Differential Regulation of Two Arms of mTORC1 Pathway Fine-Tunes Global Protein Synthesis in Resting B Lymphocytes
Abstract
1. Introduction
2. Results
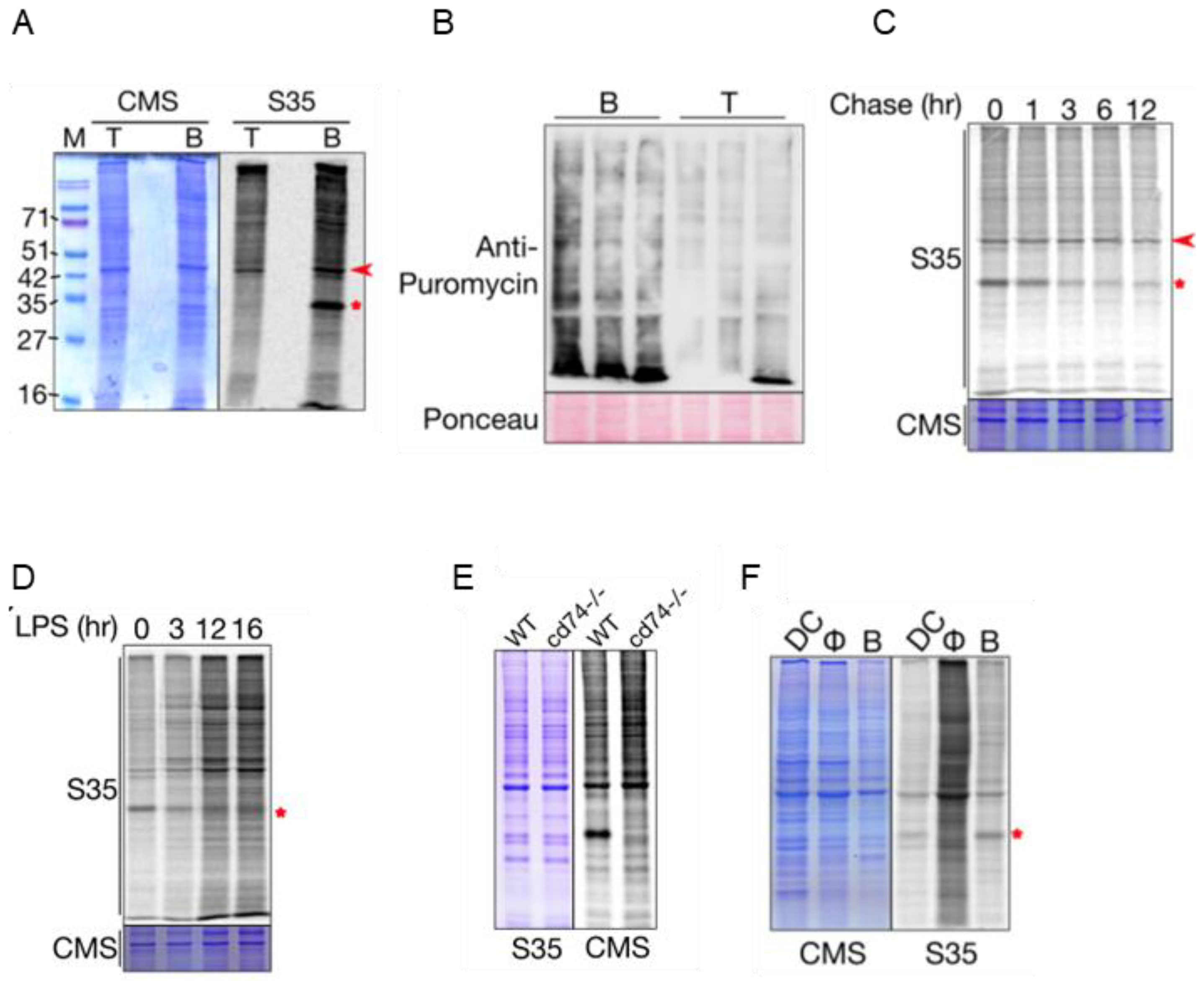
2.1. B Cells Exhibit Increased Protein Synthesis Compared with T Cells In Vivo
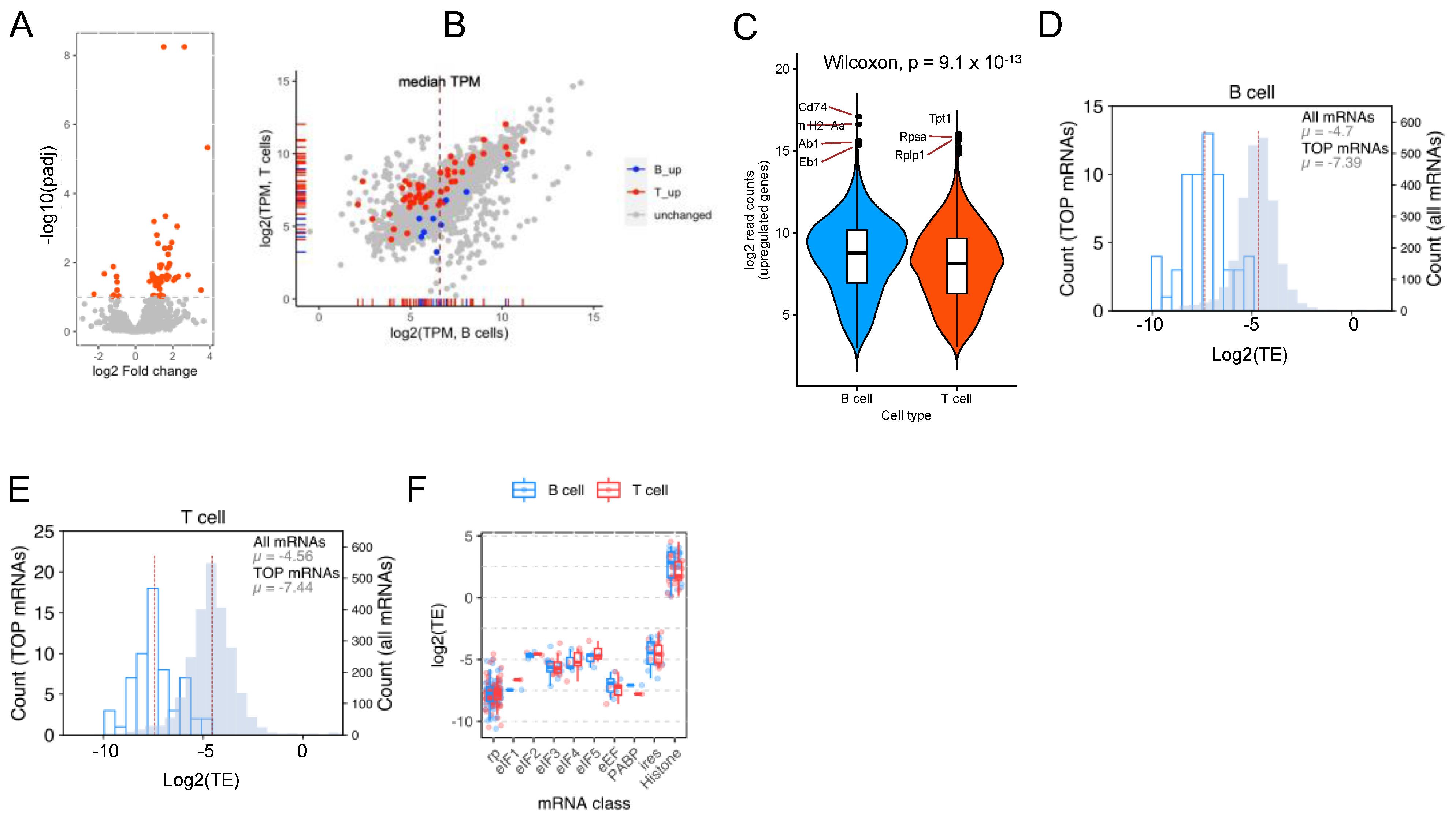
2.2. CD74 Is One of the Highly Translated mRNAs in B Cells
2.3. Genes Associated with Antigen Presentation Are Highly Translated in B Cells
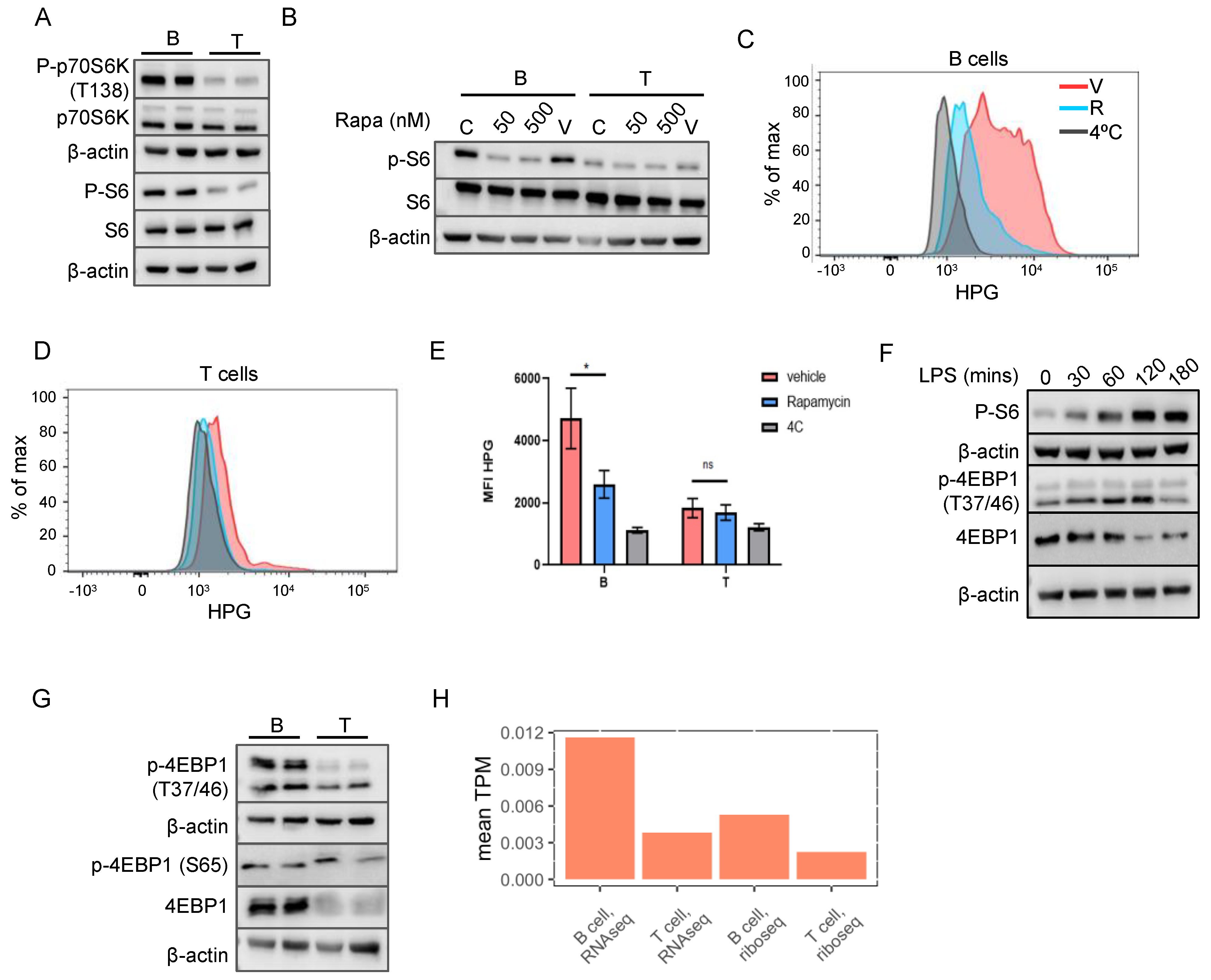
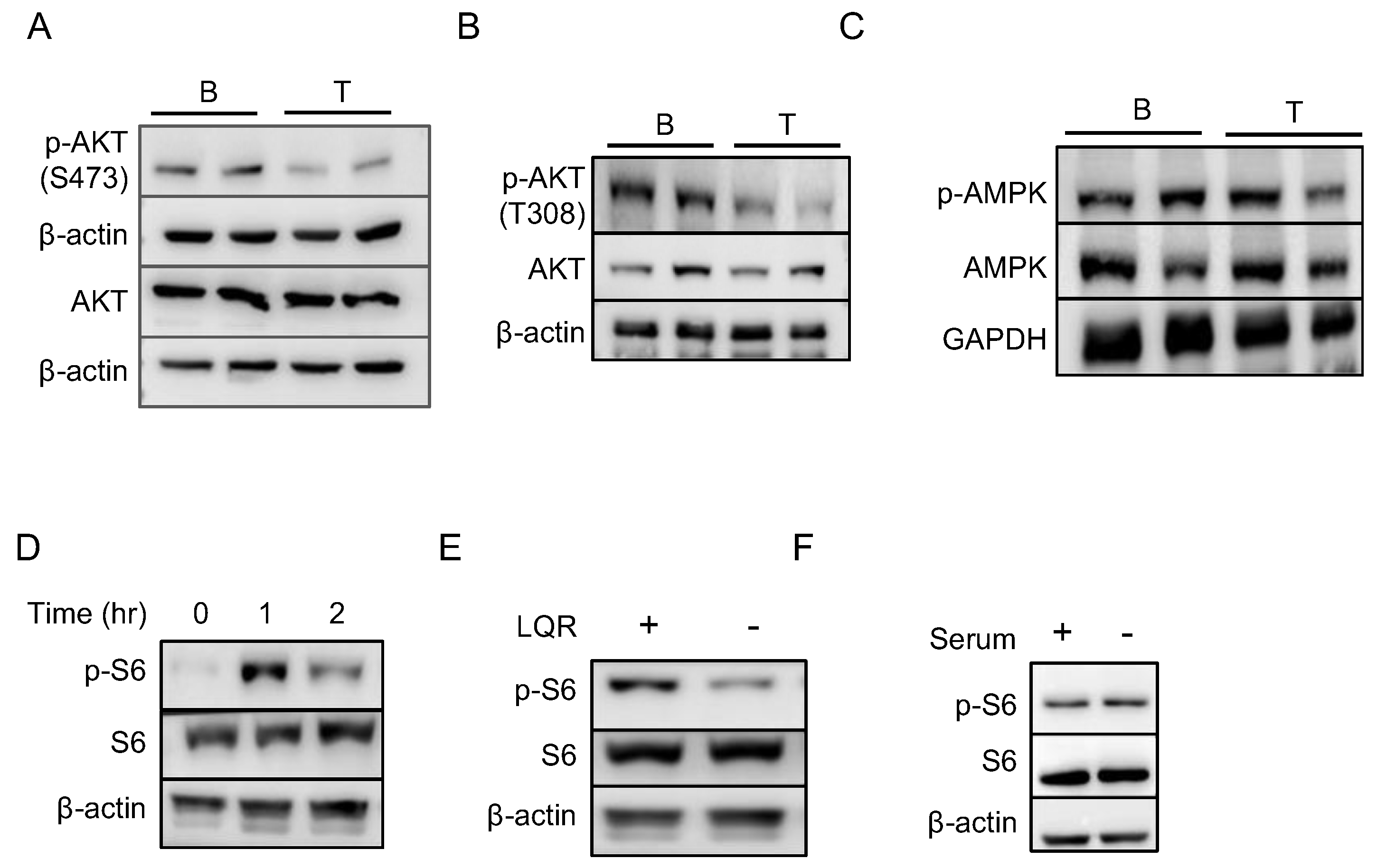
2.4. mTORC1 Pathway Is Responsible for the Increased Translation in B Cells
2.5. Both B and T Cells Exhibit Reduced Translation Efficiency of TOP mRNAs
2.6. Amino Acids Are Essential for Higher mTORC1 Activity in B Cells
3. Discussion
4. Materials and Methods
4.1. Animal Maintenance
4.2. Cell Isolation
4.3. RNAseq and Ribo-Seq
4.4. S35 Labeling of Proteins
4.5. Western Blotting
4.6. Puromycin Incorporation Assay
4.7. HPG Incorporation Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buttgereit, F.; Brand, M.D. A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 1995, 312, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Hershey, J.W.B.; Sonenberg, N.; Mathews, M.B. Principles of translational control: An overview. Cold Spring Harb. Perspect. Biol. 2012, 4, a011528. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. Regulation of mTORC1 and its impact on gene expression at a glance. J. Cell Sci. 2013, 126, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Gentilella, A.; Kozma, S.C.; Thomas, G. A liaison between mTOR signaling, ribosome biogenesis and cancer. Biochim. Biophys. Acta 2015, 1849, 812–820. [Google Scholar] [CrossRef]
- Qin, X.; Jiang, B.; Zhang, Y. 4E-BP1, a multifactor regulated multifunctional protein. Cell Cycle 2016, 15, 781–786. [Google Scholar] [CrossRef]
- Hinnebusch, A.G.; Lorsch, J.R. The mechanism of eukaryotic translation initiation: New insights and challenges. Cold Spring Harb. Perspect. Biol. 2012, 4, 1–25. [Google Scholar] [CrossRef]
- Dowling, R.J.O.; Topisirovic, I.; Alain, T.; Bidinosti, M.; Fonseca, B.D.; Petroulakis, E.; Wang, X.; Larsson, O.; Selvaraj, A.; Liu, Y.; et al. mTORC1-Mediated Cell Proliferation, But Not Cell Growth, Controlled by the 4E-BPs. Science 2010, 328, 1172–1176. [Google Scholar] [CrossRef]
- Ohanna, M.; Sobering, A.K.; Lapointe, T.; Lorenzo, L.; Praud, C.; Petroulakis, E.; Sonenberg, N.; Kelly, P.A.; Sotiropoulos, A.; Pende, M. Atrophy of S6K1−/− skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat. Cell Biol. 2005, 7, 286–294. [Google Scholar] [CrossRef]
- Powell, J.D.; Pollizzi, K.N.; Heikamp, E.B.; Horton, M.R. Regulation of immune responses by mTOR. Annu. Rev. Immunol. 2012, 30, 39–68. [Google Scholar] [CrossRef] [PubMed]
- So, L.; Lee, J.; Palafox, M.; Mallya, S.; Woxland, C.G.; Arguello, M.; Truitt, M.L.; Sonenberg, N.; Ruggero, D.; Fruman, D.A. The 4E-BP-eIF4E axis promotes rapamycin-sensitive growth and proliferation in lymphocytes. Sci. Signal. 2016, 9, ra57. [Google Scholar] [CrossRef] [PubMed]
- Meyuhas, O.; Kahan, T. The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta 2015, 1849, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Thoreen, C.C.; Chantranupong, L.; Keys, H.R.; Wang, T.; Gray, N.S.; Sabatini, D.M. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012, 485, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Jefferies, H.B.; Fumagalli, S.; Dennis, P.B.; Reinhard, C.; Pearson, R.B.; Thomas, G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997, 16, 3693–3704. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Hornstein, E.; Stolovich, M.; Levy, G.; Livingstone, M.; Templeton, D.; Avruch, J.; Meyuhas, O. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol. Cell Biol. 2001, 21, 8671–8683. [Google Scholar] [CrossRef] [PubMed]
- Khalsa, J.K.; Chawla, A.S.; Prabhu, S.B.; Vats, M.; Dhar, A.; Dev, G.; Das, N.; Mukherjee, S.; Tanwar, S.; Banerjee, H.; et al. Functionally significant metabolic differences between B and T lymphocyte lineages. Immunology 2019, 158, 104–120. [Google Scholar] [CrossRef]
- Goodman, C.A.; Hornberger, T.A. Measuring protein synthesis with SUnSET: A valid alternative to traditional techniques? Exerc. Sport Sci. Rev. 2013, 41, 107–115. [Google Scholar] [CrossRef]
- Schröder, B. The multifaceted roles of the invariant chain CD74—More than just a chaperone. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2016, 1863, 1269–1281. [Google Scholar] [CrossRef]
- Gingras, A.C.; Gygi, S.P.; Raught, B.; Polakiewicz, R.D.; Abraham, R.T.; Hoekstra, M.F.; Aebersold, R.; Sonenberg, N. Regulation of 4E-BP1 phosphorylation: A novel two step mechanism. Genes Dev. 1999, 13, 1422–1437. [Google Scholar] [CrossRef]
- Kang, S.A.; Pacold, M.E.; Cervantes, C.L.; Lim, D.; Lou, H.J.; Ottina, K.; Gray, N.S.; Turk, B.E.; Yaffe, M.B.; Sabatini, D.M. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science 2013, 341, 1236566. [Google Scholar] [CrossRef] [PubMed]
- Gingras, A.C.; Raught, B.; Gygi, S.P.; Niedzwiecka, A.; Miron, M.; Burley, S.K.; Polakiewicz, R.D.; Wyslouch-Cieszynska, A.; Aebersold, R.; Sonenberg, N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001, 15, 2852–2864. [Google Scholar] [CrossRef] [PubMed]
- Zoncu, R.; Efeyan, A.; Sabatini, D.M. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2010, 12, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Tato, I.; Bartrons, R.; Ventura, F.; Rosa, J.L. Amino acids activate mammalian target of rapamycin complex 2 (mTORC2) via PI3K/Akt signaling. J. Biol. Chem. 2011, 286, 6128–6142. [Google Scholar] [CrossRef]
- Singer, D.F.; Linderman, J.J. The relationship between antigen concentration, antigen internalization, and antigenic complexes: Modeling insights into antigen processing and presentation. J. Cell Biol. 1990, 111, 55–68. [Google Scholar] [CrossRef]
- Vascotto, F.; Le Roux, D.; Lankar, D.; Faure-André, G.; Vargas, P.; Guermonprez, P.; Lennon-Duménil, A.M. Antigen presentation by B lymphocytes: How receptor signaling directs membrane trafficking. Curr. Opin. Immunol. 2007, 19, 93–98. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]

| Antibody | Make | Catalogue Number |
|---|---|---|
| S6 | CST | 2217S |
| p-S6 (Ser 240/244) | CST | 2215S |
| 4EBP1 | CST | 9452 |
| p-4EBP1 (T37/46) | CST | 2855 |
| p-4EBP1 (S65) | CST | 9451S |
| p70S6K | CST | 9202 |
| p-P70S6K (T389) | CST | 9205 |
| AMPK | CST | 5831T |
| p-AMPK (T172) | CST | 40H9 |
| AKT | CST | 9272 |
| p-AKT (S473) | CST | 9271 |
| p-AKT (T308) | CST | 4056 |
| GAPDH | CST | 2118 |
| Beta actin | CST | 4967S |
| anti-puromycin | Sigma-Aldrich | 32160702 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dev, G.; Chawla, A.S.; Gupta, S.; Bal, V.; George, A.; Rath, S.; Arimbasseri, G.A. Differential Regulation of Two Arms of mTORC1 Pathway Fine-Tunes Global Protein Synthesis in Resting B Lymphocytes. Int. J. Mol. Sci. 2022, 23, 16017. https://doi.org/10.3390/ijms232416017
Dev G, Chawla AS, Gupta S, Bal V, George A, Rath S, Arimbasseri GA. Differential Regulation of Two Arms of mTORC1 Pathway Fine-Tunes Global Protein Synthesis in Resting B Lymphocytes. International Journal of Molecular Sciences. 2022; 23(24):16017. https://doi.org/10.3390/ijms232416017
Chicago/Turabian StyleDev, Gagan, Amanpreet Singh Chawla, Suman Gupta, Vineeta Bal, Anna George, Satyajit Rath, and G. Aneeshkumar Arimbasseri. 2022. "Differential Regulation of Two Arms of mTORC1 Pathway Fine-Tunes Global Protein Synthesis in Resting B Lymphocytes" International Journal of Molecular Sciences 23, no. 24: 16017. https://doi.org/10.3390/ijms232416017
APA StyleDev, G., Chawla, A. S., Gupta, S., Bal, V., George, A., Rath, S., & Arimbasseri, G. A. (2022). Differential Regulation of Two Arms of mTORC1 Pathway Fine-Tunes Global Protein Synthesis in Resting B Lymphocytes. International Journal of Molecular Sciences, 23(24), 16017. https://doi.org/10.3390/ijms232416017






