Anti-SOD1 Nanobodies That Stabilize Misfolded SOD1 Proteins Also Promote Neurite Outgrowth in Mutant SOD1 Human Neurons
Abstract
1. Introduction
2. Results
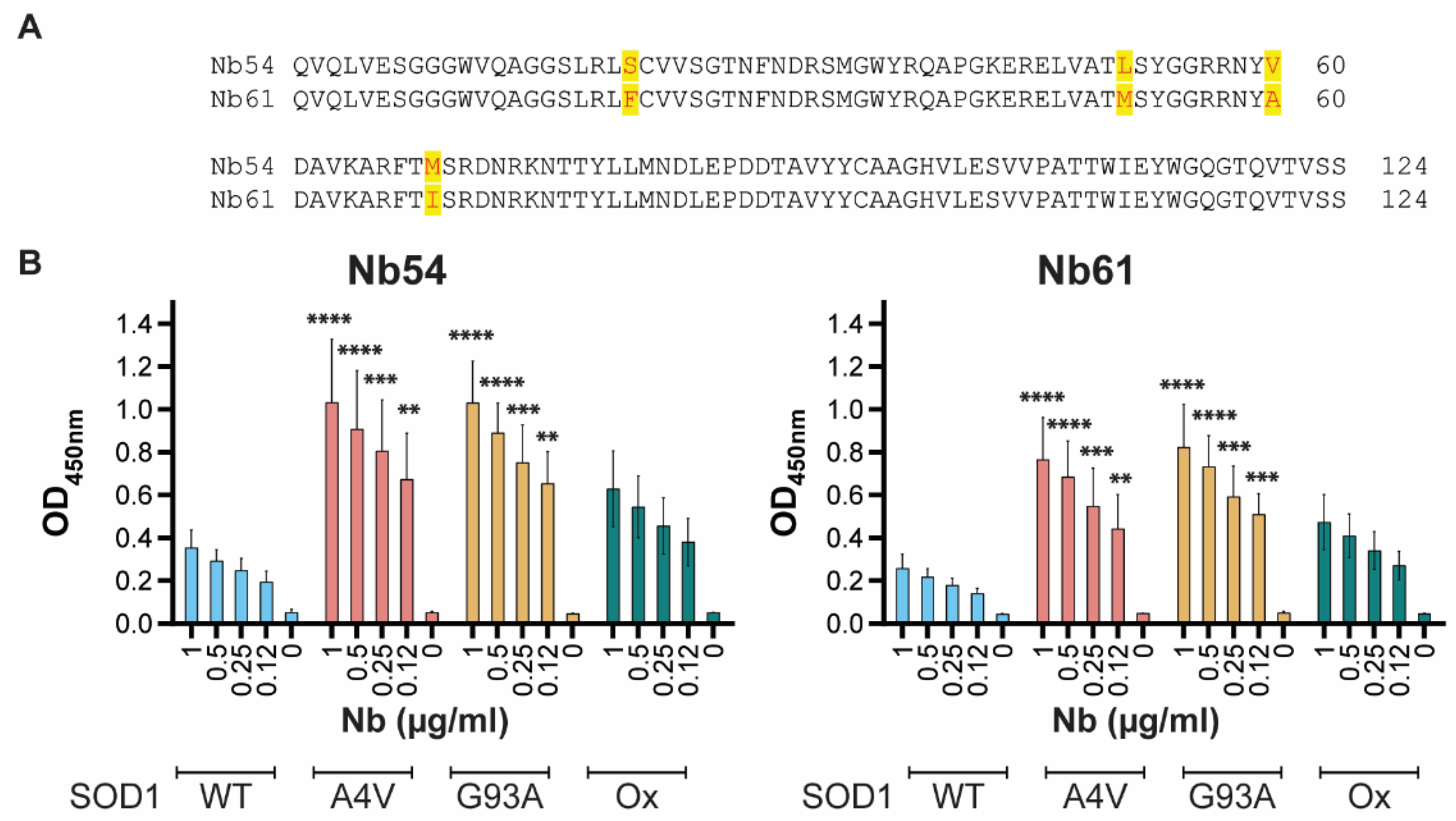
2.1. Nanobodies with Selectivity for ALS-Linked SOD1
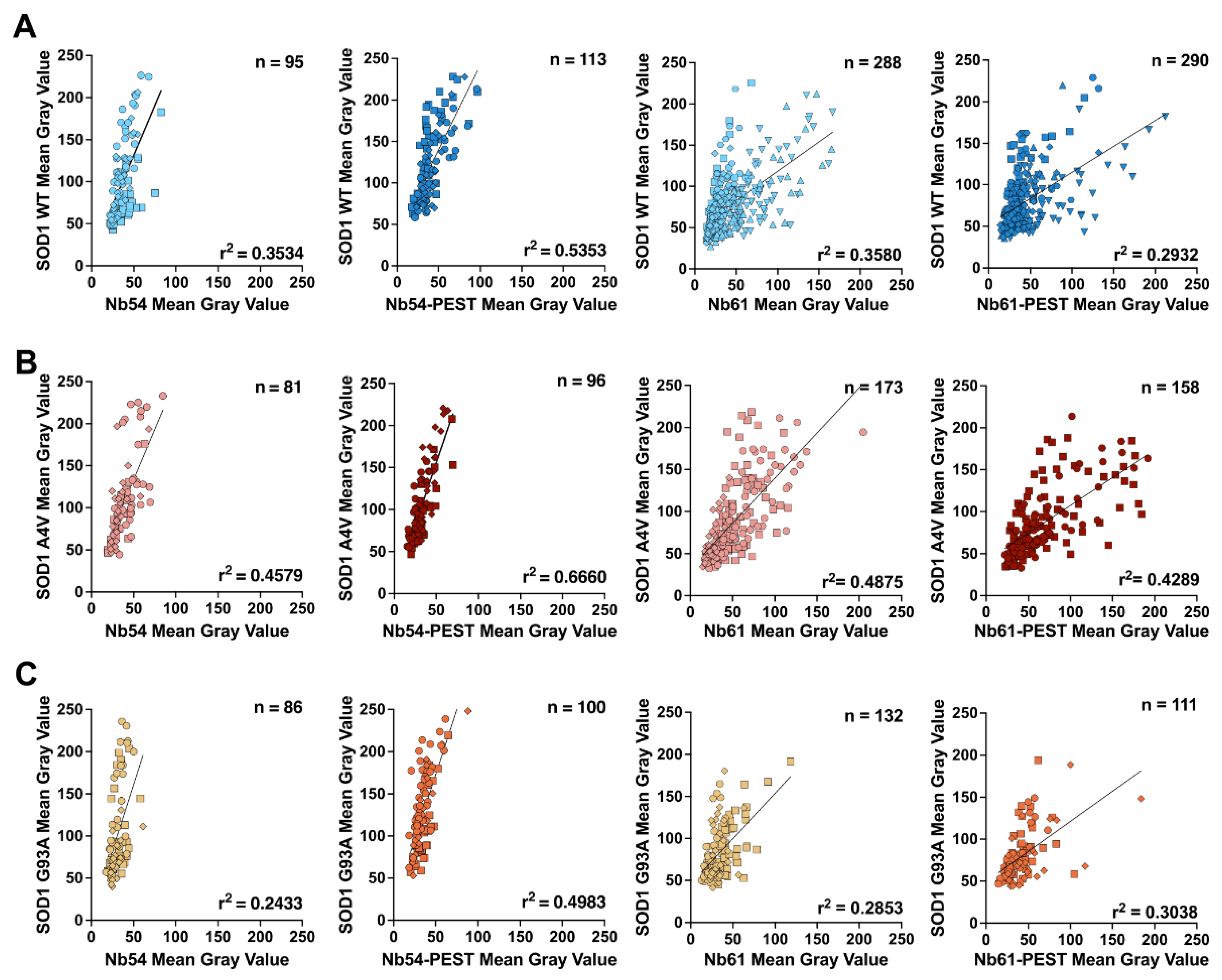
2.2. Anti-SOD1 Nanobodies Lead to Enhanced, Rather Than Reduced, Levels of Ectopic SOD1 in Cellulo
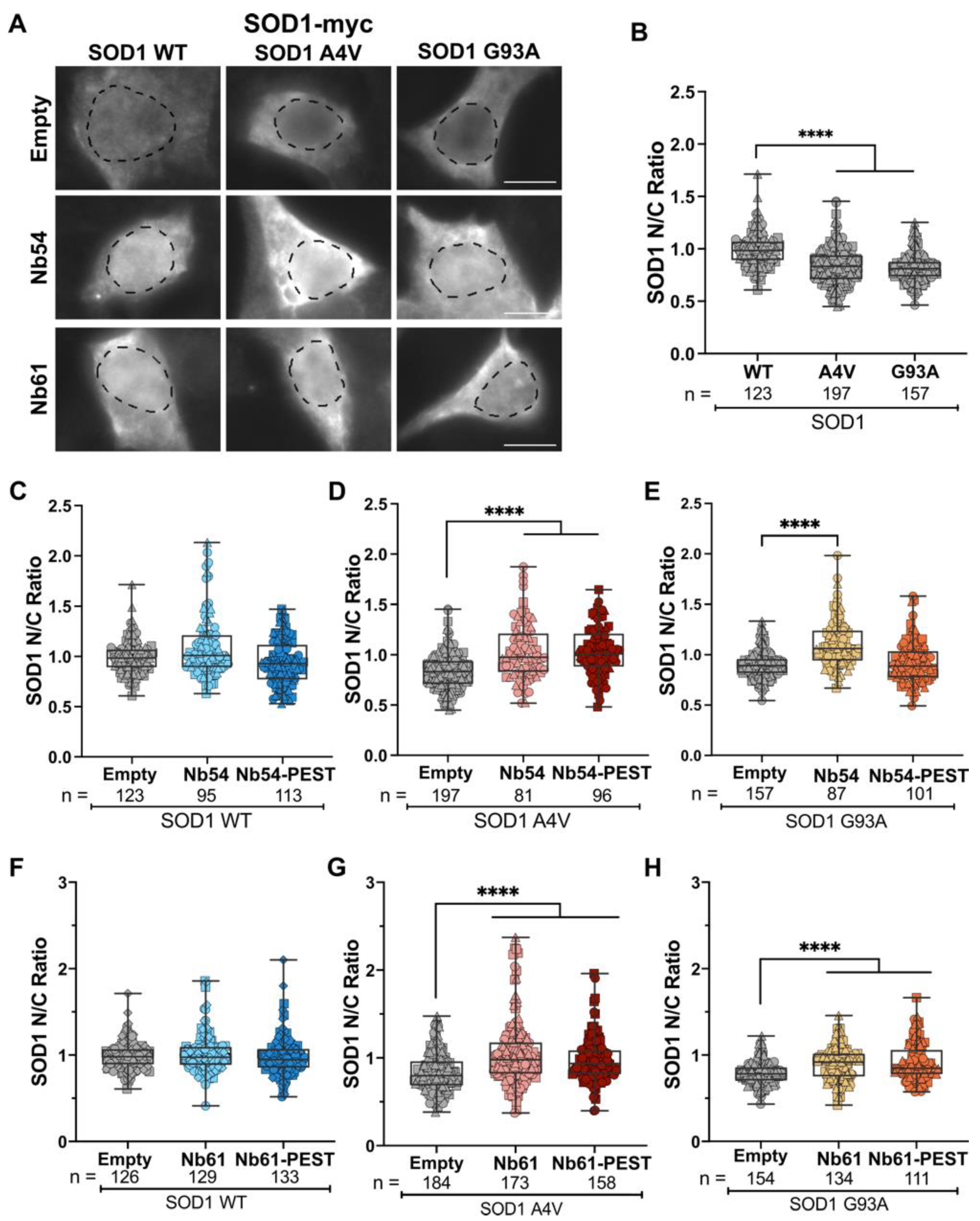
2.3. The Subcellular Localization of Mutant SOD1 Is Restored by Co-Expression of Anti-SOD1 Nanobodies
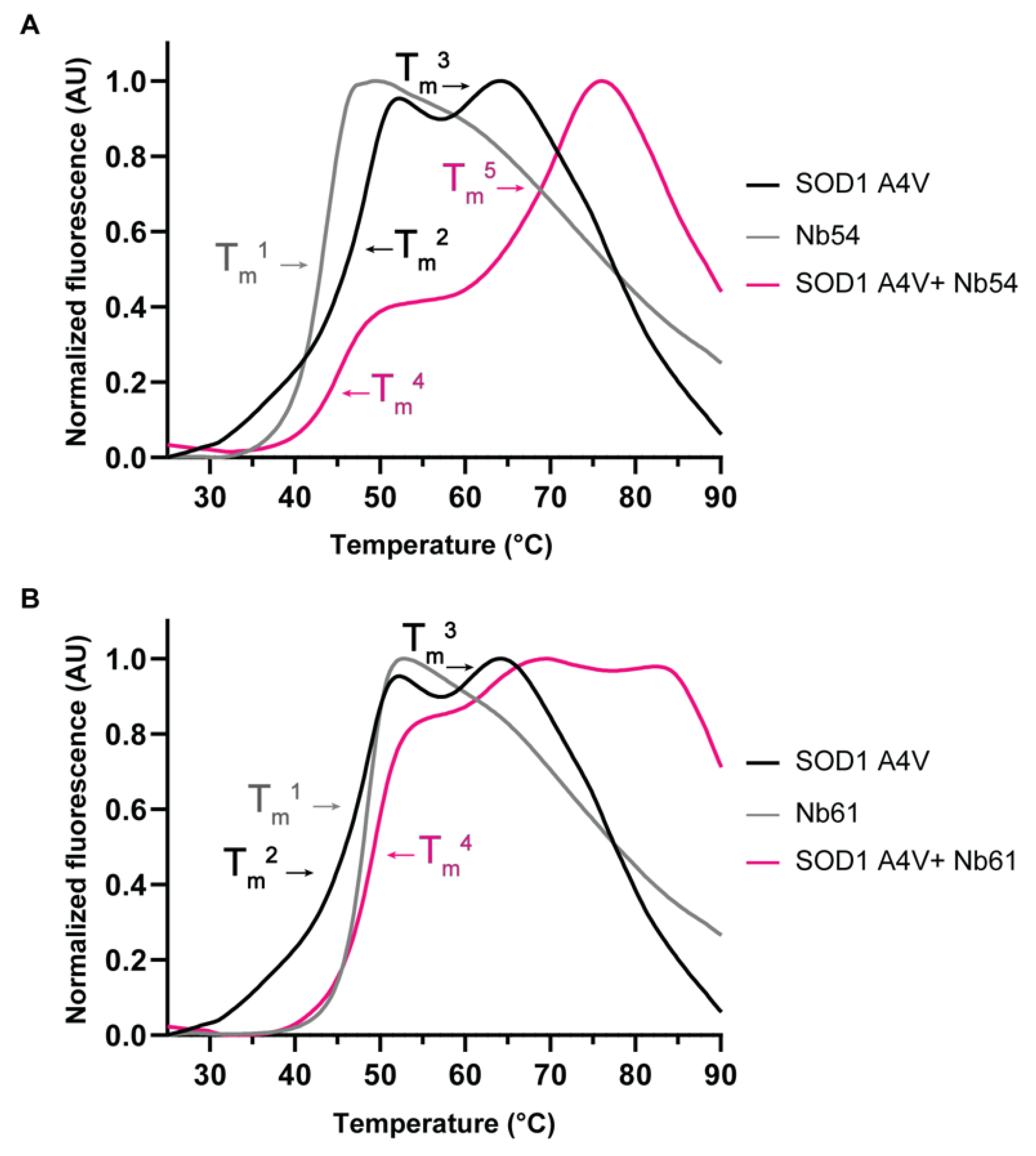
2.4. Anti-SOD1 Nanobodies Stabilize Mutant SOD1 in Cellulo
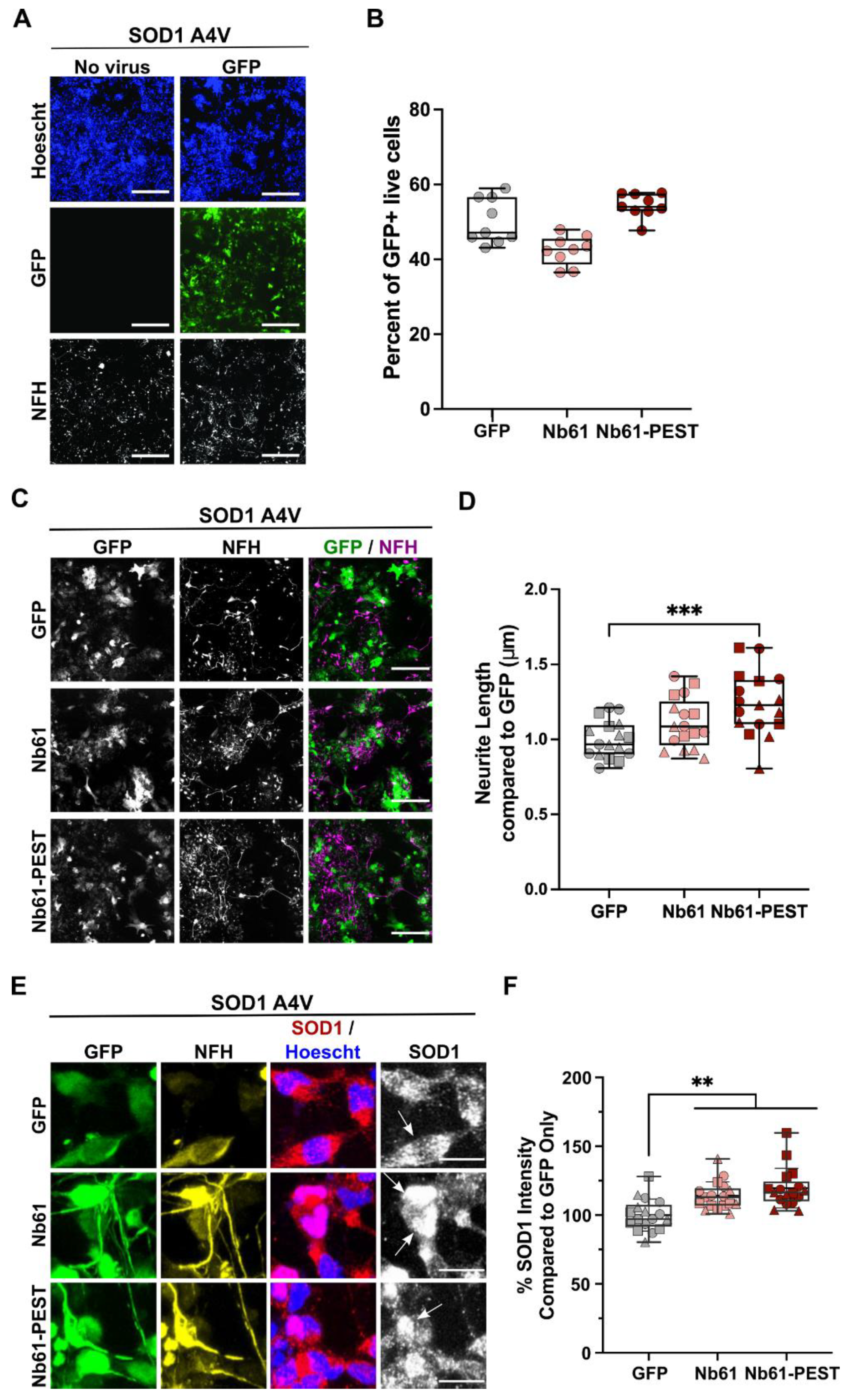
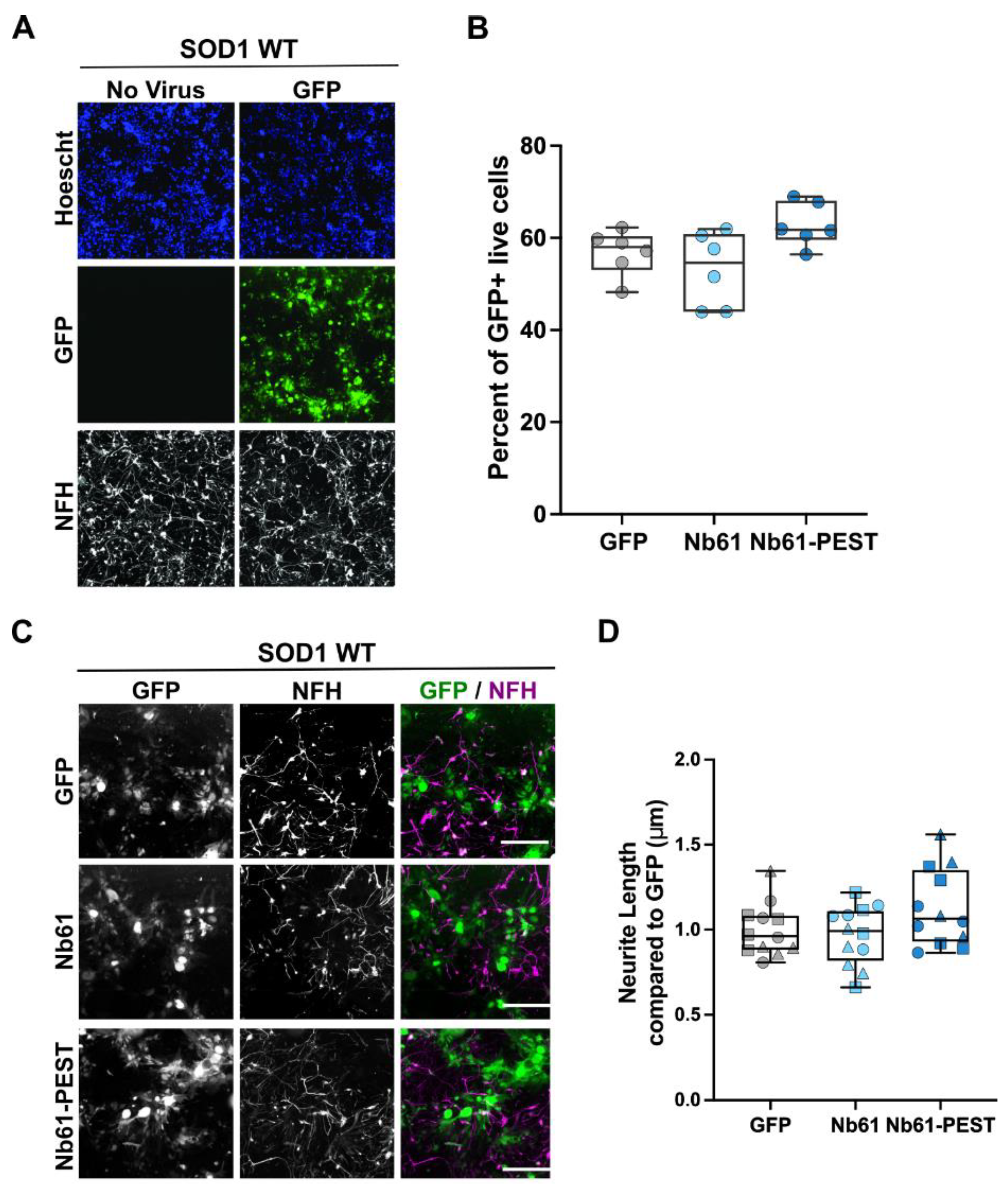
2.5. Anti-SOD1 Nanobody Expression Is Non-Toxic and Induces Enhanced Neurite Outgrowth in Human SOD1 A4V Motor Neurons
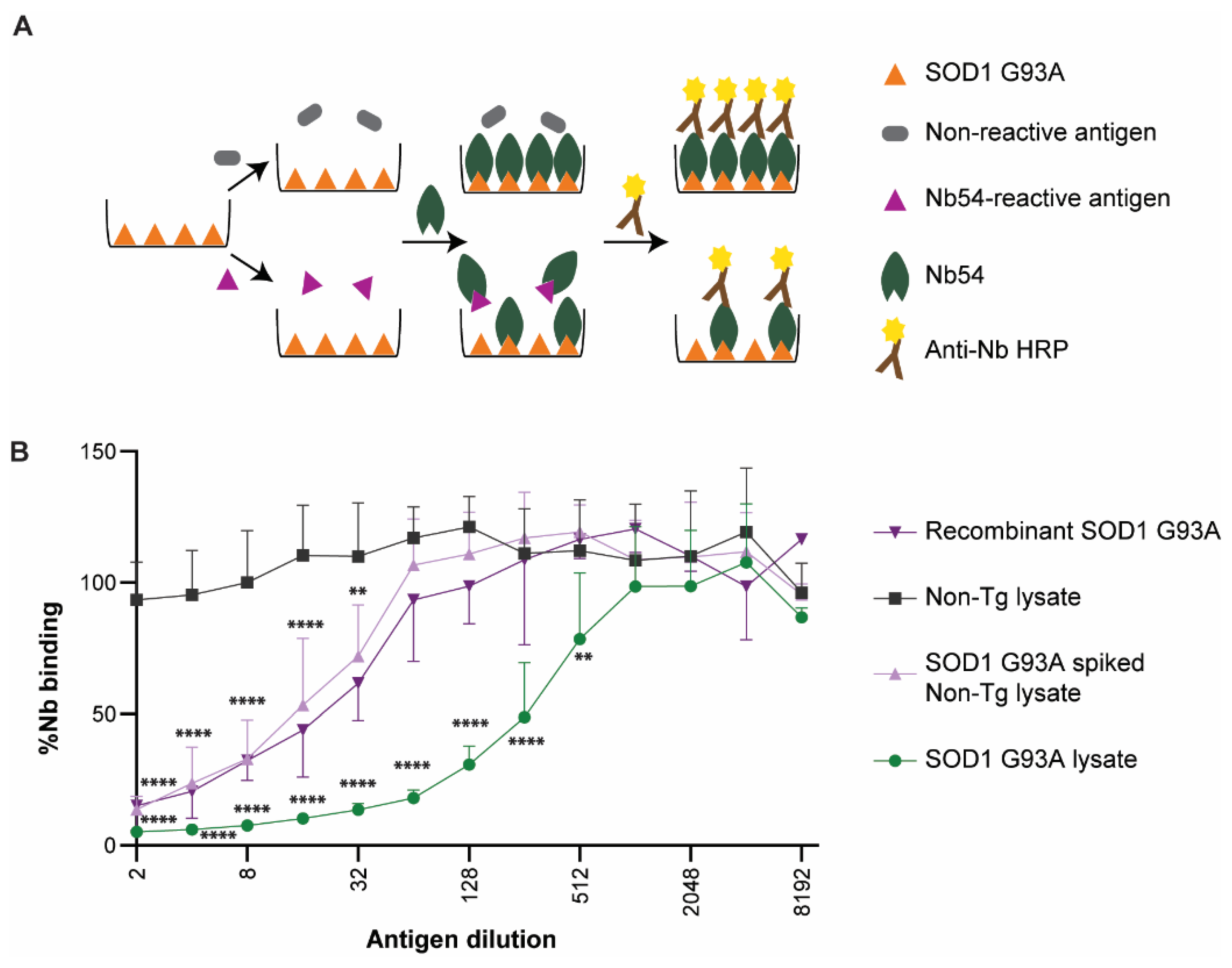
2.6. Anti-SOD1 Nanobodies Detect Human SOD1 G93A in Lysates from an ALS Mouse Model
3. Discussion
4. Materials and Methods
4.1. Generation of Anti-SOD1 Nanobodies
4.2. Plasmid Construction
4.3. Recombinant Nanobody Expression and Purification
4.4. ELISA
4.5. Transient Transfection of HEK293T Cells
4.6. Image Acquisition and Analysis of Transfected HEK293T Cells
4.6.1. HEK293T N/C Ratio
4.6.2. HEK293T Signal Intensity Analysis
4.6.3. Western Blot Analysis
4.7. iPSC Culture, Motor Neuron Differentiation, and Lentiviral Transduction
4.8. Immunofluorescence Microscopy Analysis of iPSC-Derived Motor Neurons
4.8.1. Neurite-Tracing Analysis
4.8.2. SOD1-Intensity Analysis
4.9. Differential Scanning Fluorimetry (DSF)
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, R.H.; Al-Chalabi, A. Amyotrophic Lateral Sclerosis. N. Engl. J. Med. 2017, 377, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Pardo, C.A.; Xu, Z.; Borchelt, D.R.; Price, D.L.; Sisodia, S.S.; Cleveland, D.W.; Brown, R.; Price, D.; Sisodia, S.; Cleveland, D.; et al. Superoxide dismutase is an abundant component in cell bodies, dendrites, and axons of motor neurons and in a subset of other neurons. Proc. Natl. Acad. Sci. USA 1995, 92, 954–958. [Google Scholar] [CrossRef] [PubMed]
- Trist, B.G.; Hilton, J.B.; Hare, D.J.; Crouch, P.J.; Double, K.L. Superoxide Dismutase 1 in Health and Disease: How a Frontline Antioxidant Becomes Neurotoxic. Angew. Chem. Int. Ed. Engl. 2021, 60, 9215–9246. [Google Scholar] [CrossRef] [PubMed]
- Gurney, M.E.; Pu, H.; Chiu, A.Y.; Dal Canto, M.C.; Polchow, C.Y.; Alexander, D.D.; Caliendo, J.; Hentati, A.; Kwon, Y.W.; Deng, H.X.; et al. Motor neuron degeneration in mice that express a human Cu, Zn superoxide dismutase mutation. Science 1994, 264, 1772–1775. [Google Scholar] [CrossRef]
- Rotunno, M.S.; Bosco, D.A. An emerging role for misfolded wild-type SOD1 in sporadic ALS pathogenesis. Front. Cell Neurosci. 2013, 7, 253. [Google Scholar] [CrossRef] [PubMed]
- Rotunno, M.S.; Auclair, J.R.; Maniatis, S.; Shaffer, S.A.; Agar, J.; Bosco, D.A. Identification of a misfolded region in superoxide dismutase 1 that is exposed in amyotrophic lateral sclerosis. J. Biol. Chem. 2014, 289, 28527–28538. [Google Scholar] [CrossRef]
- Wang, Q.; Johnson, J.L.; Agar, N.Y.; Agar, J.N. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS Biol. 2008, 6, e170. [Google Scholar] [CrossRef] [PubMed]
- Medinas, D.B.; Rozas, P.; Martínez Traub, F.; Woehlbier, U.; Brown, R.H.; Bosco, D.A.; Hetz, C. Endoplasmic reticulum stress leads to accumulation of wild-type SOD1 aggregates associated with sporadic amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2018, 115, 201801109. [Google Scholar] [CrossRef]
- Guareschi, S.; Cova, E.; Cereda, C.; Ceroni, M.; Donetti, E.; Bosco, D.A.; Trotti, D.; Pasinelli, P. An over-oxidized form of superoxide dismutase found in sporadic amyotrophic lateral sclerosis with bulbar onset shares a toxic mechanism with mutant SOD1. Proc. Natl. Acad. Sci. USA 2012, 109, 5074–5079. [Google Scholar] [CrossRef]
- Ezzi, S.A.; Urushitani, M.; Julien, J.P. Wild-type superoxide dismutase acquires binding and toxic properties of ALS-linked mutant forms through oxidation. J. Neurochem. 2007, 102, 170–178. [Google Scholar] [CrossRef]
- Trist, B.G.; Genoud, S.; Roudeau, S.; Rookyard, A.; Abdeen, A.; Cottam, V.; Hare, D.J.; White, M.; Altvater, J.; Fifita, J.A.; et al. Altered SOD1 maturation and post-translational modification in amyotrophic lateral sclerosis spinal cord. Brain 2022, 145, 3108–3130. [Google Scholar] [CrossRef] [PubMed]
- Mueller, C.; Berry, J.D.; McKenna-Yasek, D.M.; Gernoux, G.; Owegi, M.A.; Pothier, L.M.; Douthwright, C.L.; Gelevski, D.; Luppino, S.D.; Blackwood, M.; et al. SOD1 Suppression with Adeno-Associated Virus and MicroRNA in Familial ALS. N. Engl. J. Med. 2020, 383, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.; Cudkowicz, M.; Shaw, P.J.; Andersen, P.M.; Atassi, N.; Bucelli, R.C.; Genge, A.; Glass, J.; Ladha, S.; Ludolph, A.L.; et al. Phase 1-2 Trial of Antisense Oligonucleotide Tofersen for SOD1 ALS. N. Engl. J. Med. 2020, 383, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Boros, B.D.; Schoch, K.M.; Kreple, C.J.; Miller, T.M. Antisense Oligonucleotides for the Study and Treatment of ALS. Neurotherapeutics 2022, 19, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- van Blitterswijk, M.; Gulati, S.; Smoot, E.; Jaffa, M.; Maher, N.; Hyman, B.T.; Ivinson, A.J.; Scherzer, C.R.; Schoenfeld, D.A.; Cudkowicz, M.E.; et al. Anti-superoxide dismutase antibodies are associated with survival in patients with sporadic amyotrophic lateral sclerosis. Amyotroph Lateral. Scler. 2011, 12, 430–438. [Google Scholar] [CrossRef]
- Elchuri, S.; Oberley, T.D.; Qi, W.; Eisenstein, R.S.; Jackson Roberts, L.; Van Remmen, H.; Epstein, C.J.; Huang, T.T. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene 2005, 24, 367–380. [Google Scholar] [CrossRef]
- Kondo, T.; Reaume, A.G.; Huang, T.T.; Carlson, E.; Murakami, K.; Chen, S.F.; Hoffman, E.K.; Scott, R.W.; Epstein, C.J.; Chan, P.H. Reduction of CuZn-superoxide dismutase activity exacerbates neuronal cell injury and edema formation after transient focal cerebral ischemia. J. Neurosci. 1997, 17, 4180–4189. [Google Scholar] [CrossRef]
- Reaume, A.G.; Elliott, J.L.; Hoffman, E.K.; Kowall, N.W.; Ferrante, R.J.; Siwek, D.F.; Wilcox, H.M.; Flood, D.G.; Beal, M.F.; Brown, R.H., Jr.; et al. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat. Genet. 1996, 13, 43–47. [Google Scholar] [CrossRef]
- Shefner, J.M.; Reaume, A.G.; Flood, D.G.; Scott, R.W.; Kowall, N.W.; Ferrante, R.J.; Siwek, D.F.; Upton-Rice, M.; Brown, R.H., Jr. Mice lacking cytosolic copper/zinc superoxide dismutase display a distinctive motor axonopathy. Neurology 1999, 53, 1239–1246. [Google Scholar] [CrossRef]
- Shibata, N.; Asayama, K.; Hirano, A.; Kobayashi, M. Immunohistochemical study on superoxide dismutases in spinal cords from autopsied patients with amyotrophic lateral sclerosis. Dev. Neurosci. 1996, 18, 492–498. [Google Scholar] [CrossRef]
- Jonsson, P.A.; Ernhill, K.; Andersen, P.M.; Bergemalm, D.; Brannstrom, T.; Gredal, O.; Nilsson, P.; Marklund, S.L. Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain 2004, 127, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Bruijn, L.I.; Houseweart, M.K.; Kato, S.; Anderson, K.L.; Anderson, S.D.; Ohama, E.; Reaume, A.G.; Scott, R.W.; Cleveland, D.W. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science 1998, 281, 1851–1854. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Wang, J.; Henderson, M.J.; Yang, P.; Hagen, B.M.; Siddique, T.; Vogel, B.E.; Deng, H.X.; Fang, S. Nuclear export of misfolded SOD1 mediated by a normally buried NES-like sequence reduces proteotoxicity in the nucleus. eLife 2017, 6, e23759. [Google Scholar] [CrossRef] [PubMed]
- Saccon, R.A.; Bunton-Stasyshyn, R.K.A.; Fisher, E.M.C.; Fratta, P. Is SOD1 loss of function involved in amyotrophic lateral sclerosis? Brain A J. Neurol. 2013, 136, 2342–2358. [Google Scholar] [CrossRef]
- Trist, B.G.; Davies, K.M.; Cottam, V.; Genoud, S.; Ortega, R.; Roudeau, S.; Carmona, A.; De Silva, K.; Wasinger, V.; Lewis, S.J.G.; et al. Amyotrophic lateral sclerosis-like superoxide dismutase 1 proteinopathy is associated with neuronal loss in Parkinson’s disease brain. Acta Neuropathol. 2017, 134, 113–127. [Google Scholar] [CrossRef]
- Takeuchi, S.; Fujiwara, N.; Ido, A.; Oono, M.; Takeuchi, Y.; Tateno, M.; Suzuki, K.; Takahashi, R.; Tooyama, I.; Taniguchi, N.; et al. Induction of protective immunity by vaccination with wild-type apo superoxide dismutase 1 in mutant SOD1 transgenic mice. J. Neuropathol. Exp. Neurol. 2010, 69, 1044–1056. [Google Scholar] [CrossRef]
- Urushitani, M.; Ezzi, S.A.; Julien, J.-P. Therapeutic effects of immunization with mutant superoxide dismutase in mice models of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2007, 104, 2495–2500. [Google Scholar] [CrossRef]
- Liu, H.N.; Tjostheim, S.; Dasilva, K.; Taylor, D.; Zhao, B.; Rakhit, R.; Brown, M.; Chakrabartty, A.; McLaurin, J.; Robertson, J. Targeting of monomer/misfolded SOD1 as a therapeutic strategy for amyotrophic lateral sclerosis. J. Neurosci. 2012, 32, 8791–8799. [Google Scholar] [CrossRef]
- Gros-Louis, F.; Soucy, G.; Lariviere, R.; Julien, J.P. Intracerebroventricular infusion of monoclonal antibody or its derived Fab fragment against misfolded forms of SOD1 mutant delays mortality in a mouse model of ALS. J. Neurochem. 2010, 113, 1188–1199. [Google Scholar] [CrossRef]
- Maier, M.; Welt, T.; Wirth, F.; Montrasio, F.; Preisig, D.; McAfoose, J.; Vieira, F.G.; Kulic, L.; Späni, C.; Stehle, T.; et al. A human-derived antibody targets misfolded SOD1 and ameliorates motor symptoms in mouse models of amyotrophic lateral sclerosis. Sci. Transl. Med. 2018, 10, eaah3924. [Google Scholar] [CrossRef]
- Silva-Pilipich, N.; Smerdou, C.; Vanrell, L. A Small Virus to Deliver Small Antibodies: New Targeted Therapies Based on AAV Delivery of Nanobodies. Microorganisms 2021, 9, 1956. [Google Scholar] [CrossRef] [PubMed]
- Ghadge, G.D.; Kay, B.K.; Drigotas, C.; Roos, R.P. Single chain variable fragment antibodies directed against SOD1 ameliorate disease in mutant SOD1 transgenic mice. Neurobiol. Dis. 2019, 121, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Kriz, J.; Gravel, M.; Soucy, G.; Bareil, C.; Gravel, C.; Julien, J.P. Adeno-associated virus-mediated delivery of a recombinant single-chain antibody against misfolded superoxide dismutase for treatment of amyotrophic lateral sclerosis. Mol. Ther. 2014, 22, 498–510. [Google Scholar] [CrossRef]
- Muyldermans, S. A guide to: Generation and design of nanobodies. FEBS J. 2021, 288, 2084–2102. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, D.; Bhatt, M.; Butler, D.; De Genst, E.; Dobson, C.M.; Messer, A.; Kordower, J.H. Proteasome-targeted nanobodies alleviate pathology and functional decline in an alpha-synuclein-based Parkinson’s disease model. NPJ Park. Dis. 2018, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Pothin, E.; Lesuisse, D.; Lafaye, P. Brain Delivery of Single-Domain Antibodies: A Focus on VHH and VNAR. Pharmaceutics 2020, 12, 937. [Google Scholar] [CrossRef] [PubMed]
- Danis, C.; Dupre, E.; Zejneli, O.; Caillierez, R.; Arrial, A.; Begard, S.; Mortelecque, J.; Eddarkaoui, S.; Loyens, A.; Cantrelle, F.X.; et al. Inhibition of Tau seeding by targeting Tau nucleation core within neurons with a single domain antibody fragment. Mol. Ther. 2022, 30, 1484–1499. [Google Scholar] [CrossRef]
- Bosco, D.A.; Morfini, G.; Karabacak, N.M.; Song, Y.; Gros-Louis, F.; Pasinelli, P.; Goolsby, H.; Fontaine, B.A.; Lemay, N.; McKenna-Yasek, D.; et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 2010, 13, 1396–1403. [Google Scholar] [CrossRef]
- Deng, H.X.; Hentati, A.; Tainer, J.A.; Iqbal, Z.; Cayabyab, A.; Hung, W.Y.; Getzoff, E.D.; Hu, P.; Herzfeldt, B.; Roos, R.P.; et al. Amyotrophic lateral sclerosis and structural defects in Cu, Zn superoxide dismutase. Science 1993, 261, 1047–1051. [Google Scholar] [CrossRef]
- Molnar, K.S.; Karabacak, N.M.; Johnson, J.L.; Wang, Q.; Tiwari, A.; Hayward, L.J.; Coales, S.J.; Hamuro, Y.; Agar, J.N. A common property of amyotrophic lateral sclerosis-associated variants: Destabilization of the copper/zinc superoxide dismutase electrostatic loop. J. Biol. Chem. 2009, 284, 30965–30973. [Google Scholar] [CrossRef]
- Rechsteiner, M.; Rogers, S.W. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 1996, 21, 267–271. [Google Scholar] [CrossRef]
- Butler, D.C.; Joshi, S.N.; Genst, E.; Baghel, A.S.; Dobson, C.M.; Messer, A. Bifunctional Anti-Non-Amyloid Component alpha-Synuclein Nanobodies Are Protective In Situ. PLoS ONE 2016, 11, e0165964. [Google Scholar] [CrossRef]
- Joshi, S.N.; Butler, D.C.; Messer, A. Fusion to a highly charged proteasomal retargeting sequence increases soluble cytoplasmic expression and efficacy of diverse anti-synuclein intrabodies. MAbs 2012, 4, 686–693. [Google Scholar] [CrossRef]
- Butler, D.C.; Messer, A. Bifunctional anti-huntingtin proteasome-directed intrabodies mediate efficient degradation of mutant huntingtin exon 1 protein fragments. PLoS ONE 2011, 6, e29199. [Google Scholar] [CrossRef]
- Crapo, J.D.; Oury, T.; Rabouille, C.; Slot, J.W.; Chang, L.Y. Copper, zinc superoxide dismutase is primarily a cytosolic protein in human cells. Proc. Natl. Acad. Sci. USA 1992, 89, 10405–10409. [Google Scholar] [CrossRef]
- Rodriguez, J.A.; Valentine, J.S.; Eggers, D.K.; Roe, J.A.; Tiwari, A.; Brown, R.H.; Hayward, L.J. Familial amyotrophic lateral sclerosis-associated mutations decrease the thermal stability of distinctly metallated species of human copper/zinc superoxide dismutase. J. Biol. Chem. 2002, 277, 15932–15937. [Google Scholar] [CrossRef]
- Borchelt, D.R.; Lee, M.K.; Slunt, H.S.; Guarnieri, M.; Xu, Z.S.; Wong, P.C.; Brown, R.H., Jr.; Price, D.L.; Sisodia, S.S.; Cleveland, D.W. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc. Natl. Acad. Sci. USA 1994, 91, 8292–8296. [Google Scholar] [CrossRef]
- Boopathy, S.; Silvas, T.V.; Tischbein, M.; Jansen, S.; Shandilya, S.M.; Zitzewitz, J.A.; Landers, J.E.; Goode, B.L.; Schiffer, C.A.; Bosco, D.A. Structural basis for mutation-induced destabilization of profilin 1 in ALS. Proc. Natl. Acad. Sci. USA 2015, 112, 7984–7989. [Google Scholar] [CrossRef]
- Auclair, J.R.; Boggio, K.J.; Petsko, G.A.; Ringe, D.; Agar, J.N. Strategies for stabilizing superoxide dismutase (SOD1), the protein destabilized in the most common form of familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2010, 107, 21394–21399. [Google Scholar] [CrossRef]
- Gao, K.; Oerlemans, R.; Groves, M.R. Theory and applications of differential scanning fluorimetry in early-stage drug discovery. Biophys. Rev. 2020, 12, 85–104. [Google Scholar] [CrossRef]
- Niesen, F.H.; Berglund, H.; Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007, 2, 2212–2221. [Google Scholar] [CrossRef]
- Chen, H.; Qian, K.; Du, Z.; Cao, J.; Petersen, A.; Liu, H.; Blackbourn, L.W.t.; Huang, C.L.; Errigo, A.; Yin, Y.; et al. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell 2014, 14, 796–809. [Google Scholar] [CrossRef]
- Kiskinis, E.; Sandoe, J.; Williams, L.A.; Boulting, G.L.; Moccia, R.; Wainger, B.J.; Han, S.; Peng, T.; Thams, S.; Mikkilineni, S.; et al. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell 2014, 14, 781–795. [Google Scholar] [CrossRef]
- Rigamonti, A.; Repetti, G.G.; Sun, C.; Price, F.D.; Reny, D.C.; Rapino, F.; Weisinger, K.; Benkler, C.; Peterson, Q.P.; Davidow, L.S.; et al. Large-Scale Production of Mature Neurons from Human Pluripotent Stem Cells in a Three-Dimensional Suspension Culture System. Stem Cell Rep. 2016, 6, 993–1008. [Google Scholar] [CrossRef]
- Robberecht, W.I.M.; Rousseau, F.; Schymkowitz, J. Sinlge Domain Antibodies against SOD1 and Their Use in Medicine. WO 2014/191493 A1, 28 May 2014. [Google Scholar]
- Shvil, N.; Banerjee, V.; Zoltsman, G.; Shani, T.; Kahn, J.; Abu-Hamad, S.; Papo, N.; Engel, S.; Bernhagen, J.; Israelson, A. MIF inhibits the formation and toxicity of misfolded SOD1 amyloid aggregates: Implications for familial ALS. Cell Death Dis. 2018, 9, 107. [Google Scholar] [CrossRef]
- Sau, D.; De Biasi, S.; Vitellaro-Zuccarello, L.; Riso, P.; Guarnieri, S.; Porrini, M.; Simeoni, S.; Crippa, V.; Onesto, E.; Palazzolo, I.; et al. Mutation of SOD1 in ALS: A gain of a loss of function. Hum. Mol. Genet. 2007, 16, 1604–1618. [Google Scholar] [CrossRef]
- Trist, B.G.; Hare, D.J.; Double, K.L. A Proposed Mechanism for Neurodegeneration in Movement Disorders Characterized by Metal Dyshomeostasis and Oxidative Stress. Cell Chem. Biol. 2018, 25, 807–816. [Google Scholar] [CrossRef]
- Hossain, M.A.; Sarin, R.; Donnelly, D.P.; Miller, B.C.; Salisbury, J.P.; Conway, J.B.; Watson, S.; Winters, J.N.; Alam, N.; Sivasankar, D.; et al. Protein crosslinking as a therapeutic strategy for SOD1-related ALS. bioRxiv 2021. [Google Scholar] [CrossRef]
- Roberts, B.R.; Lim, N.K.; McAllum, E.J.; Donnelly, P.S.; Hare, D.J.; Doble, P.A.; Turner, B.J.; Price, K.A.; Lim, S.C.; Paterson, B.M.; et al. Oral treatment with Cu(II)(atsm) increases mutant SOD1 in vivo but protects motor neurons and improves the phenotype of a transgenic mouse model of amyotrophic lateral sclerosis. J. Neurosci. 2014, 34, 8021–8031. [Google Scholar] [CrossRef]
- Hilton, J.B.; Mercer, S.W.; Lim, N.K.; Faux, N.G.; Buncic, G.; Beckman, J.S.; Roberts, B.R.; Donnelly, P.S.; White, A.R.; Crouch, P.J. Cu(II)(atsm) improves the neurological phenotype and survival of SOD1(G93A) mice and selectively increases enzymatically active SOD1 in the spinal cord. Sci. Rep. 2017, 7, 42292. [Google Scholar] [CrossRef]
- Nikseresht, S.; Hilton, J.B.W.; Kysenius, K.; Liddell, J.R.; Crouch, P.J. Copper-ATSM as a Treatment for ALS: Support from Mutant SOD1 Models and Beyond. Life 2020, 10, 271. [Google Scholar] [CrossRef]
- Abel, O.; Powell, J.F.; Andersen, P.M.; Al-Chalabi, A. ALSoD: A user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum. Mutat. 2012, 33, 1345–1351. [Google Scholar] [CrossRef]
- Pleiner, T.; Bates, M.; Trakhanov, S.; Lee, C.T.; Schliep, J.E.; Chug, H.; Böhning, M.; Stark, H.; Urlaub, H.; Görlich, D. Nanobodies: Site-specific labeling for super-resolution imaging, rapid epitope-mapping and native protein complex isolation. eLife 2015, 4, e11349. [Google Scholar] [CrossRef]
- Frey, S.; Gorlich, D. A new set of highly efficient, tag-cleaving proteases for purifying recombinant proteins. J. Chromatogr. A 2014, 1337, 95–105. [Google Scholar] [CrossRef]
- Baron, D.M.; Kaushansky, L.J.; Ward, C.L.; Sama, R.R.K.; Chian, R.-J.; Boggio, K.J.; Quaresma, A.J.C.; Nickerson, J.A.; Bosco, D.A. Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol. Neurodegener. 2013, 8, 30. [Google Scholar] [CrossRef]
- Lin, Y.C.; Kumar, M.S.; Ramesh, N.; Anderson, E.N.; Nguyen, A.T.; Kim, B.; Cheung, S.; McDonough, J.A.; Skarnes, W.C.; Lopez-Gonzalez, R.; et al. Interactions between ALS-linked FUS and nucleoporins are associated with defects in the nucleocytoplasmic transport pathway. Nat. Neurosci. 2021, 24, 1077–1088. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, M.S.; Fowler-Magaw, M.E.; Kulick, D.; Boopathy, S.; Gadd, D.H.; Rotunno, M.; Douthwright, C.; Golebiowski, D.; Yusuf, I.; Xu, Z.; et al. Anti-SOD1 Nanobodies That Stabilize Misfolded SOD1 Proteins Also Promote Neurite Outgrowth in Mutant SOD1 Human Neurons. Int. J. Mol. Sci. 2022, 23, 16013. https://doi.org/10.3390/ijms232416013
Kumar MS, Fowler-Magaw ME, Kulick D, Boopathy S, Gadd DH, Rotunno M, Douthwright C, Golebiowski D, Yusuf I, Xu Z, et al. Anti-SOD1 Nanobodies That Stabilize Misfolded SOD1 Proteins Also Promote Neurite Outgrowth in Mutant SOD1 Human Neurons. International Journal of Molecular Sciences. 2022; 23(24):16013. https://doi.org/10.3390/ijms232416013
Chicago/Turabian StyleKumar, Meenakshi Sundaram, Megan E. Fowler-Magaw, Daniel Kulick, Sivakumar Boopathy, Del Hayden Gadd, Melissa Rotunno, Catherine Douthwright, Diane Golebiowski, Issa Yusuf, Zuoshang Xu, and et al. 2022. "Anti-SOD1 Nanobodies That Stabilize Misfolded SOD1 Proteins Also Promote Neurite Outgrowth in Mutant SOD1 Human Neurons" International Journal of Molecular Sciences 23, no. 24: 16013. https://doi.org/10.3390/ijms232416013
APA StyleKumar, M. S., Fowler-Magaw, M. E., Kulick, D., Boopathy, S., Gadd, D. H., Rotunno, M., Douthwright, C., Golebiowski, D., Yusuf, I., Xu, Z., Brown, R. H., Jr., Sena-Esteves, M., O'Neil, A. L., & Bosco, D. A. (2022). Anti-SOD1 Nanobodies That Stabilize Misfolded SOD1 Proteins Also Promote Neurite Outgrowth in Mutant SOD1 Human Neurons. International Journal of Molecular Sciences, 23(24), 16013. https://doi.org/10.3390/ijms232416013






