Specific S100 Proteins Bind Tumor Necrosis Factor and Inhibit Its Activity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Surface Plasmon Resonance Study of sTNF Recognition by S100 Proteins
2.2. Fluorimetric Study of sTNF-S100A11/A12 Interactions
2.3. Chemical Crosslinking
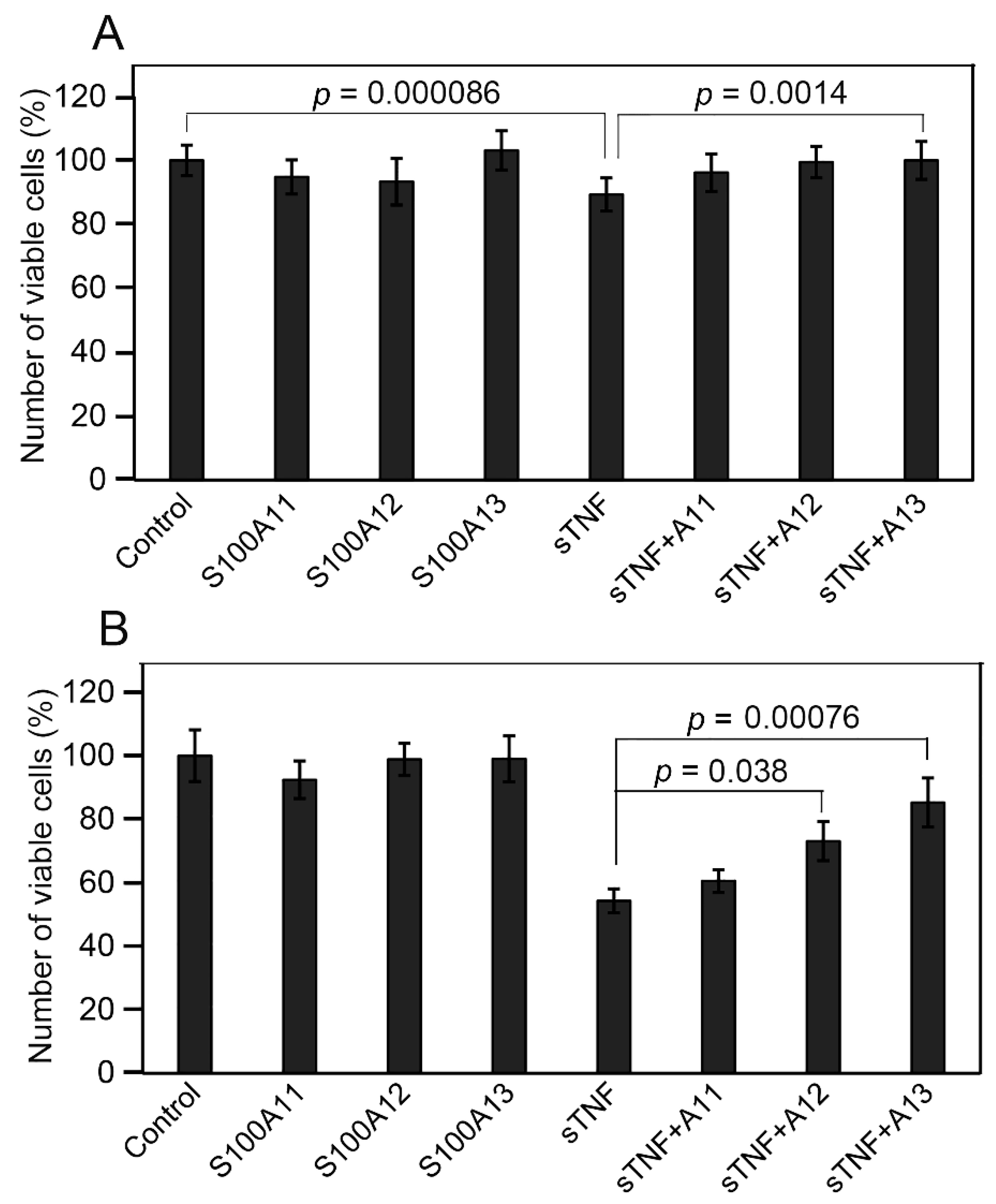
2.4. Influence of Extracellular sTNF and S100A11/A12/A13 on Viability of Huh-7 Cells
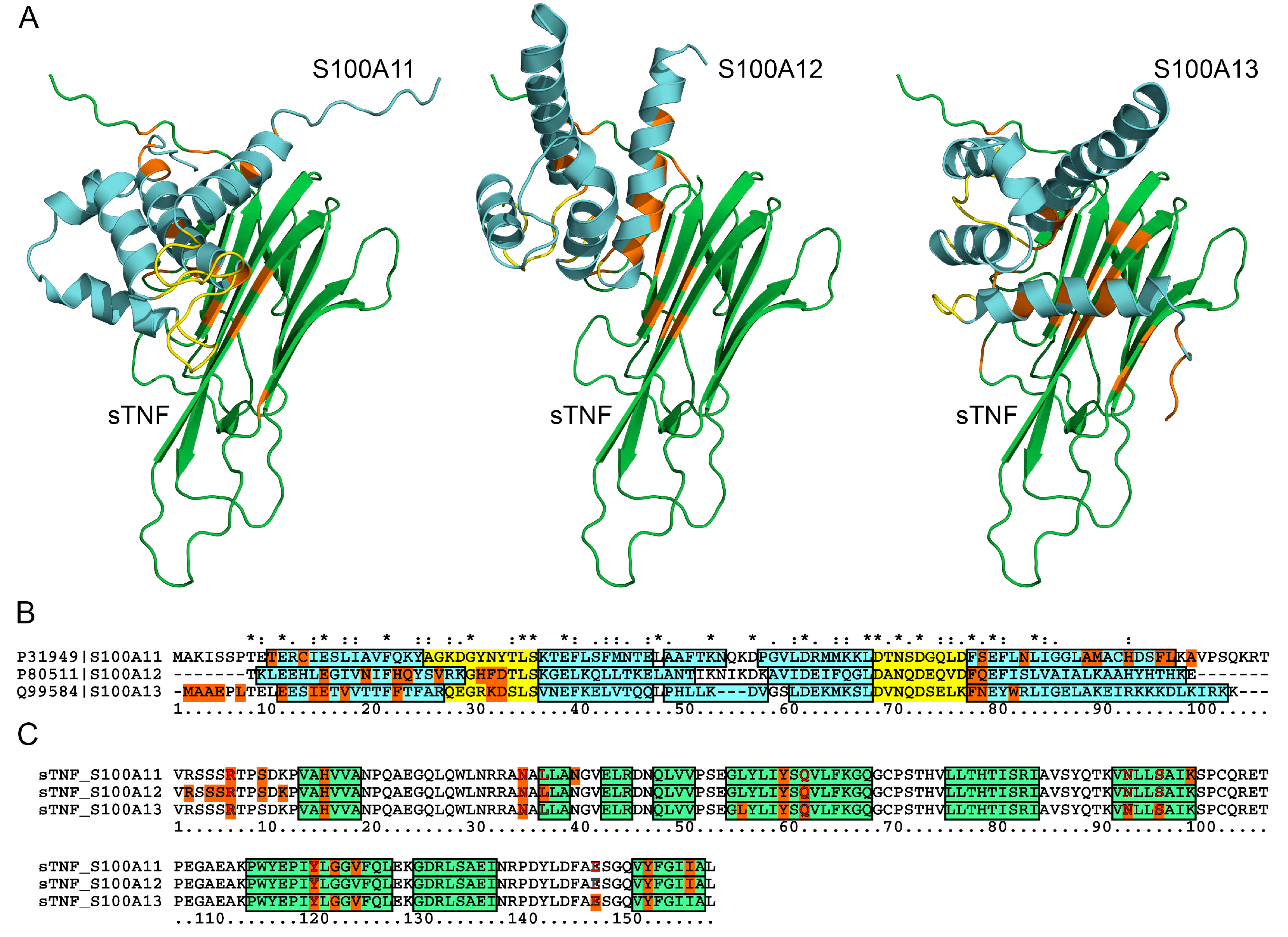
2.5. Structural Modeling of the sTNF-S100 Complexes
2.6. Human Diseases Associated to Dysregulation of TNF and S100A11/A12/A13
3. Materials and Methods
3.1. Materials
3.2. Surface Plasmon Resonance Analysis
3.3. Fluorescence Studies
3.4. Chemical Crosslinking
3.5. Cell Viability Studies
3.6. Structural Modeling of sTNF-S100 Complexes
3.7. Search of the Diseases Associated with the TNF and S100 Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Salomon, B.L. Insights into the biology and therapeutic implications of TNF and regulatory T cells. Nat. Rev. Rheumatol. 2021, 17, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, T.; Mitoma, H.; Harashima, S.-I.; Tsukamoto, H.; Shimoda, T. Transmembrane TNF-: Structure, function and interaction with anti-TNF agents. Rheumatology 2010, 49, 1215–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holbrook, J.; Lara-Reyna, S.; Jarosz-Griffiths, H.; McDermott, M.F. Tumour necrosis factor signalling in health and disease. F1000Research 2019, 8, 111. [Google Scholar] [CrossRef] [Green Version]
- Webster, J.D.; Vucic, D. The Balance of TNF Mediated Pathways Regulates Inflammatory Cell Death Signaling in Healthy and Diseased Tissues. Front. Cell Dev. Biol. 2020, 8, 365. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.; Zhang, C.; Méar, L.; Zhong, W.; Digre, A.; Katona, B.; Sjöstedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A single cell type transcriptomics map of human tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef]
- Falvo, J.V.; Tsytsykova, A.V.; Goldfeld, A.E. Transcriptional Control of the TNF Gene. Curr. Dir. Autoimmun. 2010, 11, 27–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Black, R.A.; Rauch, C.T.; Kozlosky, C.J.; Peschon, J.J.; Slack, J.L.; Wolfson, M.F.; Castner, B.J.; Stocking, K.L.; Reddy, P.; Srinivasan, S.; et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-α from cells. Nature 1997, 385, 729–733. [Google Scholar] [CrossRef]
- Corti, A.; Fassina, G.; Marcucci, F.; Barbanti, E.; Cassani, G. Oligomeric tumour necrosis factor α slowly converts into inactive forms at bioactive levels. Biochem. J. 1992, 284, 905–910. [Google Scholar] [CrossRef] [Green Version]
- Andreeva, A.; Kulesha, E.; Gough, J.; Murzin, A.G. The SCOP database in 2020: Expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 2019, 48, D376–D382. [Google Scholar] [CrossRef]
- Jones, E.Y.; Stuart, D.I.; Walker, N.P.C. Structure of tumour necrosis factor. Nature 1989, 338, 225–228. [Google Scholar] [CrossRef]
- Grell, M.; Douni, E.; Wajant, H.; Löhden, M.; Clauss, M.; Maxeiner, B.; Georgopoulos, S.; Lesslauer, W.; Kollias, G.; Pfizenmaier, K.; et al. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell 1995, 83, 793–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez, C.; Albert, I.; DeFay, K.; Zachariades, N.; Gooding, L.; Kriegler, M. A nonsecretable cell surface mutant of tumor necrosis factor (TNF) kills by cell-to-cell contact. Cell 1990, 63, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Aderka, D.; Sorkine, P.; Abu-Abid, S.; Lev, D.; Setton, A.; Cope, A.P.; Wallach, D.; Klausner, J. Shedding kinetics of soluble tumor necrosis factor (TNF) receptors after systemic TNF leaking during isolated limb perfusion. Relevance to the pathophysiology of septic shock. J. Clin. Investig. 1998, 101, 650–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalliolias, G.; Ivashkiv, L.B. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 2015, 12, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Loh, C.; Park, S.-H.; Lee, A.; Yuan, R.; Ivashkiv, L.B.; Kalliolias, G.D. TNF-induced inflammatory genes escape repression in fibroblast-like synoviocytes: Transcriptomic and epigenomic analysis. Ann. Rheum. Dis. 2019, 78, 1205–1214. [Google Scholar] [CrossRef]
- Zatterale, F.; Longo, M.; Naderi, J.; Raciti, G.A.; Desiderio, A.; Miele, C.; Beguinot, F. Chronic Adipose Tissue Inflammation Linking Obesity to Insulin Resistance and Type 2 Diabetes. Front. Physiol. 2020, 10, 1607. [Google Scholar] [CrossRef]
- Fischer, R.; Kontermann, R.E.; Pfizenmaier, K. Selective Targeting of TNF Receptors as a Novel Therapeutic Approach. Front. Cell Dev. Biol. 2020, 8, 401. [Google Scholar] [CrossRef]
- Vielhauer, V.; Mayadas, T.N. Functions of TNF and its Receptors in Renal Disease: Distinct Roles in Inflammatory Tissue Injury and Immune Regulation. Semin. Nephrol. 2007, 27, 286–308. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Hoidal, J.R.; Mukherjee, T.K. Role of TNFα in pulmonary pathophysiology. Respir. Res. 2006, 7, 125. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Q.; Li, Z.; Tao, T.; Duan, R.; Wang, X.; Su, W. TNF-α in Uveitis: From Bench to Clinic. Front. Pharmacol. 2021, 12, 740057. [Google Scholar] [CrossRef]
- Gerriets, V.; Goyal, A.; Khaddour, K. Tumor Necrosis Factor Inhibitors; StatPearls Publishing: Tampa, FL, USA, 2022. [Google Scholar]
- Evangelatos, G.; Bamias, G.; Kitas, G.D.; Kollias, G.; Sfikakis, P.P. The second decade of anti-TNF-a therapy in clinical practice: New lessons and future directions in the COVID-19 era. Rheumatol. Int. 2022, 42, 1493–1511. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Hu, K.; Li, Y.; Lu, C.; Ling, K.; Cai, C.; Wang, W.; Ye, D. Targeting TNF-α for COVID-19: Recent Advanced and Controversies. Front. Public Health 2022, 10, 833967. [Google Scholar] [CrossRef]
- Van Horssen, R.; Hagen, T.L.M.T.; Eggermont, A.M.M. TNF-α in Cancer Treatment: Molecular Insights, Antitumor Effects, and Clinical Utility. Oncologist 2006, 11, 397–408. [Google Scholar] [CrossRef] [PubMed]
- TNF Alpha Inhibitors Global Market Report 2022—By Drug (Remicade (Infliximab), Enbrel (Etanercept), Humira (Adalimumab), Cimzia (Certolizumab Pegol), Simponi (Golimumab)), By Route Of Administration (Oral, Subcutaneous, Intravenous), By Disease Type (Inflammatory Bowel Disease, Psoriatic Arthritis, Ulcerative Colitis (UC), Rheumatoid Arthritis, Ankylosing Spondylitis)—Market Size, Trends, And Global Forecast 2022–2026. Available online: https://www.thebusinessresearchcompany.com/report/tnf-alpha-inhibitor-global-market-report (accessed on 1 October 2022).
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, P.; Ali, S.A. Multifunctional Role of S100 Protein Family in the Immune System: An Update. Cells 2022, 11, 2274. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L.L.; Garrie, K.; Turner, M.D. Role of S100 proteins in health and disease. Biochim. Biophys. Acta (BBA) -Mol. Cell Res. 2020, 1867, 118677. [Google Scholar] [CrossRef]
- Sreejit, G.; Flynn, M.C.; Patil, M.; Krishnamurthy, P.; Murphy, A.J.; Nagareddy, P.R. S100 family proteins in inflammation and beyond. Adv. Clin. Chem. 2020, 98, 173–231. [Google Scholar] [CrossRef]
- Bresnick, A.R.; Weber, D.J.; Zimmer, D.B. S100 proteins in cancer. Nat. Rev. Cancer 2015, 15, 96–109. [Google Scholar] [CrossRef] [Green Version]
- Cristóvão, J.S.; Gomes, C.M. S100 Proteins in Alzheimer’s Disease. Front. Neurosci. 2019, 13, 463. [Google Scholar] [CrossRef] [Green Version]
- Holzinger, D.; Tenbrock, K.; Roth, J. Alarmins of the S100-Family in Juvenile Autoimmune and Auto-Inflammatory Diseases. Front. Immunol. 2019, 10, 182. [Google Scholar] [CrossRef]
- Sattar, Z.; Lora, A.; Jundi, B.; Railwah, C.; Geraghty, P. The S100 Protein Family as Players and Therapeutic Targets in Pulmonary Diseases. Pulm. Med. 2021, 2021, 5488591. [Google Scholar] [CrossRef] [PubMed]
- Allgöwer, C.; Kretz, A.-L.; Von Karstedt, S.; Wittau, M.; Henne-Bruns, D.; Lemke, J. Friend or Foe: S100 Proteins in Cancer. Cancers 2020, 12, 2037. [Google Scholar] [CrossRef] [PubMed]
- Bresnick, A.R. S100 proteins as therapeutic targets. Biophys. Rev. 2018, 10, 1617–1629. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, A.S.; Mayorov, S.A.; Deryusheva, E.; Avkhacheva, N.V.; Denessiouk, K.A.; Denesyuk, A.I.; Rastrygina, V.A.; Permyakov, E.A.; Permyakov, S.E. Highly specific interaction of monomeric S100P protein with interferon beta. Int. J. Biol. Macromol. 2019, 143, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Rumpret, M.; von Richthofen, H.J.; van der Linden, M.; Westerlaken, G.H.A.; Ormeño, C.T.; Low, T.Y.; Ovaa, H.; Meyaard, L. Recognition of S100 proteins by Signal Inhibitory Receptor on Leukocytes-1 negatively regulates human neutrophils. Eur. J. Immunol. 2021, 51, 2210–2217. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, A.S.; Deryusheva, E.I.; Permyakova, M.E.; Sokolov, A.S.; Rastrygina, V.A.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. Calcium-Bound S100P Protein Is a Promiscuous Binding Partner of the Four-Helical Cytokines. Int. J. Mol. Sci. 2022, 23, 12000. [Google Scholar] [CrossRef]
- Permyakov, S.E.; Denesyuk, A.I.; Denessiouk, K.A.; Permyakova, M.E.; Kazakov, A.S.; Ismailov, R.G.; Rastrygina, V.A.; Sokolov, A.S.; Permyakov, E.A. Monomeric state of S100P protein: Experimental and molecular dynamics study. Cell Calcium 2019, 80, 152–159. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Sokolov, A.S.; Rastrygina, V.A.; Solovyev, V.V.; Ismailov, R.G.; Mikhailov, R.V.; Ulitin, A.B.; Yakovenko, A.R.; Mirzabekov, T.A.; Permyakov, E.A.; et al. High-affinity interaction between interleukin-11 and S100P protein. Biochem. Biophys. Res. Commun. 2015, 468, 733–738. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Deryusheva, E.I.; Sokolov, A.S.; Permyakova, M.E.; Litus, E.A.; Rastrygina, V.A.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. Erythropoietin Interacts with Specific S100 Proteins. Biomolecules 2022, 12, 120. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Sokolov, A.S.; Permyakova, M.E.; Litus, E.A.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. Specific cytokines of interleukin-6 family interact with S100 proteins. Cell Calcium 2021, 101, 102520. [Google Scholar] [CrossRef]
- Kazakov, A.; Sofin, A.; Avkhacheva, N.; Denesyuk, A.; Deryusheva, E.; Rastrygina, V.; Sokolov, A.; Permyakova, M.; Litus, E.; Uversky, V.; et al. Interferon Beta Activity Is Modulated via Binding of Specific S100 Proteins. Int. J. Mol. Sci. 2020, 21, 9473. [Google Scholar] [CrossRef] [PubMed]
- Klingelhöfer, J.; Møller, H.D.; Sumer, E.U.; Berg, C.H.; Poulsen, M.; Kiryushko, D.; Soroka, V.; Ambartsumian, N.; Grigorian, M.; Lukanidin, E.M. Epidermal growth factor receptor ligands as new extracellular targets for the metastasis-promoting S100A4 protein. FEBS J. 2009, 276, 5936–5948. [Google Scholar] [CrossRef] [PubMed]
- Havugimana, P.C.; Goel, R.K.; Phanse, S.; Youssef, A.; Padhorny, D.; Kotelnikov, S.; Kozakov, D.; Emili, A. Scalable multiplex co-fractionation/mass spectrometry platform for accelerated protein interactome discovery. Nat. Commun. 2022, 13, 4043. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carreira, C.M.; LaVallee, T.M.; Tarantini, F.; Jackson, A.; Lathrop, J.T.; Hampton, B.; Burgess, W.H.; Maciag, T. S100A13 Is Involved in the Regulation of Fibroblast Growth Factor-1 and p40 Synaptotagmin-1 Release in Vitro. J. Biol. Chem. 1998, 273, 22224–22231. [Google Scholar] [CrossRef] [Green Version]
- Mandinova, A.; Soldi, R.; Graziani, I.; Bagalá, C.; Bellum, S.; Landriscina, M.; Tarantini, F.; Prudovsky, I.; Maciag, T. S100A13 mediates the copper-dependent stress-induced release of IL-1α from both human U937 and murine NIH 3T3 cells. J. Cell Sci. 2003, 116, 2687–2696. [Google Scholar] [CrossRef] [Green Version]
- Kazakov, A.S.; Sofin, A.D.; Avkhacheva, N.V.; Deryusheva, E.I.; Rastrygina, V.A.; Permyakova, M.E.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. Interferon-β Activity Is Affected by S100B Protein. Int. J. Mol. Sci. 2022, 23, 1997. [Google Scholar] [CrossRef]
- Gupta, A.A.; Chou, R.-H.; Li, H.; Yang, L.-W.; Yu, C. Structural insights into the interaction of human S100B and basic fibroblast growth factor (FGF2): Effects on FGFR1 receptor signaling. Biochim. Biophys. Acta -Proteins Proteom. 2013, 1834, 2606–2619. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Sokolov, A.S.; Vologzhannikova, A.A.; Permyakova, M.E.; Khorn, P.A.; Ismailov, R.G.; Denessiouk, K.A.; Denesyuk, A.I.; Rastrygina, V.A.; Baksheeva, V.E.; et al. Interleukin-11 binds specific EF-hand proteins via their conserved structural motifs. J. Biomol. Struct. Dyn. 2016, 35, 78–91. [Google Scholar] [CrossRef]
- Poiesi, C.; Albertini, A.; Ghielmi, S.; Cassani, G.; Corti, A. Kinetic analysis of TNF-α oligomer-monomer transition by surface plasmon resonance and immunochemical methods. Cytokine 1993, 5, 539–545. [Google Scholar] [CrossRef]
- Daub, H.; Traxler, L.; Ismajli, F.; Groitl, B.; Itzen, A.; Rant, U. The trimer to monomer transition of Tumor Necrosis Factor-Alpha is a dynamic process that is significantly altered by therapeutic antibodies. Sci. Rep. 2020, 10, 9265. [Google Scholar] [CrossRef] [PubMed]
- Cerezo, L.A.; Šumová, B.; Prajzlerová, K.; Veigl, D.; Damgaard, D.; Nielsen, C.H.; Pavelka, K.; Vencovský, J.; Šenolt, L. Calgizzarin (S100A11): A novel inflammatory mediator associated with disease activity of rheumatoid arthritis. Arthritis Res. Ther. 2017, 19, 79. [Google Scholar] [CrossRef] [Green Version]
- Brown, K.L.; Lubieniecka, J.M.; Armaroli, G.; Kessel, K.; Gibson, K.M.; Graham, J.; Liu, D.; Hancock, R.E.W.; Ross, C.; Benseler, S.M.; et al. S100A12 Serum Levels and PMN Counts Are Elevated in Childhood Systemic Vasculitides Especially Involving Proteinase 3 Specific Anti-neutrophil Cytoplasmic Antibodies. Front. Pediatr. 2018, 6, 341. [Google Scholar] [CrossRef]
- Dubois, C.; Marcé, D.; Faivre, V.; Lukaszewicz, A.-C.; Junot, C.; Fenaille, F.; Simon, S.; Becher, F.; Morel, N.; Payen, D. High plasma level of S100A8/S100A9 and S100A12 at admission indicates a higher risk of death in septic shock patients. Sci. Rep. 2019, 9, 15660–15667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boschetti, G.; Garnero, P.; Moussata, D.; Cuerq, C.; Préaudat, C.; Duclaux-Loras, R.; Mialon, A.; Drai, J.; Flourié, B.; Nancey, S. Accuracies of Serum and Fecal S100 Proteins (Calprotectin and Calgranulin C) to Predict the Response to TNF Antagonists in Patients with Crohnʼs Disease. Inflamm. Bowel Dis. 2015, 21, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Wittkowski, H.; Frosch, M.; Wulffraat, N.; Goldbach-Mansky, R.; Kallinich, T.; Kuemmerle-Deschner, J.; Frühwald, M.C.; Dassmann, S.; Pham, T.-H.; Roth, J.; et al. S100A12 is a novel molecular marker differentiating systemic-onset juvenile idiopathic arthritis from other causes of fever of unknown origin. Arthritis Rheum. 2008, 58, 3924–3931. [Google Scholar] [CrossRef] [PubMed]
- Foell, D.; Kane, D.; Bresnihan, B.; Vogl, T.; Nacken, W.; Sorg, C.; FitzGerald, O.; Roth, J. Expression of the pro-inflammatory protein S100A12 (EN-RAGE) in rheumatoid and psoriatic arthritis. Rheumatology 2003, 42, 1383–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulme, C.H.; Peffers, M.J.; Harrington, G.M.B.; Wilson, E.; Perry, J.; Roberts, S.; Gallacher, P.; Jermin, P.; Wright, K.T. Identification of Candidate Synovial Fluid Biomarkers for the Prediction of Patient Outcome After Microfracture or Osteotomy. Am. J. Sports Med. 2021, 49, 1512–1523. [Google Scholar] [CrossRef]
- Berberoglu, U.; Yildirim, E.; Celen, O. Serum Levels of Tumor Necrosis Factor Alpha Correlate with Response to Neoadjuvant Chemotherapy in Locally Advanced Breast Cancer. Int. J. Biol. Mark. 2004, 19, 130–134. [Google Scholar] [CrossRef]
- Li, G.; Wu, W.; Zhang, X.; Huang, Y.; Wen, Y.; Li, X.; Gao, R. Serum levels of tumor necrosis factor alpha in patients with IgA nephropathy are closely associated with disease severity. BMC Nephrol. 2018, 19, 326. [Google Scholar] [CrossRef]
- Wu, W.; Guan, Y.; Zhao, G.; Fu, X.-J.; Guo, T.-Z.; Liu, Y.-T.; Ren, X.-L.; Wang, W.; Liu, H.-R.; Li, Y.-Q. Elevated IL-6 and TNF-α Levels in Cerebrospinal Fluid of Subarachnoid Hemorrhage Patients. Mol. Neurobiol. 2015, 53, 3277–3285. [Google Scholar] [CrossRef] [PubMed]
- Bociaga-Jasik, M.; Garlicki, A.; Kalinowska-Nowak, A.; Mach, T. Concentration of proinflammatory cytokines (TNF-alpha, IL-8) in the cerebrospinal fluid and the course of bacterial meningitis. Prz. Lek. 2004, 61, 78–85. [Google Scholar]
- Koo, B.S.; Jo, S.; Kwon, E.; Shin, J.H.; Hur, J.-W.; Kim, T.-H. Effect of biologics in the level of cytokines in the synovial fluid of patients with ankylosing spondylitis. Korean J. Intern. Med. 2020, 35, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Streicher, W.W.; Lopez, M.M.; Makhatadze, G.I. Modulation of quaternary structure of S100 proteins by calcium ions. Biophys. Chem. 2010, 151, 181–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Simon, M.A.; Ecsédi, P.; Kovács, G.M.; Póti, L.; Reményi, A.; Kardos, J.; Gógl, G.; Nyitray, L. High-throughput competitive fluorescence polarization assay reveals functional redundancy in the S100 protein family. FEBS J. 2020, 287, 2834–2846. [Google Scholar] [CrossRef] [Green Version]
- Simon, M.A.; Bartus, É.; Mag, B.; Boros, E.; Roszjár, L.; Gógl, G.; Travé, G.; Martinek, T.A.; Nyitray, L. Promiscuity mapping of the S100 protein family using a high-throughput holdup assay. Sci. Rep. 2022, 12, 5904. [Google Scholar] [CrossRef]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; Del-Toro, N.; et al. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2013, 42, D358–D363. [Google Scholar] [CrossRef] [Green Version]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. TheBioGRIDdatabase: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2020, 30, 187–200. [Google Scholar] [CrossRef]
- Liedtke, C.; Plümpe, J.; Kubicka, S.; Bradham, C.A.; Manns, M.P.; Brenner, D.A.; Trautwein, C. Jun kinase modulates tumor necrosis factor-dependent apoptosis in liver cells. Hepatology 2002, 36, 315–325. [Google Scholar] [CrossRef]
- Matsushima-Nishiwaki, R.; Takai, S.; Adachi, S.; Minamitani, C.; Yasuda, E.; Noda, T.; Kato, K.; Toyoda, H.; Kaneoka, Y.; Yamaguchi, A.; et al. Phosphorylated Heat Shock Protein 27 Represses Growth of Hepatocellular Carcinoma via Inhibition of Extracellular Signal-regulated Kinase. J. Biol. Chem. 2008, 283, 18852–18860. [Google Scholar] [CrossRef] [PubMed]
- Minero, V.G.; Khadjavi, A.; Costelli, P.; Baccino, F.M.; Bonelli, G. JNK activation is required for TNFα-induced apoptosis in human hepatocarcinoma cells. Int. Immunopharmacol. 2013, 17, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Burow, M.E.; Weldon, C.B.; Tang, Y.; Navar, G.L.; Krajewski, S.; Reed, J.C.; Hammond, T.G.; Clejan, S.; Beckman, B.S. Differences in susceptibility to tumor necrosis factor alpha-induced apoptosis among MCF-7 breast cancer cell variants. Cancer Res. 1998, 58, 4940–4946. [Google Scholar] [PubMed]
- Kim, N.-M.; Koo, S.-Y.; Jeon, K.; Kim, M.H.; Lee, J.; Hong, C.Y.; Jeong, S. Rapid induction of apoptosis by combination of flavopiridol and tumor necrosis factor (TNF)-alpha or TNF-related apoptosis-inducing ligand in human cancer cell lines. Cancer Res. 2003, 63, 621–626. [Google Scholar] [PubMed]
- Berguetti, T.S.; Quintaes, L.S.P.; Pereira, T.H.; Robaina, M.C.; Cruz, A.L.S.; Maia, R.C.; de Souza, P.S. TNF-α Modulates P-Glycoprotein Expression and Contributes to Cellular Proliferation via Extracellular Vesicles. Cells 2019, 8, 500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beg, A.A.; Baltimore, D. An Essential Role for NF-κB in Preventing TNF-α-Induced Cell Death. Science 1996, 274, 782–784. [Google Scholar] [CrossRef]
- Gupta, S. A Decision Between Life and Death During TNF-α-Induced Signaling. J. Clin. Immunol. 2002, 22, 185–194. [Google Scholar] [CrossRef]
- Harper, N.; Hughes, M.; MacFarlane, M.; Cohen, G.M. Fas-associated Death Domain Protein and Caspase-8 Are Not Recruited to the Tumor Necrosis Factor Receptor 1 Signaling Complex during Tumor Necrosis Factor-induced Apoptosis. J. Biol. Chem. 2003, 278, 25534–25541. [Google Scholar] [CrossRef] [Green Version]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071–1081.e3. [Google Scholar] [CrossRef]
- Permyakov, S.E.; Ismailov, R.G.; Xue, B.; Denesyuk, A.I.; Uversky, V.N.; Permyakov, E.A. Intrinsic disorder in S100 proteins. Mol. BioSyst. 2011, 7, 2164–2180. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D. Clustal Omega. Curr. Protoc. Bioinform. 2014, 48, 3.13.1–3.13.16. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016, 45, D833–D839. [Google Scholar] [CrossRef]
- Carvalho-Silva, D.; Pierleoni, A.; Pignatelli, M.; Ong, C.K.; Fumis, L.; Karamanis, N.; Carmona, M.; Faulconbridge, A.; Hercules, A.; McAuley, E.; et al. Open Targets Platform: New developments and updates two years on. Nucleic Acids Res. 2018, 47, D1056–D1065. [Google Scholar] [CrossRef]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef] [Green Version]
- Burstein, E.A.; Emelyanenko, V.I. Log-Normal Description of Fluorescence Spectra of Organic Fluorophores. Photochem. Photobiol. 1996, 64, 316–320. [Google Scholar] [CrossRef]
- Marquardt, D.W. An Algorithm for Least-Squares Estimation of Nonlinear Parameters. J. Soc. Ind. Appl. Math. 1963, 11, 431–441. [Google Scholar] [CrossRef]
- Permyakov, E.A. Luminescent Spectroscopy of Proteins; CRC Press, Inc.: Boca Raton, FL, USA; Ann Arbor, MI, USA; London, UK; Tokyo, Japan, 1993. [Google Scholar]
- Roderburg, C.; Gautheron, J.; Luedde, T. TNF-Dependent Signaling Pathways in Liver Cancer: Promising Targets for Therapeutic Strategies? Dig. Dis. 2012, 30, 500–507. [Google Scholar] [CrossRef]
- Jing, Y.; Sun, K.; Liu, W.; Sheng, D.; Zhao, S.; Gao, L.; Wei, L. Tumor necrosis factor-α promotes hepatocellular carcinogenesis through the activation of hepatic progenitor cells. Cancer Lett. 2018, 434, 22–32. [Google Scholar] [CrossRef]
- Jang, H.S.K.A.Y.-H.C.M.-K.; Kim, H.; Chung, Y.-H. Clinical Aspects of Tumor Necrosis Factor-α Signaling in Hepatocellular Carcinoma. Curr. Pharm. Des. 2014, 20, 2799–2808. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Y.; Zhang, M.; Hu, J.; Li, Z.; Han, B. The high expression of TNF-α and NF-κB in tumor microenvironment predicts good prognosis of patients with BCLC-0-B hepatocellular carcinoma. Transl. Cancer Res. 2019, 8, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Liu, L.; Xue, T.; Jing, C.; Xu, X.; Wu, Y.; Wang, M.; Xie, X.; Zhang, B. Comprehensive Analysis of the Prognosis and Correlations With Immune Infiltration of S100 Protein Family Members in Hepatocellular Carcinoma. Front. Genet. 2021, 12, 648156. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Ye, B.; Ao, J.; Zhu, X.; Zhang, Y.; Chai, Z.; Wang, C.; Sun, H. High expression of S100A12 on intratumoral stroma cells indicates poor prognosis following surgical resection of hepatocellular carcinoma. Oncol. Lett. 2018, 16, 5398–5404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gohar, F.; Anink, J.; Moncrieffe, H.; Van Suijlekom-Smit, L.W.; Prince, F.H.; van Rossum, M.A.; Dolman, K.M.; Hoppenreijs, E.P.; Cate, R.T.; Ursu, S.; et al. S100A12 Is Associated with Response to Therapy in Juvenile Idiopathic Arthritis. J. Rheumatol. 2018, 45, 547–554. [Google Scholar] [CrossRef]
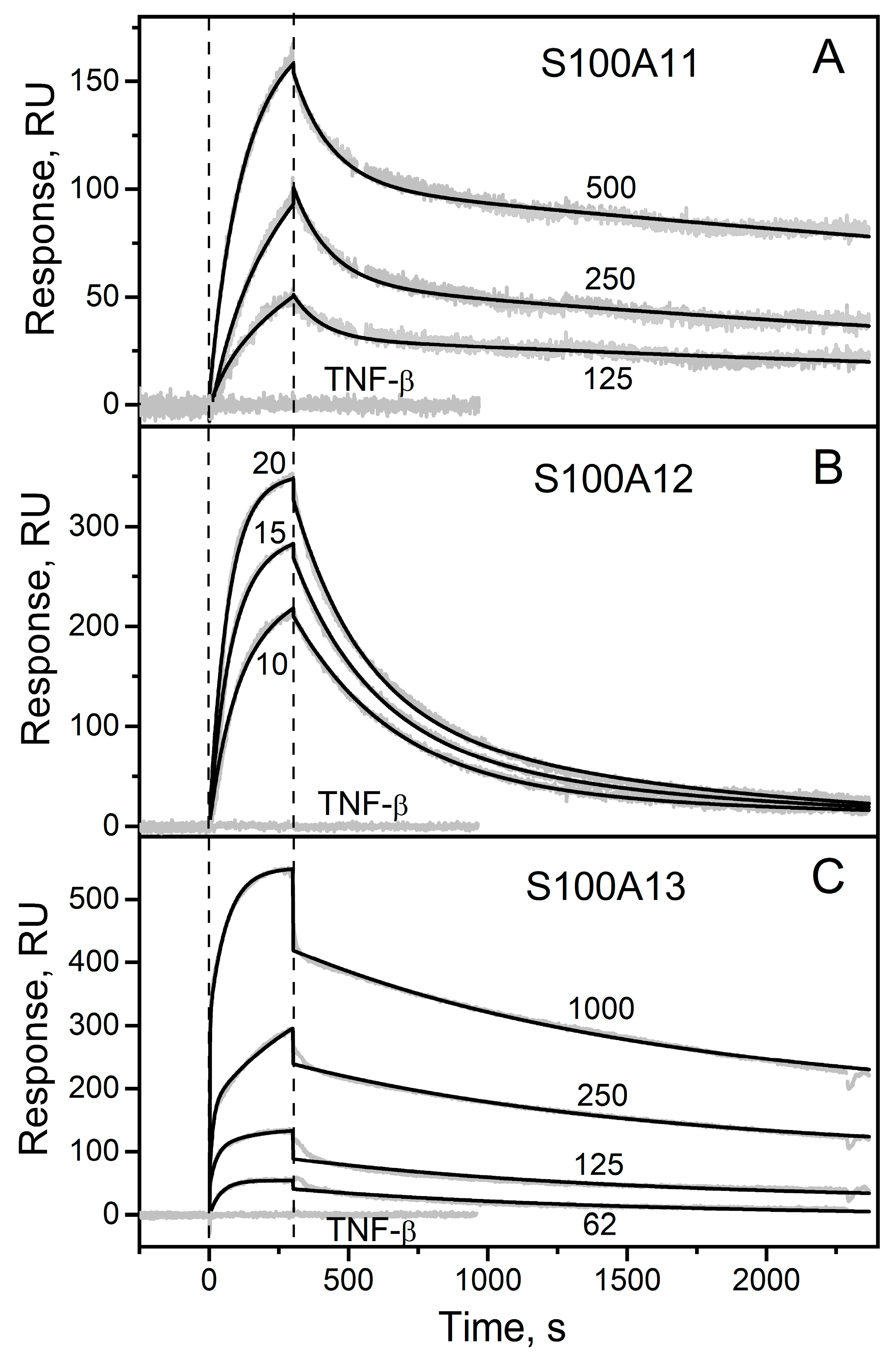


| Analyte | kd1, s−1 | Kd1, nM | kd2, s−1 | Kd2, nM |
|---|---|---|---|---|
| S100A11 | (1.8 ± 0.5) × 10−4 | 28 ± 16 | (8.4 ± 0.3) × 10−3 | 695 ± 73 |
| S100A12 | (1.8 ± 1.4) × 10−3 | 2.3 ± 1.7 | (2.3 ± 1.7) × 10−3 | 8.4 ± 0.5 |
| S100A13 | (9.0 ± 1.6) × 10−4 | 2.4 ± 0.7 | (7.3 ± 2.5) × 10−3 | 4.3 ± 2.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kazakov, A.S.; Zemskova, M.Y.; Rystsov, G.K.; Vologzhannikova, A.A.; Deryusheva, E.I.; Rastrygina, V.A.; Sokolov, A.S.; Permyakova, M.E.; Litus, E.A.; Uversky, V.N.; et al. Specific S100 Proteins Bind Tumor Necrosis Factor and Inhibit Its Activity. Int. J. Mol. Sci. 2022, 23, 15956. https://doi.org/10.3390/ijms232415956
Kazakov AS, Zemskova MY, Rystsov GK, Vologzhannikova AA, Deryusheva EI, Rastrygina VA, Sokolov AS, Permyakova ME, Litus EA, Uversky VN, et al. Specific S100 Proteins Bind Tumor Necrosis Factor and Inhibit Its Activity. International Journal of Molecular Sciences. 2022; 23(24):15956. https://doi.org/10.3390/ijms232415956
Chicago/Turabian StyleKazakov, Alexey S., Marina Y. Zemskova, Gleb K. Rystsov, Alisa A. Vologzhannikova, Evgenia I. Deryusheva, Victoria A. Rastrygina, Andrey S. Sokolov, Maria E. Permyakova, Ekaterina A. Litus, Vladimir N. Uversky, and et al. 2022. "Specific S100 Proteins Bind Tumor Necrosis Factor and Inhibit Its Activity" International Journal of Molecular Sciences 23, no. 24: 15956. https://doi.org/10.3390/ijms232415956








