Novel Virulence Factors Deciphering Klebsiella pneumoniae KpC4 Infect Maize as a Crossing-Kingdom Pathogen: An Emerging Environmental Threat
Abstract
:1. Introduction
2. Results
2.1. Genome Features
2.2. The Survival Mechanism of Strain KpC4 in Plants
2.3. Carbohydrate Metabolism
2.4. Aromatic Compounds Degradation
2.5. Survival against Plant Defenses
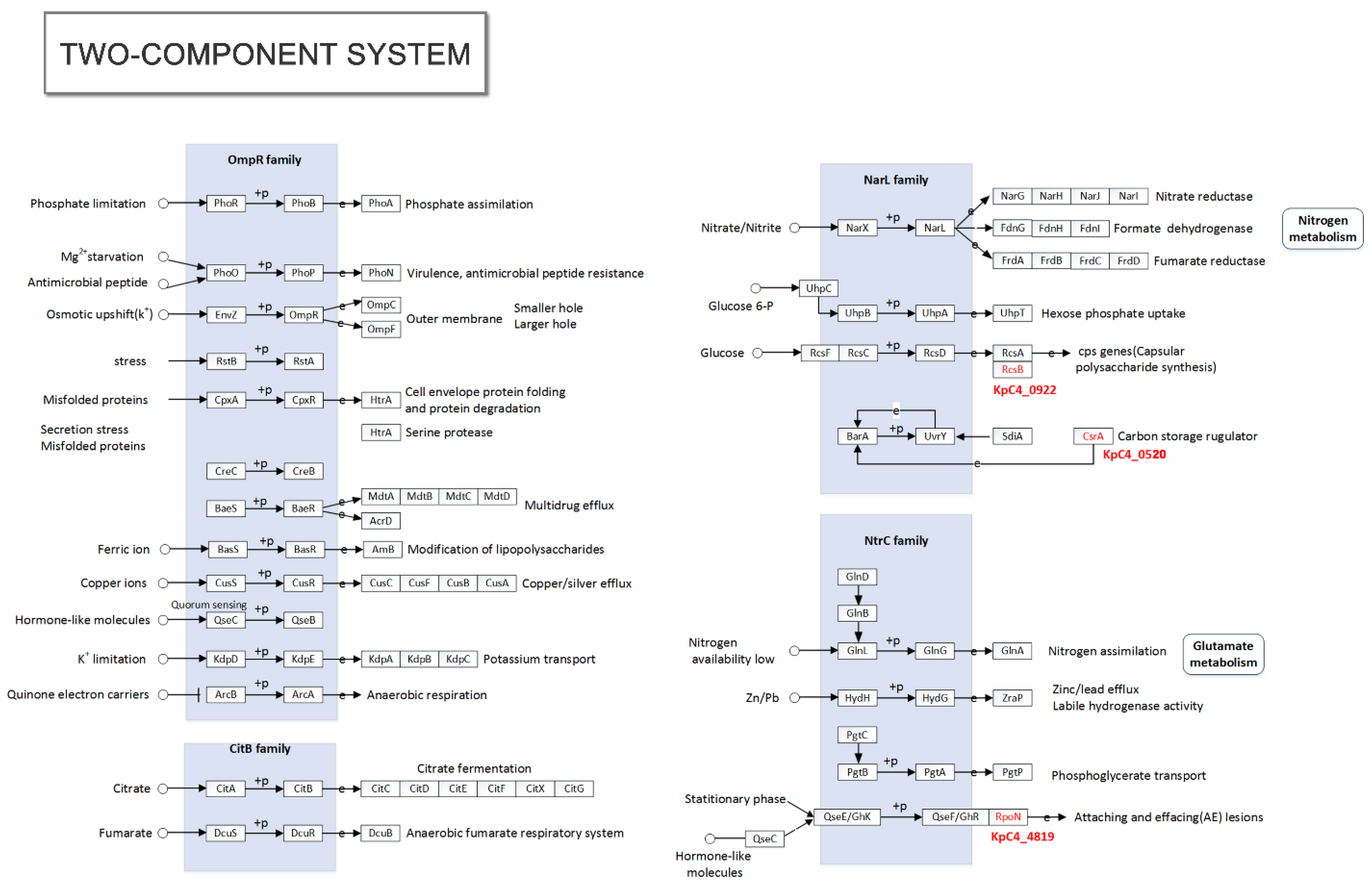
2.5.1. Evade the Plant’s Defense System
2.5.2. Plant-Induced and Associated Genes
2.5.3. Plant Disease-Causing Genes
2.6. Pathogenicity of KpC4
2.6.1. Capsule
2.6.2. Lipopolysaccharide
2.6.3. Adhesin
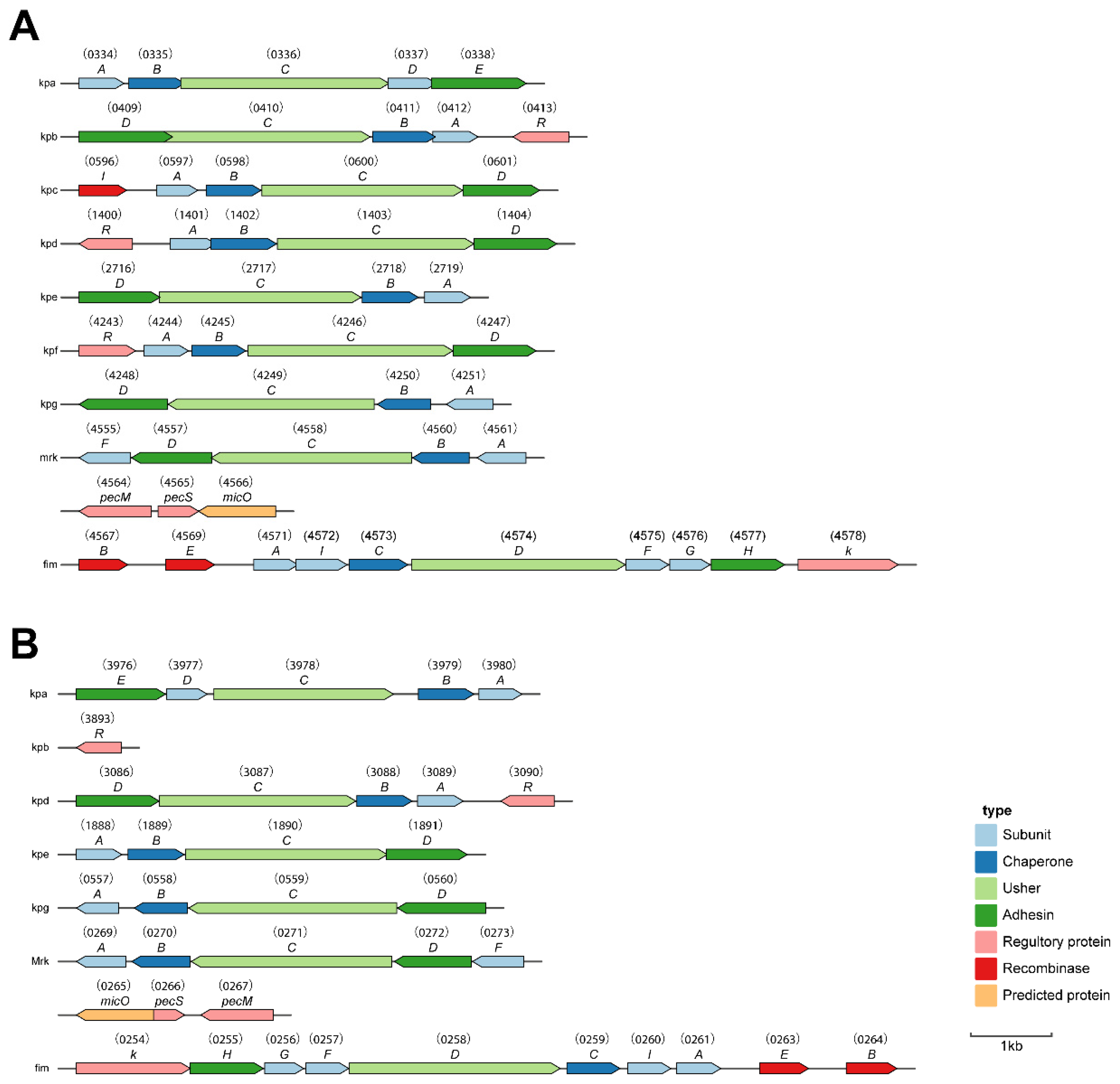
2.7. Siderophores and Transporters
2.8. Comparative Genome Analysis
2.9. Potential Drug Target of K. pneumoniae KpC4
3. Discussion
4. Materials and Methods
4.1. Strain Isolation and Verification
4.2. Isolation and Purification of DNA for Library Production
4.3. Genome Sequencing
4.4. Gene Prediction and Annotation
4.5. Prediction and Functional Analysis of Disease-Related Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cubero, M.; Grau, I.; Tubau, F.; Pallarés, R.; Domínguez, M.Á.; Linares, J.; Ardanuy, C. Molecular epidemiology of Klebsiella pneumoniae strains causing bloodstream infections in adults. Microb. Drug Resist. 2018, 24, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.S.; Huang, J.C.; Chen, S.T.; Sun, J.H.; Wang, C.C.; Lin, S.F.; Hsu, R.S.; Lin, J.D.; Huang, S.Y.; Huang, Y.Y. Characteristics of Klebsiella pneumoniae bacteremia in community-acquired and nosocomial infections in diabetic patients. Chang Gung Med. J. 2010, 33, 532. [Google Scholar] [PubMed]
- Piperaki, E.-T.; Syrogiannopoulos, G.A.; Tzouvelekis, L.S.; Daikos, G.L. Klebsiella pneumoniae: Virulence, biofilm and antimicrobial resistance. Pediatr. Infect. Dis. J. 2017, 36, 1002–1005. [Google Scholar] [CrossRef] [PubMed]
- Cubero, M.; Grau, I.; Tubau, F.; Pallarés, R.; Dominguez, M.A.; Liñares, J.; Ardanuy, C. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007–2013). Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2016, 22, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, S.; Zhang, J.; Zhao, Y.; Yu, L.; Wang, Y.; Wang, Y.; Bao, M.; Fu, Y.; Li, C.; Zhang, X. Outbreak of KPC-2 Carbapenem-resistant Klebsiella pneumoniae ST76 and Carbapenem-resistant K2 Hypervirulent Klebsiella pneumoniae ST375 strains in northeast China: Molecular and virulent characteristics. BMC Infect. Dis. 2020, 20, 1–14. [Google Scholar] [CrossRef]
- Holt, K.E.; Wertheim, H.; Zadoks, R.N.; Baker, S.; Whitehouse, C.A.; Dance, D.; Jenney, A.; Connor, T.R.; Hsu, L.Y.; Severin, J. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA 2015, 112, E3574–E3581. [Google Scholar] [CrossRef] [Green Version]
- Shilpi, S.; Bhadauria, N.; Sharma, P.D.; Paul, P.K. Biofilm formation and survival of Serratia fonticola, Klebsiella pneumoniae and Chryseobacterium jejuense on tomato phylloplane. Pertanika J. Trop. Agric. Sci. 2019, 42, 1097–1110. [Google Scholar]
- Sylvester, W.; Son, R.; Lew, K.; Rukayadi, Y. Antibacterial activity of Java turmeric (Curcuma xanthorrhiza Roxb.) extract against Klebsiella pneumoniae isolated from several vegetables. Int. Food Res. J. 2015, 22, 1770–1776. [Google Scholar]
- Huang, M.; Li, L.; Wu, Y.; Ho, H.; He, P.F.; Li, G.; He, P.B.; Xiong, G.R.; Yuan, Y.; He, Y. Pathogenicity of Klebsiella pneumonia (KpC4) infecting maize and mice. J. Integr. Agric. 2016, 15, 1510–1520. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.H.; Huang, M.; He, P.F.; Ho, H.H.; Wu, Y.X.; He, Y.Q. A new bacterial leaf spot of banana caused by Klebsiella pneumoniae subsp. pneumoniae in Yunnan province of China. Acta Phytopathol. Sin. 2016, 46, 536–542. [Google Scholar]
- Ajayasree, T. Survival of Klebsiella pneumoniae strain borkar in pomegranate orchard soil and its tolerance to temperature and pH. J. Appl. Biotechnol. Bioeng. 2018, 5, 299–301. [Google Scholar]
- Huang, M.; He, P.F.; Munir, S.; Wu, Y.; Li, X.; He, P.B.; He, Y. Ecology and etiology of bacterial top rot in maize caused by Klebsiella pneumoniae KpC4. Microb. Pathog. 2020, 139, 103906–103917. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Lan, Y.; Wang, S.; Liu, Q.; Jian, Z.; Li, Y.; Chen, X.; Yan, Q.; Liu, W. Epidemiology and molecular characteristics of the type VI secretion system in Klebsiella pneumoniae isolated from bloodstream infections. J. Clin. Lab. Anal. 2020, 34, e23459. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Wu, Y.; He, P. Advances in humans and animals opportunistic pathogens from environment infecting plants by crossing kingdoms. Acta Microbiol. Sin. 2016, 56, 188–197. (In Chinese) [Google Scholar]
- Pereira, R.P.I.; Christina, P.R.; de Almeida, L.G.P.; Lima, N.C.B.; Raquel, G.; Vivan, A.C.P.; Xavier, D.E.; Barcellos, F.G.; Marsileni, P.; Vespero, E.C. Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC Genom. 2014, 15, 1–16. [Google Scholar]
- Wu, K.M.; Li, L.H.; Yan, J.J.; Tsao, N.; Liao, T.L.; Tsai, H.C.; Fung, C.P.; Chen, H.J.; Liu, Y.M.; Wang, J.T. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J. Bacteriol. 2009, 191, 4492–4501. [Google Scholar] [CrossRef] [PubMed]
- DE Fouts, T.H.; De Boy, R.T.; Daugherty, S.; Ren, Q.; Badger, J.H.; Durkin, A.S.; Huot, H.; Shrivastava, S.; Kothari, S.; Dodson, R.J.; et al. Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet. 2008, 4, e1000141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Kaufman, M.G.; Miazgowicz, K.L.; Bagdasarian, M.; Walker, E.D. Molecular characterization of a cold-active recombinant xylanase from Flavobacterium johnsoniae and its applicability in xylan hydrolysis. Bioresour. Technol. 2013, 128, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Masai, E.; Sasaki, M.; Minakawa, Y.; Abe, T.; Sonoki, T.; Miyauchi, K.; Katayama, Y.; Fukuda, M. A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J. Bacteriol. 2004, 186, 2757–2765. [Google Scholar] [CrossRef] [Green Version]
- Danhorn, T.; Fuqua, C. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 2007, 61, 401–422. [Google Scholar] [CrossRef]
- Eulberg, D.; Lakner, S.; Golovleva, L.A.; Schlömann, M. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: Evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J. Bacteriol. 1998, 180, 1072–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priefert, H.; Rabenhorst, J.; Steinbüchel, A. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J. Bacteriol. 1997, 179, 2595–2607. [Google Scholar] [CrossRef] [Green Version]
- Hammond-Kosack, K.E.; Jones, J. Resistance gene-dependent plant defense responses. Plant Cell 1996, 8, 1773–1791. [Google Scholar] [PubMed] [Green Version]
- Zeidler, D.; Zähringer, U.; Gerber, I.; Dubery, I.; Hartung, T.; Bors, W.; Hutzler, P.; Durner, J. Innate immunity in Arabidopsis thaliana: Lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sci. USA 2004, 101, 15811–15816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vicente, J.B.; Teixeira, M. Redox and spectroscopic properties of the Escherichia coli nitric oxide-detoxifying system involving flavorubredoxin and its NADH-oxidizing redox partner. J. Biol. Chem. 2005, 280, 34599–34608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burse, A.; Weingart, H.; Ullrich, M.S. The phytoalexin-inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora. Mol. Plant-Microbe Interact. 2004, 17, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Q.; Zhang, M.; Dong, Z.; Sun, W.; Cao, X. Inducible gene expression systems in plants. Plant Gene Trait. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Boch, J.; Joardar, V.; Gao, L.; Robertson, T.L.; Lim, M.; Kunkel, B.N. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol. Microbiol. 2002, 44, 73–88. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.; Allen, C. Ralstonia solanacearum genes induced during growth in tomato: An inside view of bacterial wilt. Mol. Microbiol. 2004, 53, 1641–1660. [Google Scholar] [CrossRef]
- Landini, P.; Volkert, M.R. Transcriptional activation of the Escherichia coli adaptive response gene aidB is mediated by binding of methylated Ada protein. Evidence for a new consensus sequence for Ada-binding sites. J. Biol. Chem. 1995, 270, 8285–8289. [Google Scholar] [CrossRef] [Green Version]
- Maria, M.; Stefania, V.; Lucia, B.; Monika, S. Genotoxic effects of the hydroxycinnamic acid derivates—Caffeic, chlorogenic and cichoric. Biologia 2005, 60, 275–279. [Google Scholar]
- Park, S.J.; Gunsalus, R.P. Oxygen, iron, carbon, and superoxide control of the fumarase fumA and fumC genes of Escherichia coli: Role of the arcA, fnr, and soxR gene products. J. Bacteriol. 1995, 177, 6255–6262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, Y.; Yumoto, N.; Tokushige, M.; Fukui, K.; Ohyanishiguchi, H. Purification and characterization of two types of fumarase from Escherichia coli. J. Biochem. 1991, 109, 728. [Google Scholar] [CrossRef] [PubMed]
- Bereswill, S.; Geider, K. Characterization of the rcsB gene from Erwinia amylovora and its influence on exoploysaccharide synthesis and virulence of the fire blight pathogen. J. Bacteriol. 1997, 179, 1354–1361. [Google Scholar] [CrossRef] [Green Version]
- Peng, R.; Kong, X.; Zhou, X. Genotyping and drug resistance of biofilm forming strains of Klebsiella Pneumoniae. J. Clin. Transfus. Lab. Med. 2017, 19, 456–459. [Google Scholar]
- Whitfield, C.; Roberts, I. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 1999, 31, 1307–1319. [Google Scholar] [CrossRef]
- Rahn, A.; Drummelsmith, J.; Whitfield, C. Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: Relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J. Bacteriol. 1999, 181, 2307–2313. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Fang, H.; Yang, H.; Lin, T.; Hsieh, P.; Tsai, F.; Keynan, Y.; Wang, J. Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J. Clin. Microbiol. 2008, 46, 2231–2240. [Google Scholar] [CrossRef]
- Pannen, D.; Fabisch, M.; Gausling, L.; Schnetz, K. Interaction of the RcsB response regulator with auxiliary transcription regulators in Escherichia coli. J. Biol. Chem. 2016, 291, 2357. [Google Scholar] [CrossRef] [Green Version]
- De, M.S.; Yu, J.; Fookes, M.; Mcateer, S.P.; Llobet, E.; Finn, S.; Spence, S.; Monahan, A.; Kissenpfennig, A.; Ingram, R.J. Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog. 2015, 11, e1004766. [Google Scholar]
- Raetz, C.R.; Guan, Z.; Ingram, B.O.; Six, D.A.; Song, F.; Wang, X.; Zhao, J. Discovery of new biosynthetic pathways: The lipid A story. J. Lipid Res. 2009, 50, S103–S108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regué, M.; Climent, N.; Abitiu, N.; Coderch, N.; Merino, S.; Izquierdo, L.; Altarriba, M.; Tomás, J.M. Genetic characterization of the Klebsiella pneumoniae waa gene cluster, involved in core lipopolysaccharide biosynthesis. J. Bacteriol. 2001, 183, 3564–3573. [Google Scholar] [CrossRef] [Green Version]
- Merino, S.; Altarriba, M.; Izquierdo, L.; Nogueras, M.M.; Regué, M.; Tomás, J.M. Cloning and sequencing of the Klebsiella pneumoniae O5 wb gene cluster and its role in pathogenesis. Infect. Immun. 2000, 68, 2435–2440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosen, D.A.; Hilliard, J.K.; Tiemann, K.M.; Todd, E.M.; Morley, S.C.; Hunstad, D.A. Klebsiella pneumoniae FimK promotes virulence in murine pneumonia. J. Infect. Dis. 2016, 213, 649–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Struve, C.; Bojer, M.; Krogfelt, K.A. Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect. Immun. 2008, 76, 4055–4065. [Google Scholar] [CrossRef] [Green Version]
- Ong, C.L.; Beatson, S.A.; Totsika, M.; Forestier, C.; Mcewan, A.G.; Schembri, M.A. Molecular analysis of type 3 fimbrial genes from Escherichia coli, Klebsiella and Citrobacter species. BMC Microbiol. 2010, 10, 183. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.C.; Huang, Y.J.; Fung, C.P.; Peng, H.L. Regulation of the Klebsiella pneumoniae Kpc fimbriae by the site-specific recombinase KpcI. Microbiology 2010, 156 Pt 7, 1983–1992. [Google Scholar] [CrossRef] [Green Version]
- Chuang, Y.; Fang, C.; Lai, S.; Chang, S.; Wang, J. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 2006, 193, 645–654. [Google Scholar] [CrossRef] [Green Version]
- Martínezromero, E.; Silvasanchez, J.; Barrios, H.; Rodríguezmedina, N.; Martínezbarnetche, J.; Téllezsosa, J.; Gómezbarreto, R.E.; Garzaramos, U. Draft genome sequences of Klebsiella variicola plant isolates. Genome Announc. 2015, 3, e01015-15. [Google Scholar] [CrossRef]
- Agladze, K.; Wang, X.; Romeo, T. Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J. Bacteriol. 2005, 187, 8237–8246. [Google Scholar] [CrossRef] [Green Version]
- Torres, A.G.; Jeter, C.; Langley, W.; Matthysse, A.G. Differential binding of Escherichia coli O157:H7 to alfalfa, human epithelial cells, and plastic is mediated by a variety of surface structures. Appl. Environ. Microbiol. 2005, 71, 8008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.A.; Isaacson, R.E. Identification of Escherichia coli genes that are specifically expressed in a murine model of Septicemic Infection. Infect. Immun. 2002, 70, 3404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balestrino, D.; Haagensen, J.A.J.; Rich, C.; Forestier, C. Characterization of type 2 quorum sensing in Klebsiella pneumoniae and relationship with biofilm formation. J. Bacteriol. 2005, 187, 2870–2880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Fang, C.; Lee, C.; Shun, C.; Wang, J. Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection [with discussion]. J. Infect. Dis. 2005, 192, 117–128. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Syu, W.; Ho, W.; Lin, C.; Tsai, S.; Wang, S. SitA contributes to the virulence of Klebsiella pneumoniae in a mouse infection model. Microbes Infect. 2014, 16, 161–170. [Google Scholar] [CrossRef]
- Lawlor, M.; O’Connor, C.; Miller, V.L. Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun. 2007, 75, 1463–1472. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, P.; Lin, T.; Lee, C.; Tsai, S.; Wang, J. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 2008, 197, 1717. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Liu, M.; Cui, Y.; Wang, L.; Zhang, Y.; Qiu, J.; Yang, R.; Liu, C.; Zhou, D. A novel PCR-based genotyping scheme for clinical Klebsiella pneumoniae. Future Microbiol. 2014, 9, 21–32. [Google Scholar] [CrossRef]
- Chi, X.; Berglund, B.; Zou, H.; Zheng, B.; Börjesson, S.; Ji, X.; Ottoson, J.; Lundborg, C.S.; Li, X.; Nilsson, L.E. Characterization of clinically relevant strains of extended-spectrum β-lactamase-producing Klebsiella pneumoniae occurring in environmental sources in a rural area of China by using whole-genome sequencing. Front. Microbiol. 2019, 10, 211–213. [Google Scholar] [CrossRef]
- Puspanadan, S.; Af Sah-Hejri, L.; Loo, Y.; Nillian, E.; Kuan, C.; Goh, S.; Chang, W.; Lye, Y.; John, Y.; Rukayadi, Y. Detection of Klebsiella pneumoniae in raw vegetables using most probable number-polymerase chain reaction (MPN-PCR). Int. Food Res. J. 2012, 19, 1757–1762. [Google Scholar]
- Rabins, L.; Bhattacharya Kumar, A.; Vijayan, V.J. Characterization of Klebsiella pneumoniae from fresh vegetables marketed in Puducherry. Int. J. Curr. Res. 2017, 8, 40598–40603. [Google Scholar]
- Nicolau Korres, A.M.; Aquije, G.M.d.F.V.; Buss, D.S.; Ventura, J.A.; Fernandes, P.M.B.; Fernandes, A.A.R. Comparison of biofilm and attachment mechanisms of a phytopathological and clinical isolate of Klebsiella pneumoniae subsp. pneumoniae. Sci. World J. 2013, 2013, 925375–925380. [Google Scholar]
- Atmani, S.M.; Messai, Y.; Alouache, S.; Fernandez, R.; Estepa, V.; Torres, C.; Bakour, R. Virulence characteristics and genetic background of ESBL-producing Klebsiella pneumoniae isolates from wastewater. Fresenius Environ. Bull. 2015, 24, 103–112. [Google Scholar]
- Davis, G.S.; Waits, K.; Nordstrom, L.; Weaver, B.; Aziz, M.; Gauld, L.; Grande, H.; Bigler, R.; Horwinski, J.; Porter, S. Intermingled Klebsiella pneumoniae populations between retail meats and human urinary tract infections. Clin. Infect. Dis. 2015, 61, 892–899. [Google Scholar] [CrossRef] [Green Version]
- Matsen, J.M.; Spindler, J.A.; Blosser, R.O. Characterization of Klebsiella isolates from natural receiving waters and comparison with human isolates. Appl. Microbiol. 1974, 28, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Struve, C.; Krogfelt, K.A. Pathogenic potential of environmental Klebsiella pneumoniae isolates. Environ. Microbiol. 2004, 6, 584–590. [Google Scholar] [CrossRef]
- Gross, D.C.; Lichens-Park, A.; Kole, C. Genomics of Erwinia amylovora and related Erwinia species associated with pome fruit trees. In Genomics of Plant-Associated Bacteria; Springer: Berlin/Heidelberg, Germany, 2014; Volume 9783642553783, pp. 1–36. [Google Scholar]
- Pristovek, P.; Sengupta, K.; Löhr, F.; Schfer, B.; von Trebra, W.M.; Rüterjans, H.; Bernhard, F. Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box. J. Biol. Chem. 2003, 278, 17752–17759. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Korban, S.S.; Zhao, Y. The Rcs phosphorelay system is essential for pathogenicity in Erwinia amylovora. Mol. Plant Pathol. 2010, 10, 277–290. [Google Scholar] [CrossRef]
- Van Baarlen, P.; Van Belkum, A.; Summerbell, R.C.; Crous, P.W.; Thomma, B.P. Molecular mechanisms of pathogenicity: How do pathogenic microorganisms develop cross-kingdom host jumps? FEMS Microbiol. Rev. 2007, 31, 239–277. [Google Scholar] [CrossRef] [Green Version]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Ogawa, W.; Li, D.; Yu, P.; Begum, A.; Mizushima, T.; Kuroda, T.; Tsuchiya, T. Multidrug resistance in Klebsiella pneumoniae MGH78578 and cloning of genes responsible for the resistance. Biol. Pharm. Bull. 2005, 28, 1505–1509. [Google Scholar] [CrossRef] [PubMed]
- Garzaramos, U.; Silvasánchez, J.; Martínezromero, E.; Tinoco, P.; Pinagonzales, M.; Barrios, H.; Martínezbarnetche, J.; Gómezbarreto, R.E.; Tellezsosa, J. Development of a Multiplex-PCR probe system for the proper identification of Klebsiella variicola. BMC Microbiol. 2015, 15, 1–14. [Google Scholar]
- Pereira, R.P.I.; Cristina, P.R.; Carolina, V.E.; Marsileni, P.; Goda, Z.L.F.; Almeida, L.G.P.; Gerber, A.L.; Vasconcelos, A.T.R.; Cristina, G.A.; Fabiana, N.M. Pyrosequencing-based analysis reveals a novel capsular gene cluster in a KPC-producing Klebsiella pneumoniae clinical isolate identified in Brazil. BMC Microbiol. 2012, 12, 173–184. [Google Scholar]
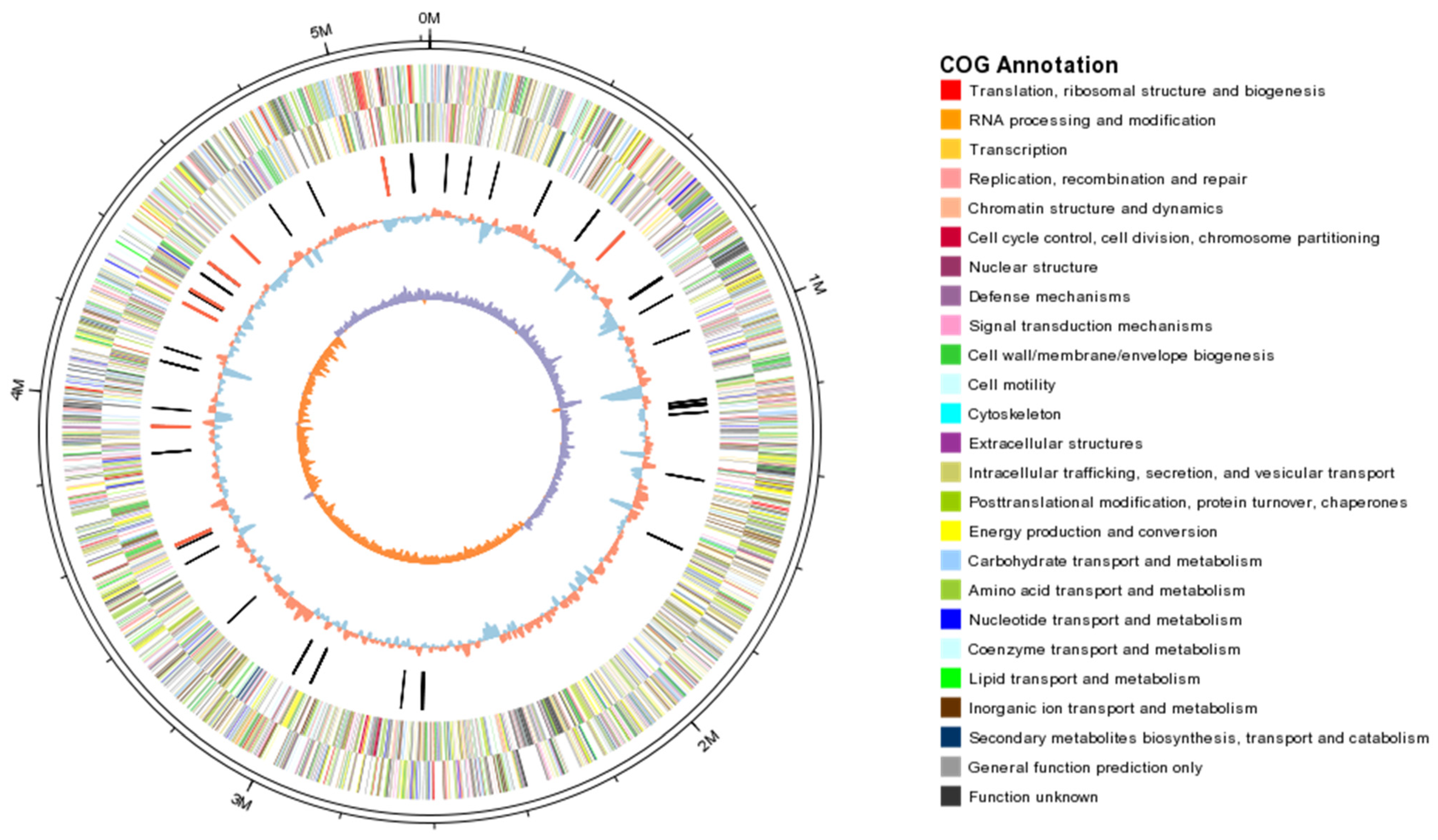
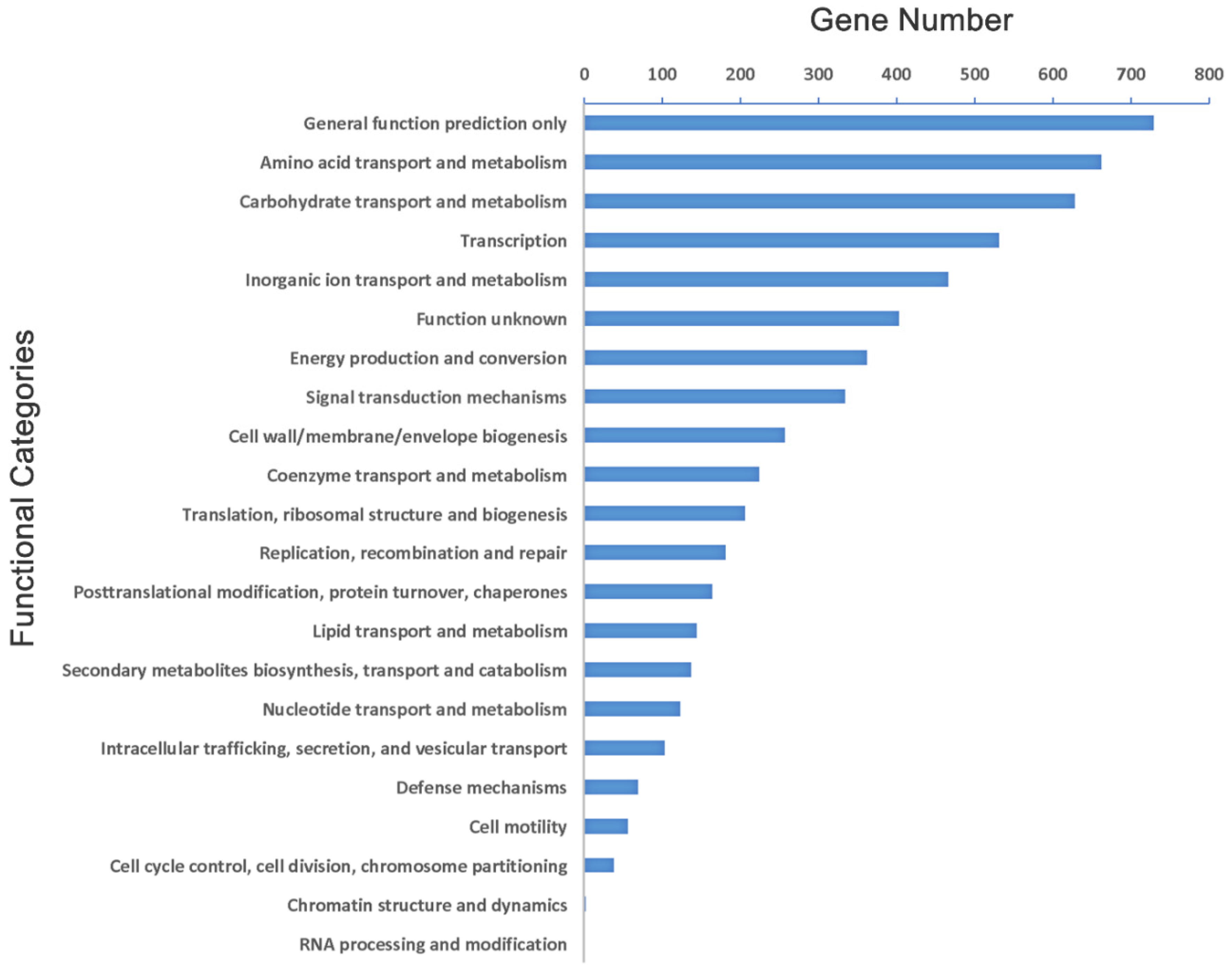

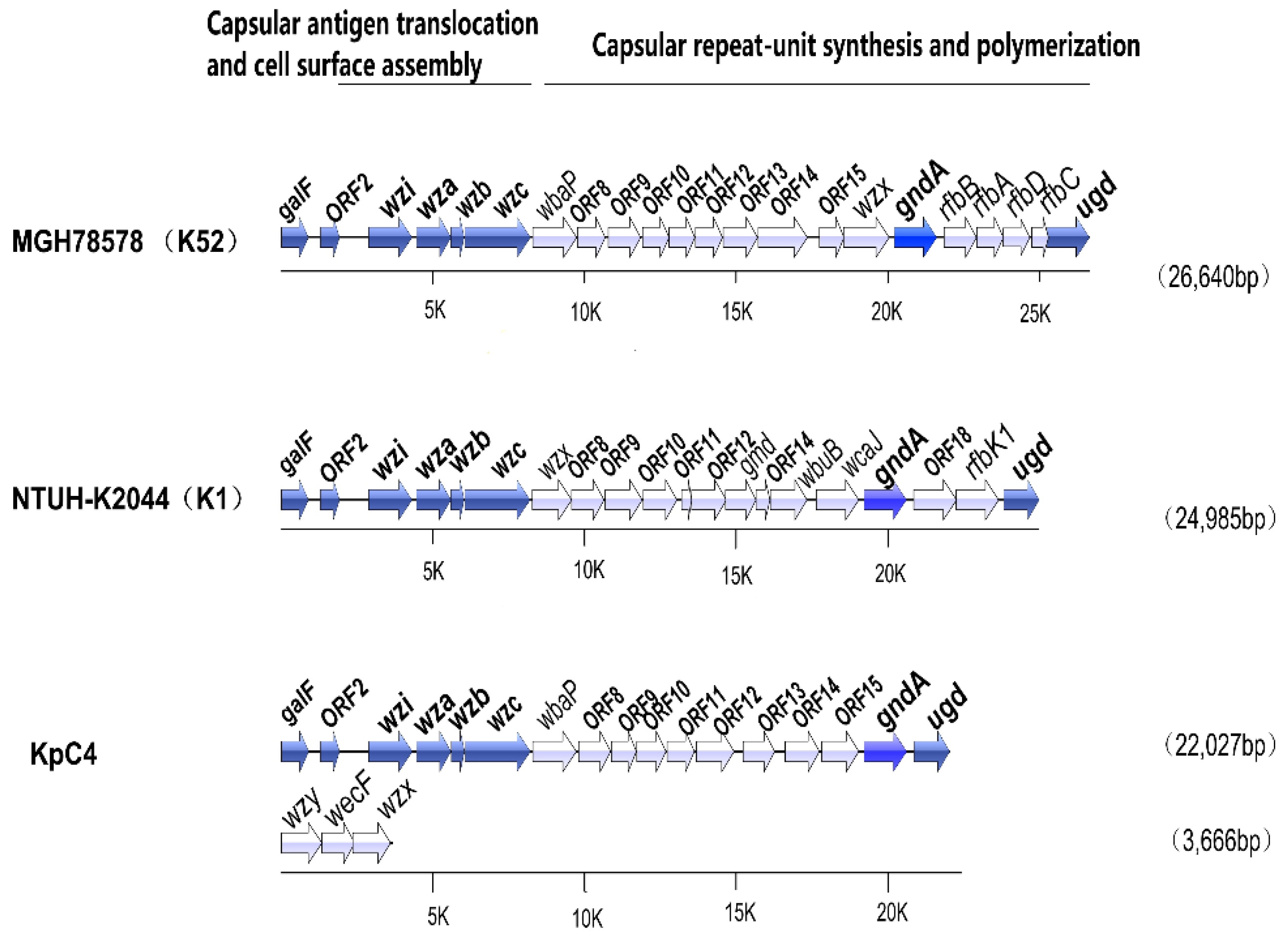
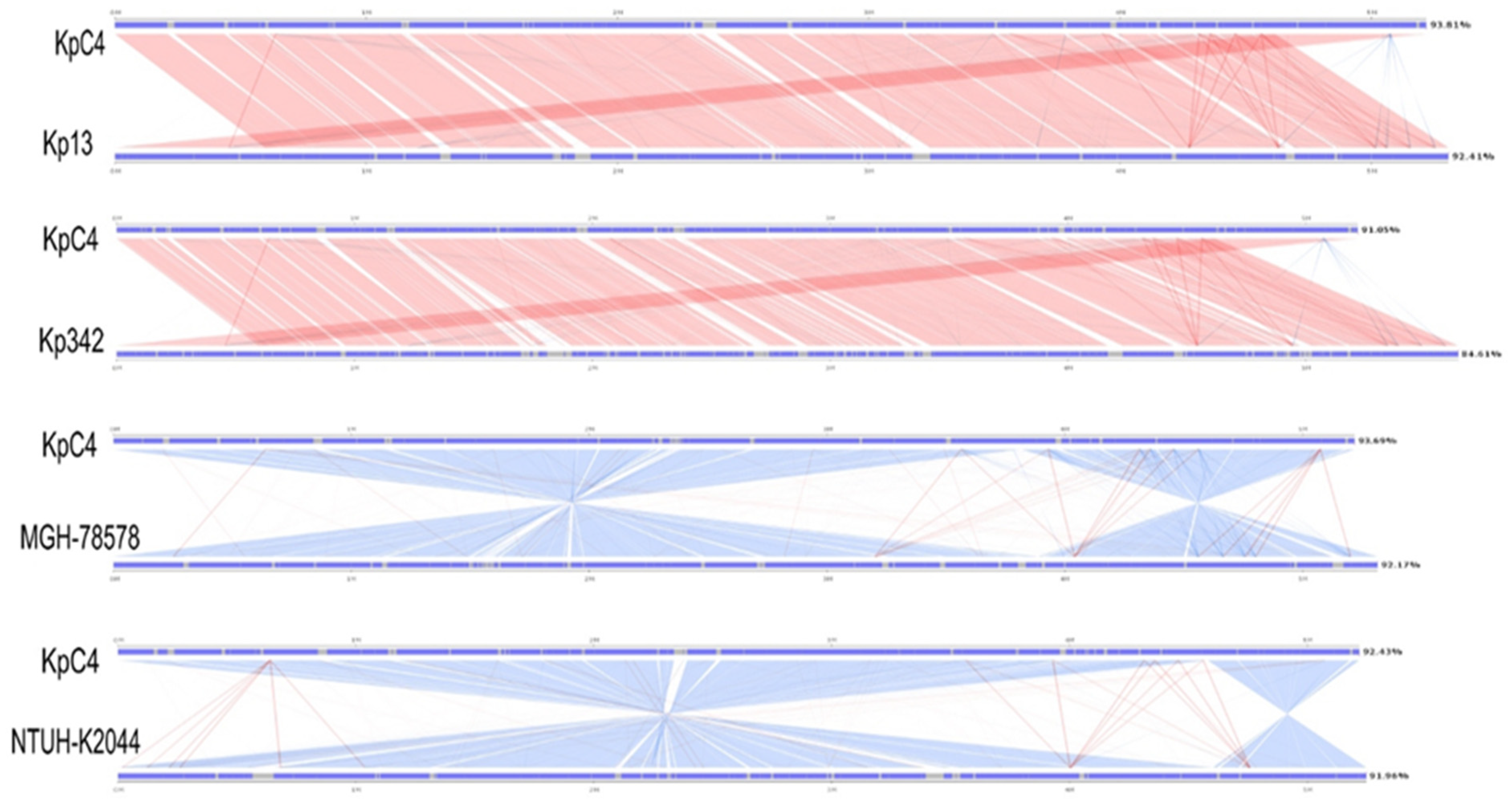
| Kp Strains | Size (bp) | GC Content (%) | No. of CDSs | Mean CDS Size (bp) | Coding Density (%) | No. of tRNAs | Reference |
|---|---|---|---|---|---|---|---|
| KpC4 | 5,218,784 | 58.87 | 4912 | 920 | 86.25 | 86 | This study |
| MGH 78578 | 5,315,120 | 57.5 | 4776 | 958 | 86.1 | 86 | [15] |
| Kp13 | 5,307,003 | 57.5 | 5288 | 896 | 89.3 | 86 | [15] |
| NTUH-K2044 | 5,248,520 | 57.7 | 5130 | 939 | 89.4 | 86 | [16] |
| Kv342 (K. variicola) | 5,641,239 | 57.3 | 5425 | 915 | 88.0 | 88 | [17] |
| Kp52.145 | 5,438,894 | 56.4 | 5314 | - | 88.0 | 85 | [17] |
| Query_id | Gene | Identity (%) | Function | Pathogen Species | Accession | Disease Name | Experimental Host |
|---|---|---|---|---|---|---|---|
| KpC4_4208 | metJ | 96.19 | Repressor of the methionine biosynthesis regulon | Pectobacterium atrosepticum | Q6CZA0 | Rotting of tubers; blackleg disease of the plant stem | Potato |
| KpC4_4753 | rsmB | 70.09 | Modulates plant cell wall degrading enzymes | Pectobacterium atrosepticum | Q6D000 | Rotting of tubers; blackleg disease of the plant stem | Potato |
| KpC4_4136 | corA | 89.87 | Magnesium/nickel/cobalt transporter | Pectobacterium carotovorum | E1AP33 | Soft rot disease | Celery/carrot |
| KpC4_3550 | RsmA | 79.41 | Post-transcriptional regulator | P. wasabiae | D0KML5 | Soft rot | Tobacco/potato |
| KpC4_0446 | rpoS | 95.76 | Regulation of stress and starvation response | E. amylovora | D4HX24 | Fire blight | Pear/loquat |
| KpC4_0922 | rcsB | 92.09 | Promote transcription of the genes for capsule synthesis | E. amylovora | P96320 | Fire blight | Pear |
| KpC4_4860 | nlpI | 86.05 | Tetratricopeptide lipoprotein | E. amylovora | D4ICB6 | Fire blight | Pear |
| KpC4_3140 | AcrB | 79.57 | Multidrug efflux pump | E. amylovora | Q7WTQ9 | Fire blight | Apple |
| KpC4_4819 | rpoN | 84.07 | Sigma factor, regulating essential virulence gene | E. amylovora | D4HUY5 | Fire blight | Apple |
| KpC4_4766 | AcrB | 79.57 | Multidrug efflux pump | E. amylovora | Q7WTQ9 | Fire blight | Apple |
| KpC4_4696 | argD | 78.71 | N-acetylornithine aminotransferase enzyme | E. amylovora | D4I307 | Fire blight | Pear/apple |
| KpC4_0520 | rsmAXoo | 82.14 | RNA-binding protein | Xanthomonas oryzae pv. Oryzae | E2J5T5 | Bacterial leaf blight | Rice |
| KpC4_2459 | iutA | 75.93 | Siderophore-mediated iron acquisition | Pantoea stewartii | H3RJF2 | Stewart wilt of sweet corn | Maize |
| Gene Cluster | ORF No. | ORF Name | ORF Location | ORF Homolog Characteristics |
|---|---|---|---|---|
| waa | coaD | KpC4_4461 | Phosphopantetheine adenylyltransferase | |
| 1 | waaE | KpC4_4462 | Glucosyl transferase | |
| 2 | waaA | KpC4_4463 | Kdo transferase | |
| 3 | ORF4 | KpC4_4464 | Glycosyl transferase | |
| 4 | wabH | KpC4_4465 | Glycosyl transferase | |
| 5 | wabG | KpC4_4466 | Glucuronic acid transferase | |
| 6 | waaQ | KpC4_4467 | Heptosyl III transferase | |
| 7 | wabN | KpC4_4468 | Deacetylase | |
| 8 | ORF8 | KpC4_4469 | LPS 1,2-N-acetylglucosaminetransferase | |
| 9 | waaL | KpC4_4470 | O-antigen ligase | |
| 10 | ORF10 | KpC4_4471 | Glycosyltransferase | |
| 11 | waaC | KpC4_4472 | Heptosyltransferase I | |
| 12 | waaF | KpC4_4473 | Heptosyltransferase II | |
| 13 | hldD | KpC4_4474 | ADP-L-glycero-D-manno-heptose-6-epimerase | |
| kbl | KpC4_4475 | 2-amino-3-ketobutyrate coenzyme A ligase | ||
| Lpx | lpxA | KpC4_3397 | UDP-N-acetylglucosamine acyltransferase | |
| lpxB | KpC4_3396 | Lipid-A-disaccharide synthase | ||
| lpxC | KpC4_3497 | UDP-3-O-acyl N-acetylglucosamine deacetylase | ||
| lpxD | KpC4_3399 KpC4_4042 | UDP-3-O-(3-hydroxymyristoyl)-glucosamine n-acyltransferase | ||
| lpxK | KpC4_2629 | Tetraacyldisaccharide 4′-kinase |
| Category | System | Gene | Role | CDS ∆ | Find In | ||||
|---|---|---|---|---|---|---|---|---|---|
| KpC4 | 78578 | K2044 | Kp13 | 342 | |||||
| Feo | Feo | feoABC | Fe2+ transport | KpC4_4660 | + | − | + | − | − |
| ABC transporter | Sit | sitABCD | Fe2+ transport | KpC4_0468 | + | + | + | + | + |
| Kfu | kfuABC | Fe3+ transport | KpC4_2569 | ||||||
| Fec | fecBDE | Ferric citrate transport | KpC4_1411 | ||||||
| fpbABC | Fe3+ transport | − | − | + | − | + | |||
| Hemophore-based | Hmu | hmuRSTUV | Heme utilization | KpC4_0458 | + | + | + | + | + |
| Siderophore-based | Fep | fepABCG | Enterobactin transport | KpC4_4005 | + | + | + | + | + |
| Ent | EntA-F | Enterobactin synthesis | KpC4_2951 | + | + | + | + | + | |
| Fhu | fhuA-C | Ferrichrome transport | KpC4_3427 | ||||||
| IroA | iroN | Salmochelin transport | KpC4_2314 | − | − | + | − | − | |
| iroBCDE | Salmochelin synthesis | KpC4_1881 | ‡ | − | + | − | − | ||
| Aerobactin | iutA | Aerobactin transport | KpC4_2459 | + | ± | + | ± | ± | |
| iucABCD | Aerobactin synthesis | − | − | + | − | − | |||
| Yersinia HPI | ybtPQXS, ybtA-irp2-irp1- ybtUTE-fyuA | Yersiniabactin synthesis and transport | − | − | + | + | − | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, M.; He, P.; He, P.; Wu, Y.; Munir, S.; He, Y. Novel Virulence Factors Deciphering Klebsiella pneumoniae KpC4 Infect Maize as a Crossing-Kingdom Pathogen: An Emerging Environmental Threat. Int. J. Mol. Sci. 2022, 23, 16005. https://doi.org/10.3390/ijms232416005
Huang M, He P, He P, Wu Y, Munir S, He Y. Novel Virulence Factors Deciphering Klebsiella pneumoniae KpC4 Infect Maize as a Crossing-Kingdom Pathogen: An Emerging Environmental Threat. International Journal of Molecular Sciences. 2022; 23(24):16005. https://doi.org/10.3390/ijms232416005
Chicago/Turabian StyleHuang, Min, Pengfei He, Pengbo He, Yixin Wu, Shahzad Munir, and Yueqiu He. 2022. "Novel Virulence Factors Deciphering Klebsiella pneumoniae KpC4 Infect Maize as a Crossing-Kingdom Pathogen: An Emerging Environmental Threat" International Journal of Molecular Sciences 23, no. 24: 16005. https://doi.org/10.3390/ijms232416005
APA StyleHuang, M., He, P., He, P., Wu, Y., Munir, S., & He, Y. (2022). Novel Virulence Factors Deciphering Klebsiella pneumoniae KpC4 Infect Maize as a Crossing-Kingdom Pathogen: An Emerging Environmental Threat. International Journal of Molecular Sciences, 23(24), 16005. https://doi.org/10.3390/ijms232416005






