Throughput Sequencing. PLoS ONE 2013, 8, e58700. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, C.; Li, Y.; Lam, T.-W.; Yiu, S.-M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Note, P. Spagedi: A Versatile Computer Program to Analyse Spatial. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Lippert, C.; Listgarten, J.; Liu, Y.; Kadie, C.M.; Davidson, R.I.; Heckerman, D. FaST linear mixed models for genome-wide association studies. Nat. Methods 2011, 8, 833–835. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.-Y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef]
- Govta, N.; Polda, I.; Sela, H.; Cohen, Y.; Beckles, D.M.; Korol, A.B.; Fahima, T.; Saranga, Y.; Krugman, T. Genome-Wide As-sociation Study in Bread Wheat Identifies Genomic Regions Associated with Grain Yield and Quality under Contrasting Water Availability. Int. J. Mol. Sci. 2022, 23, 10575. [Google Scholar] [CrossRef]
- Turner, S.D. Qqman: An R Package for Visualizing GWAS Results Using Q-Q and Manhattan Plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- Quijada, P.A.; Udall, J.A.; Lambert, B.; Osborn, T.C. Quantitative Trait Analysis of Seed Yield and Other Complex Traits in Hybrid Spring Rapeseed (Brassica Napus L.): 1. IdentiWcation of Genomic Regions from Winter Germplasm. Theor. Appl. Genet. 2006, 113, 549–561. [Google Scholar] [CrossRef]
- Basunanda, P.; Radoev, M.; Ecke, W.; Friedt, W.; Becker, H.C.; Snowdon, R.J. Comparative mapping of quantitative trait loci involved in heterosis for seedling and yield traits in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 2009, 120, 271–281. [Google Scholar] [CrossRef]
- Shi, Y.; Zhang, X.; Xu, Z.-Y.; Li, L.; Zhang, C.; Schläppi, M. Influence of EARLI1-like genes on flowering time and lignin synthesis of Arabidopsis thaliana. Plant Biol. 2011, 13, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Shu, C.; Chen, L.; Xu, J.; Wu, J.; Liu, K. Identification of a major QTL for silique length and seed weight in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 2012, 125, 285–296. [Google Scholar] [CrossRef]
- Qi, L.; Mao, L.; Sun, C.; Pu, Y.; Fu, T.; Ma, C.; Shen, J.; Tu, J.; Yi, B.; Wen, J. Interpreting the Genetic Basis of Silique Traits in Brassica Napus Using a Joint QTL Network. Plant Breed. 2014, 133, 52–60. [Google Scholar] [CrossRef]
- Fu, Y.; Wei, D.; Dong, H.; He, Y.; Cui, Y.; Mei, J.; Wan, H.; Li, J.; Snowdon, R.; Friedt, W.; et al. Comparative Quan-titative Trait Loci for Silique Length and Seed Weight in Brassica Napus. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef]
- Geng, X.; Jiang, C.; Yang, J.; Wang, L.; Wu, X.; Wei, W. Rapid Identification of Candidate Genes for Seed Weight Using the SLAF-Seq Method in Brassica napus. PLoS ONE 2016, 11, e0147580. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, X.; Wang, H.; Tian, J.; Li, B.; Chen, L.; Chao, H.; Long, Y.; Xiang, J.; Gan, J.; et al. Genome-Wide Identification of QTL for Seed Yield and Yield-Related Traits and Construction of a High-Density Consensus Map for QTL Comparison in Brassica napus. Front. Plant Sci. 2016, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Wang, M.; Long, Y.; Huang, Y.; Shi, L.; Zhang, C.; Liu, X.; Fitt, B.D.L.; Xiang, J.; Mason, A.S.; et al. Incorporating pleiotropic quantitative trait loci in dissection of complex traits: Seed yield in rapeseed as an example. Theor. Appl. Genet. 2017, 130, 1569–1585. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, Y.; Li, S.; Ge, X.; Li, Z. High Density Linkage Map Construction and QTL Detection for Three Silique-Related Traits in Orychophragmus violaceus Derived Brassica napus Population. Front. Plant Sci. 2017, 8, 1512. [Google Scholar] [CrossRef]
- Ye, J.; Yang, Y.; Chen, B.; Shi, J.; Luo, M.; Zhan, J.; Wang, X.; Liu, G.; Wang, H. An integrated analysis of QTL mapping and RNA sequencing provides further insights and promising candidates for pod number variation in rapeseed (Brassica napus L.). BMC Genom. 2017, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Tan, C.; Li, Y.; He, Y.; Wei, S.; Cui, Y.; Chen, Y.; Wei, D.; Fu, Y.; He, Y.; et al. Genome-Wide Association Study Reveals Both Overlapping and Independent Genetic Loci to Control Seed Weight and Silique Length in Brassica napus. Front. Plant Sci. 2018, 9, 921. [Google Scholar] [CrossRef]
- Raboanatahiry, N.; Chao, H.; Dalin, H.; Pu, S.; Yan, W.; Yu, L.; Wang, B.; Li, M. QTL Alignment for Seed Yield and Yield Related Traits in Brassica napus. Front. Plant Sci. 2018, 9, 1127. [Google Scholar] [CrossRef]
- Li, B.; Gao, J.; Chen, J.; Wang, Z.; Shen, W.; Yi, B.; Wen, J.; Ma, C.; Shen, J.; Fu, T.; et al. Identification and fine mapping of a major locus controlling branching in Brassica napus. Theor. Appl. Genet. 2020, 133, 771–783. [Google Scholar] [CrossRef]
- Wang, H.; Zaman, Q.U.; Huang, W.; Mei, D.; Liu, J.; Wang, W.; Ding, B.; Hao, M.; Fu, L.; Cheng, H.; et al. QTL and Candidate Gene Identification for Silique Length Based on High-Dense Genetic Map in Brassica napus L. Front. Plant Sci. 2019, 10, 1579. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yu, K.; Pang, C.; Wu, X.; Shi, R.; Sun, C.; Zhang, W.; Chen, F.; Zhang, J.; Wang, X. QTL Analysis of Five Silique-Related Traits in Brassica napus L. Across Multiple Environments. Front. Plant Sci. 2021, 12, 766271. [Google Scholar] [CrossRef] [PubMed]
| Trait | Year | Mean ± SD | Range | Variance | CV (%) |
|---|---|---|---|---|---|
| SL (cm) | 2018 | 5.74 ± 0.91 | 4.32–10.86 | 0.83 | 15.85 |
| 2019 | 6.56 ± 1.13 | 4.34–12.71 | 1.28 | 17.23 | |
| 2020 | 7.72 ± 1.06 | 5.19–12.06 | 1.12 | 13.73 | |
| SS (number) | 2018 | 15.76 ± 2.60 | 9.00–22.93 | 6.75 | 16.50 |
| 2019 | 22.84 ± 3.90 | 9.94–34.73 | 15.19 | 17.08 | |
| 2020 | 26.50 ± 4.05 | 12.67–35.10 | 16.41 | 15.28 | |
| SMI (number) | 2018 | 50.02 ± 11.69 | 28.40–88.27 | 136.58 | 23.37 |
| 2019 | 61.02 ± 12.30 | 26.33–111.67 | 151.27 | 20.16 | |
| 2020 | 71.01 ± 14.23 | 25.25–111.67 | 202.44 | 20.04 | |
| TSW (g) | 2018 | 5.07 ± 0.86 | 3.07–7.28 | 0.75 | 16.96 |
| 2019 | 4.02 ± 0.83 | 2.20–7.27 | 0.70 | 20.65 | |
| 2020 | 3.11 ± 0.72 | 1.69–5.45 | 0.52 | 23.15 | |
| SYP (g) | 2018 | 16.20 ± 5.03 | 4.10–33.24 | 25.26 | 31.05 |
| 2019 | 26.12 ± 9.77 | 6.67–50.00 | 95.45 | 37.40 | |
| 2020 | 22.42 ± 8.53 | 5.37–47.00 | 72.80 | 38.05 | |
| DWP (g) | 2018 | 77.60 ± 15.90 | 38.67–127.33 | 252.69 | 20.49 |
| 2019 | 99.48 ± 27.33 | 46.67–183.33 | 747.01 | 27.47 | |
| 2020 | 144.31 ± 28.69 | 80.00–215.00 | 823.28 | 19.88 | |
| GV (%) | 11.14 ± 14.86 | 0.00–72.00 | 220.90 | 133.00 | |
| GR (%) | 14.13 ± 17.00 | 0.00–76.50 | 288.89 | 120.00 | |
| RSDI (%) | 85.87 ± 17.00 | 23.50–100.00 | 288.89 | 20.00 |
| Yield-Related Traits | Salt-Related Traits | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Trait | QTL | Interval (bp) | Peak SNP (bp) | Trait | Chr | SNP | SNP Position (bp) | Rep1 | Rep2 |
| A02 | DWP/SMI | pqA2.1 | 1,897,176–3,836,751 | 2,397,176/3,336,751 | GV/GR/RSDI | A02 | rsA02_2496879 | 2,496,879 | √ | √ |
| GR | A02 | rsA02_3530820 | 3,530,820 | √ | ||||||
| SL | qSL18–A2.1 | 7,907,803–8,907,803 | 8,407,803 | GV/GR | A02 | rsA02_8228124 | 8,228,124 | √ | √ | |
| qSL19–A2.2 | 8,051,356–9,051,356 | 8,551,356 | ||||||||
| A05 | SL | qSL18–A5.2 | 6,194,267–7,194,267 | 6,694,267 | GV/GR/RSDI | A05 | rsA05_6670181 | 6,670,181 | √ | √ |
| GV/GR/RSDI | A05 | rsA05_6692257 | 6,692,257 | √ | √ | |||||
| GV | A05 | rsA05_6692275 | 6,692,275 | √ | √ | |||||
| SMI | qSMI20–A5.1 | 18,168,998–19,168,998 | 18,668,998 | GR | A05 | rsA05_18700305 | 18,700,305 | √ | ||
| A09 | SL | qSL18–A9.6 | 31,410,855–32,410,855 | 31,910,855 | GR | A09 | rsA09_31840807 | 31,840,807 | √ | |
| C04 | SL | qSL18–C4.2 | 5,051,524–6,051,524 | 5,551,524 | RSDI | C04 | rsC04_5933538 | 5,933,538 | √ | |
| RSDI | C04 | rsC04_5933539 | 5,933,539 | √ | ||||||
| RSDI | C04 | rsC04_5933579 | 5,933,579 | √ | ||||||
| RSDI | C04 | rsC04_5933820 | 5,933,820 | √ | ||||||
| C09 | DWP/SS | pqC9.1 | 12,405,354–13,405,354 | 12,521,248/12,521,335/12,724,461/12,976,490/12,976,522/12,905,354 | GR | C09 | rsC09_12521248 | 12,521,248 | √ | |
| GV/GR | C09 | rsC09_12521335 | 12,521,335 | √ | √ | |||||
| GV | C09 | rsC09_12645583 | 12,645,583 | √ | ||||||
| GV | C09 | rsC09_12645586 | 12,645,586 | √ | ||||||
| GV/GR/RSDI | C09 | rsC09_12949548 | 12,949,548 | √ | √ | |||||
| GV/GR/RSDI | C09 | rsC09_12949561 | 12,949,561 | √ | √ | |||||
| Gene Name | Chr | Site_strat (bp) | Site_end (bp) | Distance (bp) | SNP Location (bp) | Orthologous Gene ID in Arabidopsis | Gene Name in Arabidopsis | Function Description in Arabidopsis |
|---|---|---|---|---|---|---|---|---|
| BnaA06g33990D | A06 | 22,513,737 | 22,516,788 | 5′_42149 | 22,471,588, 22,471,644 | AT2G02820 | MYB88 | a putative transcription factor (MYB88) |
| BnaA03g33890D | A03 | 16,409,029 | 16,410,113 | 5′_33265 | 16,375,764 | AT3G15500 | ANAC55 | NAC domain-containing protein 55 |
| BnaC06g34470D | C06 | 33,929,460 | 33,934,599 | 3′_86296 | 33,843,164, 33,843,483, 33,843,558 | AT1G73660 | M3KDELTA6 | serine/threonine–protein kinase EDR1 |
| BnaA02g14490D | A02 | 8,200,831 | 8,202,439 | 3′_25685 | 8,228,124 | AT1G69700 | HVA22C | HVA22–like protein c |
| BnaA03g50340D | A03 | 26,148,221 | 26,149,401 | 5′_23636 | 26,173,037 | AT2G18960 | OST2 | a plasma membrane proton ATPase |
| BnaA03g50610D | A03 | 26,258,416 | 26,258,990 | 5′_85379 | 26,173,037 | AT4G33730 | ATCAPE1 | CAP-derived peptide1 |
| BnaA02g05520D | A02 | 2,548,591 | 2,548,857 | 3′_51712 | 2,496,879 | AT5G22270 | SIED1 | salt-induced and EIN3/EIL1-dependent 1 |
| BnaA02g05590D | A02 | 2,569,927 | 2,571,416 | 3′_73048 | 2,496,879 | AT5G22360 | VAMP714 | Vesicle-associated membrane protein 714 |
| BnaA05g11750D | A05 | 6,644,918 | 6,646,115 | 3′_46142 | 6,670,181, 6,692,257, 6,692,275 | AT4G18780 | LEW2 | cellulose synthase A catalytic subunit 8 |
| BnaA05g11880D | A05 | 6,747,961 | 6,752,424 | 3′_55704 | 6,670,181, 6,692,257, 6,692,275 | AT2G30580 | DRIP2 | C3HC4 RING-domain-containing ubiquitin E3 ligase |
| BnaA05g20580D | A05 | 15,949,088 | 15,952,214 | 3′_99304 | 16,051,518 | AT3G19490 | NHD1 | member of Na +/H+ antiporter-putative family |
| BnaA05g20640D | A05 | 15,986,304 | 15,989,708 | 5′_61810 | 16,051,518 | AT3G19420 | PEN2 | phosphatase with low tyrosine phosphatase activity |
| BnaA10g22560D | A10 | 15,173,521 | 15,178,533 | 5′_10326 | 15,188,859 | AT5G09410 | CAMTA1 | Calmodulin-binding transcription activator 1 |
| BnaA10g22820D | A10 | 15,266,296 | 15,267,019 | 3′_77437 | 15,188,859 | AT5G08620 | STRS2 | DEAD2013box ATP-dependent RNA helicase 25 |
| BnaA10g22850D | A10 | 15,278,666 | 15,281,149 | 3′_89807 | 15,188,859 | AT5G08590 | SNRK2.1 | serine/threonine-protein kinase SRK2G |
| BnaA10g22880D | A10 | 15,287,668 | 15,290,955 | 5′_98809 | 15,188,859 | AT5G08560 | WDR26 | a WD-40-repeat-containing protein |
| BnaC02g31110D | C02 | 33,207,103 | 33,208,273 | 5′_11078 | 33,196,025, 33,196,054 | AT5G44650 | CEST | chloroplast protein-enhancing stress tolerance |
| BnaC09g15950D | C09 | 12,945,604 | 12,947,996 | 5′_1552 | 12,949,548, 12,949,561 | AT1G56570 | PGN | putative pentatricopeptide repeat-containing protein |
| BnaC09g16050D | C09 | 13,037,634 | 13,039,703 | 5′_88086 | 12,949,548, 12,949,561 | AT1G02205 | CER1 | protein ECERIFERUM 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Li, P.; Zhang, J.; Li, Y.; Xu, A.; Huang, Z. Genome-Wide Association Studies of Salt Tolerance at the Seed Germination Stage and Yield-Related Traits in Brassica napus L. Int. J. Mol. Sci. 2022, 23, 15892. https://doi.org/10.3390/ijms232415892
Zhang Y, Li P, Zhang J, Li Y, Xu A, Huang Z. Genome-Wide Association Studies of Salt Tolerance at the Seed Germination Stage and Yield-Related Traits in Brassica napus L. International Journal of Molecular Sciences. 2022; 23(24):15892. https://doi.org/10.3390/ijms232415892
Chicago/Turabian StyleZhang, Yan, Ping Li, Jie Zhang, Yaqi Li, Aixia Xu, and Zhen Huang. 2022. "Genome-Wide Association Studies of Salt Tolerance at the Seed Germination Stage and Yield-Related Traits in Brassica napus L." International Journal of Molecular Sciences 23, no. 24: 15892. https://doi.org/10.3390/ijms232415892
APA StyleZhang, Y., Li, P., Zhang, J., Li, Y., Xu, A., & Huang, Z. (2022). Genome-Wide Association Studies of Salt Tolerance at the Seed Germination Stage and Yield-Related Traits in Brassica napus L. International Journal of Molecular Sciences, 23(24), 15892. https://doi.org/10.3390/ijms232415892






