First Identification of a Large Set of Serine Hydrolases by Activity-Based Protein Profiling in Dibutyl Phthalate-Exposed Zebrafish Larvae
Abstract
1. Introduction
2. Results
2.1. Survival and Developmental Abnormalities in Control and DBP-Exposed Embryos and Larvae
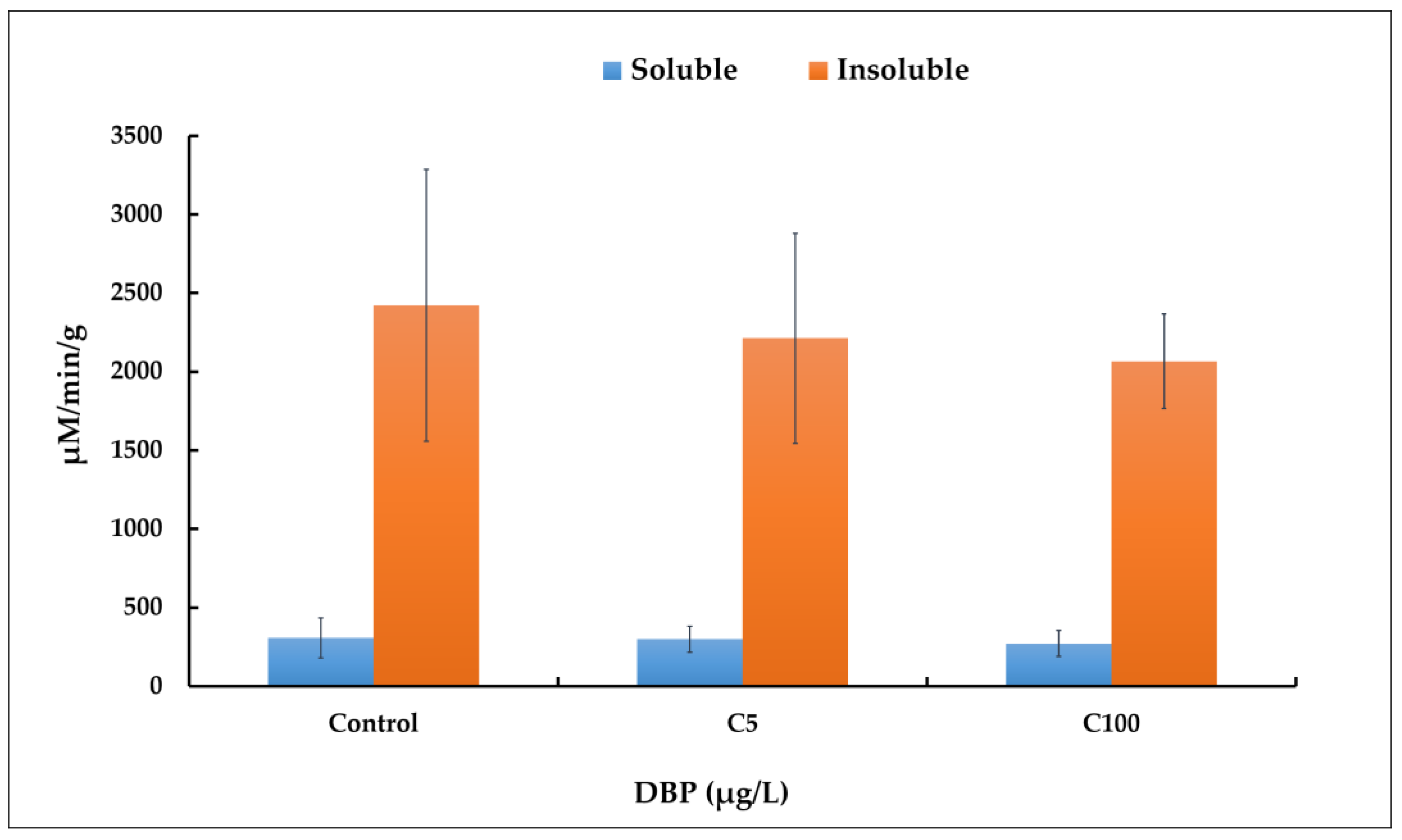
2.2. AChE Activity Assay
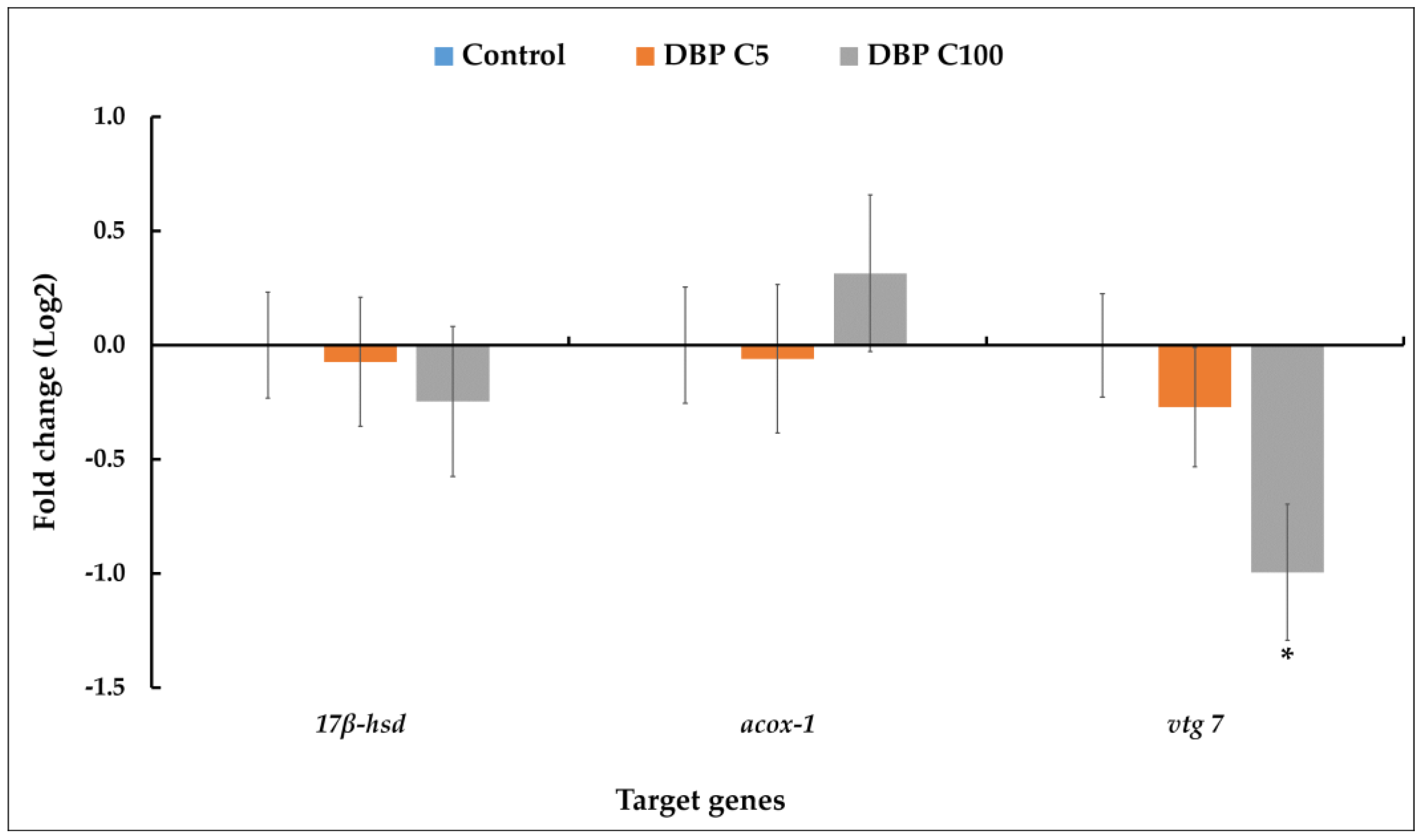
2.3. qRT-PCR Expression of Relevant Genes
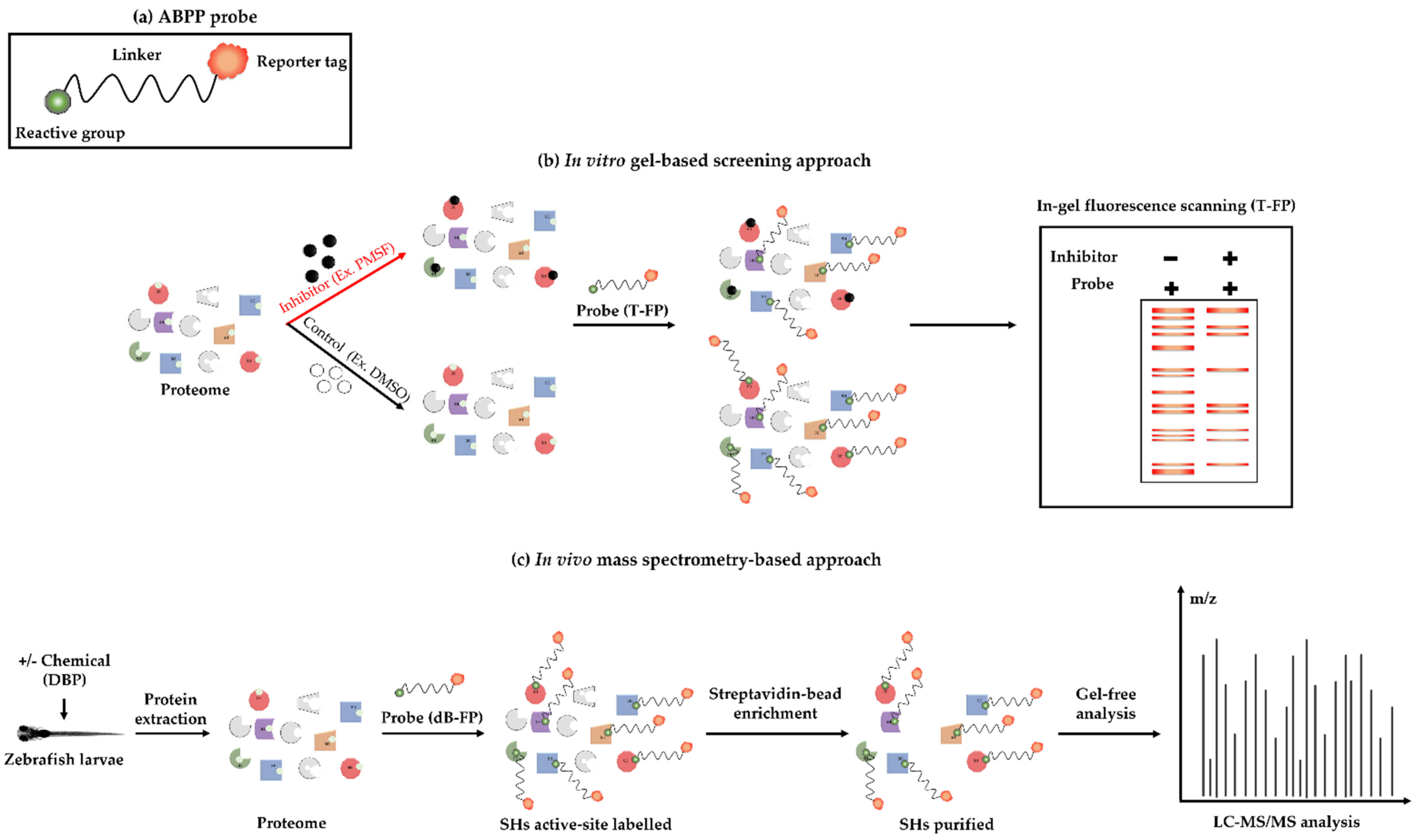
2.4. Validation of the Use of ABPP Probes for Profiling SHs in the Zebrafish Larvae Proteome
2.5. Identification of SHs in the Proteome of Zebrafish Larvae Exposed to DBP
2.6. Functional Annotation of SHs
3. Discussion
4. Materials and Methods
Mass Spectrometry
- -
- (1) Absent in no probe (DMSO) samples, (2) in labelled samples with probe (dB-FP), presence of unique peptides in at least three out of four replicates with at least two of four of the replicates showing LFQ ratios.
- -
- When proteins are present in no-probe samples, (3) a significant difference p < 0.05 (t-test, 2-tailed distribution) * between labelled samples with probe (dB-FP) and no-probe (DMSO) samples, (4) an abundance of at least two times higher in labelled samples with probe than in no-probe (DMSO) samples.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Net, S.; Sempéré, R.; Delmont, A.; Paluselli, A.; Ouddane, B. Occurrence, Fate, Behavior and Ecotoxicological State of Phthalates in Different Environmental Matrices. Environ. Sci. Technol. 2015, 49, 4019–4035. [Google Scholar] [CrossRef] [PubMed]
- Peijnenburg, W.J.G.M. Phthalates. In Encyclopedia of Ecology; Jørgensen, S.E., Fath, B.D., Eds.; Academic Press: Oxford, UK, 2008; pp. 2733–2738. ISBN 978-0-08-045405-4. [Google Scholar]
- Liang, D.-W.; Zhang, T.; Fang, H.H.P.; He, J. Phthalates Biodegradation in the Environment. Appl. Microbiol. Biotechnol. 2008, 80, 183. [Google Scholar] [CrossRef]
- ECHA. Available online: https://echa.europa.eu/ (accessed on 7 February 2022).
- Gazouli, M.; Yao, Z.-X.; Boujrad, N.; Corton, J.C.; Culty, M.; Papadopoulos, V. Effect of Peroxisome Proliferators on Leydig Cell Peripheral-Type Benzodiazepine Receptor Gene Expression, Hormone-Stimulated Cholesterol Transport, and Steroidogenesis: Role of the Peroxisome Proliferator-Activator Receptor α. Endocrinology 2002, 143, 2571–2583. [Google Scholar] [CrossRef] [PubMed]
- Mathieu-Denoncourt, J.; Wallace, S.J.; de Solla, S.R.; Langlois, V.S. Plasticizer Endocrine Disruption: Highlighting Developmental and Reproductive Effects in Mammals and Non-Mammalian Aquatic Species. Gen. Comp. Endocrinol. 2015, 219, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Wójtowicz, A.K.; Szychowski, K.A.; Wnuk, A.; Kajta, M. Dibutyl Phthalate (DBP)-Induced Apoptosis and Neurotoxicity Are Mediated via the Aryl Hydrocarbon Receptor (AhR) but Not by Estrogen Receptor Alpha (ERα), Estrogen Receptor Beta (ERβ), or Peroxisome Proliferator-Activated Receptor Gamma (PPARγ) in Mouse Cortical Neurons. Neurotox. Res. 2017, 31, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Jobling, S.; Reynolds, T.; White, R.; Parker, M.G.; Sumpter, J.P. A Variety of Environmentally Persistent Chemicals, Including Some Phthalate Plasticizers, Are Weakly Estrogenic. Environ. Health Perspect 1995, 103, 582–587. [Google Scholar] [CrossRef]
- Cooke, P.S.; Simon, L.; Denslow, N.D. Endocrine Disruptors. In Haschek and Rousseaux’s Handbook of Toxicologic Pathology; Elsevier: Amsterdam, The Netherlands, 2013; pp. 1123–1154. ISBN 978-0-12-415759-0. [Google Scholar]
- Shen, O.; Wu, W.; Du, G.; Liu, R.; Yu, L.; Sun, H.; Han, X.; Jiang, Y.; Shi, W.; Hu, W.; et al. Thyroid Disruption by Di-n-Butyl Phthalate (DBP) and Mono-n-Butyl Phthalate (MBP) in Xenopus Laevis. PLoS ONE 2011, 6, e19159. [Google Scholar] [CrossRef]
- Lee, E.; Ahn, M.Y.; Kim, H.J.; Kim, I.Y.; Han, S.Y.; Kang, T.S.; Hong, J.H.; Park, K.L.; Lee, B.M.; Kim, H.S. Effect of Di(n-Butyl) Phthalate on Testicular Oxidative Damage and Antioxidant Enzymes in Hyperthyroid Rats. Environ. Toxicol. 2007, 22, 245–255. [Google Scholar] [CrossRef]
- Lin, Q.; Chen, S.; Chao, Y.; Huang, X.; Wang, S.; Qiu, R. Carboxylesterase-Involved Metabolism of Di-n-Butyl Phthalate in Pumpkin (Cucurbita Moschata) Seedlings. Environ. Pollut. 2017, 220, 421–430. [Google Scholar] [CrossRef]
- Mahajan, R.; Verma, S.; Kushwaha, M.; Singh, D.; Akhter, Y.; Chatterjee, S. Biodegradation of Di-n-butyl Phthalate by Psychrotolerant Sphingobium Yanoikuyae Strain P4 and Protein Structural Analysis of Carboxylesterase Involved in the Pathway. Int. J. Biol. Macromol. 2019, 122, 806–816. [Google Scholar] [CrossRef]
- de Lima, D.; Roque, G.M.; de Almeida, E.A. In Vitro and In Vivo Inhibition of Acetylcholinesterase and Carboxylesterase by Metals in Zebrafish (Danio Rerio). Mar. Environ. Res. 2013, 91, 45–51. [Google Scholar] [CrossRef]
- Velki, M.; Meyer-Alert, H.; Seiler, T.-B.; Hollert, H. Enzymatic Activity and Gene Expression Changes in Zebrafish Embryos and Larvae Exposed to Pesticides Diazinon and Diuron. Aquat. Toxicol. 2017, 193, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Acey, R.A.; Bailey, S.; Healy, P.; Jo, C.; Unger, T.F.; Hudson, R.A. A Butyrylcholinesterase in the Early Development of the Brine Shrimp (Artemia Salina) Larvae: A Target for Phthalate Ester Embryotoxicity? Biochem. Biophys. Res. Commun. 2002, 299, 659–662. [Google Scholar] [CrossRef] [PubMed]
- Ghorpade, N.; Mehta, V.; Khare, M.; Sinkar, P.; Krishnan, S.; Rao, C.V. Toxicity Study of Diethyl Phthalate on Freshwater Fish Cirrhina Mrigala. Ecotoxicol. Environ. Saf. 2002, 53, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Jee, J.H.; Koo, J.G.; Keum, Y.H.; Park, K.H.; Choi, S.H.; Kang, J.C. Effects of Dibutyl Phthalate and Di-Ethylhexyl Phthalate on Acetylcholinesterase Activity in Bagrid Catfish, Pseudobagrus Fulvidraco (Richardson). J. Appl. Ichthyol. 2009, 25, 771–775. [Google Scholar] [CrossRef]
- Xu, H.; Shao, X.; Zhang, Z.; Zou, Y.; Chen, Y.; Han, S.; Wang, S.; Wu, X.; Yang, L.; Chen, Z. Effects of Di-n-Butyl Phthalate and Diethyl Phthalate on Acetylcholinesterase Activity and Neurotoxicity Related Gene Expression in Embryonic Zebrafish. Bull Environ. Contam Toxicol 2013, 91, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Ge, S.-S.; Zhao, Y.-C.; Chen, C.; Yang, S. Activity-Based Protein Profiling: An Efficient Approach to Study Serine Hydrolases and Their Inhibitors in Mammals and Microbes. RSC Adv. 2016, 6, 113327–113343. [Google Scholar] [CrossRef]
- Bachovchin, D.A.; Cravatt, B.F. The Pharmacological Landscape and Therapeutic Potential of Serine Hydrolases. Nat. Rev. Drug. Discov. 2012, 11, 52–68. [Google Scholar] [CrossRef]
- Adibekian, A.; Martin, B.R.; Wang, C.; Hsu, K.-L.; Bachovchin, D.A.; Niessen, S.; Hoover, H.; Cravatt, B.F. Click-Generated Triazole Ureas as Ultrapotent in Vivo–Active Serine Hydrolase Inhibitors. Nat. Chem. Biol. 2011, 7, 469–478. [Google Scholar] [CrossRef]
- Long, J.Z.; Cravatt, B.F. The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease. Chem. Rev. 2011, 111, 6022–6063. [Google Scholar] [CrossRef]
- Simon, G.M.; Cravatt, B.F. Activity-Based Proteomics of Enzyme Superfamilies: Serine Hydrolases as a Case Study. J. Biol. Chem. 2010, 285, 11051–11055. [Google Scholar] [CrossRef] [PubMed]
- Moellering, R.E.; Cravatt, B.F. How Chemoproteomics Can Enable Drug Discovery and Development. Chem. Biol. 2012, 19, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Patricelli, M.P.; Cravatt, B.F. Activity-Based Protein Profiling: The Serine Hydrolases. Proc. Natl. Acad. Sci. USA 1999, 96, 14694–14699. [Google Scholar] [CrossRef]
- Cravatt, B.F.; Wright, A.T.; Kozarich, J.W. Activity-Based Protein Profiling: From Enzyme Chemistry to Proteomic Chemistry. Annu. Rev. Biochem. 2008, 77, 383–414. [Google Scholar] [CrossRef] [PubMed]
- Szegletes, T.; Bálint, T.; Szegletes, Z.; Nemcsók, J. In Vivo Effects of Deltamethrin Exposure on Activity and Distribution of Molecular Forms of Carp AChE. Ecotoxicol. Environ. Saf. 1995, 31, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bernard, V.; Girard, E.; Hrabovska, A.; Camp, S.; Taylor, P.; Plaud, B.; Krejci, E. Distinct Localization of Collagen Q and PRiMA Forms of Acetylcholinesterase at the Neuromuscular Junction. Mol. Cell Neurosci. 2011, 46, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Kantae, V.; Krekels, E.H.J.; Ordas, A.; González, O.; van Wijk, R.C.; Harms, A.C.; Racz, P.I.; van der Graaf, P.H.; Spaink, H.P.; Hankemeier, T. Pharmacokinetic Modeling of Paracetamol Uptake and Clearance in Zebrafish Larvae: Expanding the Allometric Scale in Vertebrates with Five Orders of Magnitude. Zebrafish 2016, 13, 504–510. [Google Scholar] [CrossRef]
- Vliegenthart, A.D.B.; Tucker, C.S.; Del Pozo, J.; Dear, J.W. Zebrafish as Model Organisms for Studying Drug-Induced Liver Injury. Br. J. Clin. Pharmacol. 2014, 78, 1217–1227. [Google Scholar] [CrossRef]
- Santoro, M.M. Zebrafish as a Model to Explore Cell Metabolism. Trends Endocrinol. Metab. 2014, 25, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Patton, E.E.; Zon, L.I.; Langenau, D.M. Zebrafish Disease Models in Drug Discovery: From Preclinical Modelling to Clinical Trials. Nat. Rev. Drug. Discov. 2021, 20, 611–628. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Dong, X.; Zhang, Z.; Yang, M.; Wu, X.; Liu, H.; Lao, Q.; Li, C. Assessment of Immunotoxicity of Dibutyl Phthalate Using Live Zebrafish Embryos. Fish Shellfish Immunol. 2015, 45, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Zarragoitia, M.; Trant, J.M.; Cajaraville, M.P. Effects of Dibutylphthalate and Ethynylestradiol on Liver Peroxisomes, Reproduction, and Development of Zebrafish (Danio Rerio). Environ. Toxicol. Chem. 2006, 25, 2394. [Google Scholar] [CrossRef] [PubMed]
- Dang, Z. Interpretation of Fish Biomarker Data for Identification, Classification, Risk Assessment and Testing of Endocrine Disrupting Chemicals. Environ. Int. 2016, 92–93, 422–441. [Google Scholar] [CrossRef]
- Ortiz-Zarragoitia, M.; Cajaraville, M.P. Effects of Selected Xenoestrogens on Liver Peroxisomes, Vitellogenin Levels and Spermatogenic Cell Proliferation in Male Zebrafish. Comp. Biochem. Physiol. Part C: Toxicol. Pharmacol. 2005, 141, 133–144. [Google Scholar] [CrossRef]
- Zhou, J.; Cai, Z.-H.; Xing, K.-Z. Potential Mechanisms of Phthalate Ester Embryotoxicity in the Abalone Haliotis Diversicolor Supertexta. Environ. Pollut. 2011, 159, 1114–1122. [Google Scholar] [CrossRef]
- Xie, H.Q.; Ma, Y.; Fu, H.; Xu, T.; Luo, Y.; Liu, Y.; Chen, Y.; Xu, L.; Xia, Y.; Zhao, B. New Perspective on the Regulation of Acetylcholinesterase via the Aryl Hydrocarbon Receptor. J. Neurochem. 2021, 158, 1254–1262. [Google Scholar] [CrossRef]
- Uren-Webster, T.M.; Lewis, C.; Filby, A.L.; Paull, G.C.; Santos, E.M. Mechanisms of Toxicity of Di(2-Ethylhexyl) Phthalate on the Reproductive Health of Male Zebrafish. Aquat. Toxicol. 2010, 99, 360–369. [Google Scholar] [CrossRef]
- Lapinskas, P.J.; Brown, S.; Leesnitzer, L.M.; Blanchard, S.; Swanson, C.; Cattley, R.C.; Corton, J.C. Role of PPARα in Mediating the Effects of Phthalates and Metabolites in the Liver. Toxicology 2005, 207, 149–163. [Google Scholar] [CrossRef]
- Mindnich, R.; Möller, G.; Adamski, J. The Role of 17 Beta-Hydroxysteroid Dehydrogenases. Mol. Cell. Endocrinol. 2004, 218, 7–20. [Google Scholar] [CrossRef]
- Khan, M.N.; Renaud, R.L.; Leatherland, J.F. Steroid Metabolism by Embryonic Tissues of Arctic Charr,Salvelinus Alpinus. Gen. Comp. Endocrinol. 1997, 105, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Dang, Z. Fish Biomarkers for Regulatory Identification of Endocrine Disrupting Chemicals. Environ. Pollut. 2014, 185, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Qiu, X.; Meng, S.; Xu, H.; Wu, X.; Yang, M. Proteomic Profile and Toxicity Pathway Analysis in Zebrafish Embryos Exposed to Bisphenol A and Di-n-Butyl Phthalate at Environmentally Relevant Levels. Chemosphere 2018, 193, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, J.; Liu, W.; Sheng, G.D.; Tu, Y.; Ma, Y. Separation and Aquatic Toxicity of Enantiomers of the Pyrethroid Insecticide Lambda-Cyhalothrin. Environ. Toxicol. Chem. 2008, 27, 174–181. [Google Scholar] [CrossRef]
- Embry, M.R.; Belanger, S.E.; Braunbeck, T.A.; Galay-Burgos, M.; Halder, M.; Hinton, D.E.; Léonard, M.A.; Lillicrap, A.; Norberg-King, T.; Whale, G. The Fish Embryo Toxicity Test as an Animal Alternative Method in Hazard and Risk Assessment and Scientific Research. Aquat. Toxicol. 2010, 97, 79–87. [Google Scholar] [CrossRef]
- Simmons, D.B.D.; Benskin, J.P.; Cosgrove, J.R.; Duncker, B.P.; Ekman, D.R.; Martyniuk, C.J.; Sherry, J.P. Omics for Aquatic Ecotoxicology: Control of Extraneous Variability to Enhance the Analysis of Environmental Effects: Variability of Omics Data in Aquatic Ecotoxicology. Environ. Toxicol. Chem. 2015, 34, 1693–1704. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The Zebrafish Reference Genome Sequence and Its Relationship to the Human Genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Zheng, W.; Lin, Q.; Issah, M.A.; Liao, Z.; Shen, J. Identification of PLA2G7 as a Novel Biomarker of Diffuse Large B Cell Lymphoma. BMC Cancer 2021, 21, 927. [Google Scholar] [CrossRef]
- Lewis, L.D. An In Vivo Proof of Principle Trial to Determine Whether the Nutritional Supplement Conjugated Linoleic Acid (CLA, ClarinolTM) Can Modulate the Lipogenic Pathway in Breast Cancer Tissue; ClinicalTrials.gov: Bethesda, MD, USA, 2019.
- Columbia University. MRCP With Secretin Stimulation for the Evaluation of Pancreatic Endocrine and Exocrine Function Following Surgical Resection for Pancreatic Adenocarcinoma; ClinicalTrials.gov: Bethesda, MD, USA, 2015.
- GlaxoSmithKline. A Two Part, Multicenter Phase IIa, Placebo Controlled Study, to Examine the Safety, Tolerability, and Effects of GSK256073 on Lipids in Subjects With Dyslipidemia; ClinicalTrials.gov: Bethesda, MD, USA, 2019.
- Albers, G.W. TIA Triage Trial and Evaluation of Vascular-Specific Inflammatory BiomarkerLp-PLA2 as a Stratification Tool for TIA Triage and Stroke Risk; ClinicalTrials.gov: Bethesda, MD, USA, 2019.
- Tierbach, A.; Groh, K.J.; Schönenberger, R.; Schirmer, K.; Suter, M.J.-F. Glutathione S-Transferase Protein Expression in Different Life Stages of Zebrafish (Danio Rerio). Toxicol. Sci. 2018, 162, 702–712. [Google Scholar] [CrossRef]
- Cravedi, J.-P.; Perdu-durand, E. The Phthalate Diesters DEHP and DBP Do Not Induce Lauric Acid Hydroxylase Activity in Rainbow Trout. Mar. Environ. Res. 2002, 54, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Otte, J.C.; Schultz, B.; Fruth, D.; Fabian, E.; van Ravenzwaay, B.; Hidding, B.; Salinas, E.R. Intrinsic Xenobiotic Metabolizing Enzyme Activities in Early Life Stages of Zebrafish (Danio Rerio). Toxicol Sci 2017, 159, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Pang, S.; Sun, X.; Gao, J.; Chen, J.; Chen, X.; Li, X.; Wang, C. Evaluation of Acute and Developmental Effects of Difenoconazole via Multiple Stage Zebrafish Assays. Environ. Pollut. 2013, 175, 147–157. [Google Scholar] [CrossRef]
- Ellman, G.L.; Courtney, K.D.; Andres, V.; Featherstone, R.M. A New and Rapid Colorimetric Determination of Acetylcholinesterase Activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Maradonna, F.; Evangelisti, M.; Gioacchini, G.; Migliarini, B.; Olivotto, I.; Carnevali, O. Assay of Vtg, ERs and PPARs as Endpoint for the Rapid in Vitro Screening of the Harmful Effect of Di-(2-Ethylhexyl)-Phthalate (DEHP) and Phthalic Acid (PA) in Zebrafish Primary Hepatocyte Cultures. Toxicol. Vitr. 2013, 27, 84–91. [Google Scholar] [CrossRef]
- Zhu, Y.; Hua, R.; Zhou, Y.; Li, H.; Quan, S.; Yu, Y. Chronic Exposure to Mono-(2-Ethylhexyl)-Phthalate Causes Endocrine Disruption and Reproductive Dysfunction in Zebrafish. Environ. Toxicol. Chem. 2016, 35, 2117–2124. [Google Scholar] [CrossRef]
- Lang, X.; Wang, L.; Zhang, Z. Stability Evaluation of Reference Genes for Real-Time PCR in Zebrafish (Danio Rerio) Exposed to Cadmium Chloride and Subsequently Infected by Bacteria Aeromonas Hydrophila. Aquat. Toxicol. 2016, 170, 240–250. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Carlson, A.; Sinitcyn, P.; Mann, M.; Cox, J. Visualization of LC-MS/MS Proteomics Data in MaxQuant. Proteomics 2015, 15, 1453–1456. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus Computational Platform for Comprehensive Analysis of (Prote)Omics Data. Nat Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]

| Gene | Protein Name | Uniprot Accession | Mass (kDa) | S/I | SP/mSH | Unique Peptides | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 100 µg/L DBP | 5 µg/L DBP | Control (DMSO) | |||||||||
| No probe | dB-FP | No probe | dB-FP | No probe | dB-FP | ||||||
| aadac | Arylacetamide deacetylase | E7F2W1_DANRE | 46.923 | S | mSH | 0, 0, 0, 0 | 9, 6, 7, 12 | 0, 0, 0, 0 | 8, 7, 8, 13 | 0, 0, 0, 0 | 9, 7, 9, 13 |
| abhd10a | Abhydrolase domain-containing 10, depalmitoylase a | A0A0R4ILM1_DANRE | 31.639 | S | mSH | 0, 0, 0, 0 | 8, 3, 6, 9 | 0, 0, 0, 0 | 6, 8, 8, 11 | 0, 0, 0, 0 | 6, 3, 6, 8 |
| abhd10b | Transgelin 3b | E7F9Y7_DANRE | 31.94 | S | mSH | 0, 0, 0, 0 | 11, 8, 9, 11 | 0, 0, 0, 0 | 12, 11, 12, 12 | 0, 0, 0, 0 | 12, 8, 11, 11 |
| abhd17ab | Abhydrolase domain-containing 17A, depalmitoylase b | A0A2R8PY04_DANRE | 32.846 | S | SP | 0, 0, 0, 0 | 4, 1, 3, 5 | 0, 0, 0, 0 | 6, 5, 4, 5 | 0, 0, 0, 0 | 6, 1, 5, 5 |
| abhd17b | Abhydrolase domain-containing 17B, depalmitoylase (Fragment) | A3KPQ6_DANRE | 32.28 | S | SP | 0, 0, 0, 0 | 4, 4, 5, 6 | 0, 0, 0, 0 | 5, 5, 5, 8 | 0, 0, 0, 0 | 5, 4, 4, 7 |
| abhd6a | Abhydrolase domain-containing 6, acylglycerol lipase a | E7F881_DANRE | 38.103 | S | mSH | 0, 0, 0, 0 | 5, 3, 5, 9 | 0, 0, 0, 0 | 5, 9, 4, 9 | 0, 0, 0, 0 | 7, 3, 4, 7 |
| acot14 | Acyl-CoA thioesterase 14 | Q5RHG4_DANRE | 49.161 | S | mSH | 0, 0, 0, 0 | 4, 1, 3, 4 | 0, 0, 0, 0 | 4, 4, 4, 6 | 0, 0, 0, 0 | 3, 2, 3, 5 |
| acot18 | Acyl-CoA thioesterase 18 (Fragment) | Q5RH36_DANRE | 48.765 | S | mSH | 0, 0, 0, 0 | 5, 5, 5, 8 | 0, 0, 0, 0 | 4, 7, 6, 11 | 0, 0, 0, 0 | 6, 6, 4, 9 |
| acot20 | Acyl-CoA thioesterase 20 | Q5SPG8_DANRE | 48.601 | S | mSH | 0, 0, 0, 0 | 10, 4, 10, 15 | 0, 0, 0, 0 | 9, 9, 13, 16 | 0, 0, 0, 0 | 8, 3, 8, 12 |
| afmid | Kynurenine formamidase (Fragment) | A0A0R4INQ3_DANRE | 30.694 | S | mSH | 0, 0, 0, 0 | 6, 2, 7, 5 | 0, 0, 0, 0 | 4, 6, 9, 6 | 0, 0, 0, 0 | 5, 5, 7, 6 |
| apeh | Acyl-peptide hydrolase | F1QTY6_DANRE | 83.262 | S | SP | 0, 0, 0, 0 | 6, 3, 3, 6 | 0, 0, 0, 0 | 5, 6, 9, 9 | 0, 0, 0, 0 | 5, 3, 10, 7 |
| ccdc57 | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase | E7F5V3_DANRE | 274.19 | S | mSH | 0, 0, 0, 0 | 23, 11, 21, 40 | 0, 0, 0, 0 | 21, 21, 26, 26 | 0, 0, 0, 0 | 25, 9, 28, 28 |
| I | mSH | 0, 0, 0, 0 | 7, 7, 5, 4 | 0, 0, 0, 0 | 6, 7, 10, 3 | 0, 0, 0, 0 | 7, 5, 6, 3 | ||||
| cel.1 | Carboxylic ester hydrolase | F1R1T7_DANRE | 60.783 | S | mSH | 0, 0, 0, 0 | 7, 5, 5, 6 | 0, 0, 0, 0 | 3, 5, 6, 7 | 0, 0, 0, 0 | 3, 5, 4, 7 |
| cel.2 | Carboxylic ester hydrolase | F6P131_DANRE | 60.555 | S | mSH | 0, 0, 0, 0 | 5, 5, 4, 8 | 0, 0, 0, 0 | 4, 5, 6, 10 | 0, 0, 0, 0 | 2, 4, 6, 8 |
| cela1.6 | Chymotrypsin-like elastase family member 1, tandem duplicate 6 | A0A2R8RMN0_DANRE | 29.475 | S | SP | 0, 4, 1, 3 | 5, 7, 3, 3 | 0, 1, 1, 3 | 2, 7, 3, 5 | 0, 4, 1, 3 | 2, 7, 1, 5 |
| ces2a | Carboxylic ester hydrolase | Q1LYL6_DANRE | 60.611 | S | mSH | 0, 0, 0, 0 | 7, 8, 8, 7 | 0, 0, 0, 0 | 8, 10, 9, 7 | 0, 0, 0, 0 | 8, 7, 8, 7 |
| ces2a | Carboxylic ester hydrolase (Fragment) | Q6GMJ1_DANRE | 60.634 | S | mSH | 0, 0, 0, 0 | 4, 5, 7, 5 | 0, 0, 0, 0 | 6, 5, 8, 4 | 0, 0, 0, 0 | 7, 4, 5, 5 |
| ces2b | Carboxylic ester hydrolase | A0A0R4IMR1_DANRE | 29.286 | S | mSH | 0, 0, 0, 0 | 4, 5, 5, 3 | 0, 0, 0, 0 | 4, 4, 4, 3 | 0, 0, 0, 0 | 4, 4, 4, 3 |
| ces2b | Carboxylesterase 2b (Fragment) | A0A0R4IU19_DANRE | 104.95 | S | mSH | 0, 0, 0, 0 | 21, 23, 17, 16 | 0, 0, 0, 0 | 20, 25, 20, 19 | 0, 0, 0, 0 | 19, 18, 20, 20 |
| 104.95 | I | mSH | 0, 0, 0, 0 | 4, 7, 3, 4 | 0, 0, 0, 0 | 1, 3, 7, 3 | 0, 0, 0, 0 | 5, 5, 3, 2 | |||
| ces3 | Carboxylic ester hydrolase | A0A2R8QSI8_DANRE | 59.431 | S | mSH | 0, 0, 0, 0 | 2, 2, 2, 2 | 0, 0, 0, 0 | 2, 2, 2, 2 | 0, 0, 0, 0 | 1, 2, 2, 2 |
| ces3 | Carboxylic ester hydrolase | Q1LUZ9_DANRE | 60.297 | S | mSH | 0, 0, 0, 0 | 4, 3, 4, 4 | 0, 0, 0, 0 | 4, 3, 4, 4 | 0, 0, 0, 0 | 3, 2, 4, 4 |
| cpvl | Carboxypeptidase vitellogenic-like | Q7ZU43_DANRE | 54.514 | S | SP | 0, 0, 0, 0 | 13, 11, 12, 15 | 0, 0, 0, 0 | 15, 13, 15, 13 | 0, 0, 0, 0 | 13, 10, 13, 12 |
| ctrb1 | Chymotrypsinogen B1 | F1QFX9_DANRE | 28.245 | S | SP | 0, 0, 0, 0 | 1, 5, 1, 1 | 0, 0, 0, 0 | 0, 4, 2, 1 | 0, 0, 0, 0 | 0, 5, 0, 2 |
| Ctsa | Carboxypeptidase | A0A0R4ILY1_DANRE | 53.007 | S | SP | 0, 0, 1, 1 | 8, 5, 7, 6 | 0, 0, 0, 1 | 6, 7, 8, 6 | 0, 0, 0, 0 | 8, 5, 6, 5 |
| I | SP | 0, 0, 0, 0 | 4, 4, 3, 3 | 0, 0, 0, 0 | 3, 1, 2, 3 | 0, 0, 0, 1 | 3, 1, 3, 1 | ||||
| dpp4 | Dipeptidyl peptidase 4 | B5DDZ4_DANRE | 84.667 | S | mSH | 0, 0, 0, 0 | 14, 12, 11, 6 | 0, 0, 0, 0 | 12, 13, 12, 10 | 0, 0, 0, 0 | 12, 10, 12, 12 |
| dpp9 | Dipeptidyl-peptidase 9 | A0A2R8QM73_DANRE | 98.325 | S | SP | 0, 0, 0, 0 | 18, 8, 13, 7 | 0, 0, 0, 0 | 17, 13, 18, 11 | 0, 0, 0, 1 | 16, 7, 17, 13 |
| I | SP | 0, 0, 0, 0 | 5, 6, 6, 4 | 0, 0, 0, 0 | 6, 4, 9, 6 | 0, 0, 0, 0 | 7, 6, 4, 5 | ||||
| ela2 | Elastase 2 | A0A0R4IXD6_DANRE | 28.887 | S | SP | 0, 2, 0, 0 | 3, 5, 1, 2 | 0, 1, 0, 0 | 0, 5, 2, 2 | 0, 0, 0, 1 | 0, 4, 1, 3 |
| esd | S-formylglutathione hydrolase | Q567K2_DANRE | 31.171 | S | SP | 0, 0, 0, 0 | 6, 4, 5, 6 | 0, 0, 0, 0 | 5, 6, 8, 6 | 0, 0, 0, 1 | 3, 3, 3, 7 |
| faah | Fatty acid amide hydrolase | F1RCW3_DANRE | 65.153 | S | SP | 0, 0, 0, 0 | 1, 1, 3, 11 | 0, 0, 0, 0 | 2, 6, 4, 6 | 0, 0, 0, 0 | 2, 0, 6, 7 |
| faah2b | Fatty-acid amide hydrolase 2-B | F1QM44_DANRE | 57.151 | S | SP | 0, 0, 0, 0 | 12, 8, 8, 15 | 0, 0, 0, 0 | 12, 11, 13, 13 | 0, 0, 0, 0 | 14, 7, 13, 17 |
| fap | Dipeptidyl peptidase 4 | B0R1C4_DANRE | 85.806 | S | SP | 0, 0, 0, 0 | 25, 28, 27, 23 | 0, 0, 0, 0 | 28, 34, 30, 28 | 0, 0, 0, 0 | 29, 25, 26, 29 |
| I | SP | 0, 0, 0, 0 | 9, 14, 15, 16 | 0, 0, 0, 0 | 12, 12, 14, 16 | 0, 0, 0, 0 | 10, 16, 15, 7 | ||||
| Lcat | Lecithin-cholesterol acyltransferase | A0A0R4IDL2_DANRE | 49.139 | S | mSH | 0, 0, 0, 0 | 8, 6, 10, 10 | 0, 0, 0, 0 | 7, 10, 10, 10 | 0, 0, 0, 0 | 8, 5, 9, 9 |
| lypla1 | Lysophospholipase 1 | Q568J5_DANRE | 21.218 | S | mSH | 0, 0, 0, 0 | 2, 1, 1, 4 | 0, 0, 0, 0 | 1, 3, 1, 4 | 0, 0, 0, 0 | 1, 2, 1, 4 |
| lypla2 | Lysophospholipase 2 | Q6PBW8_DANRE | 25.067 | S | mSH | 0, 0, 0, 0 | 8, 8, 11, 10 | 0, 0, 0, 0 | 11, 11, 10, 10 | 0, 0, 0, 0 | 10, 10, 10, 10 |
| I | mSH | 0, 0, 0, 0 | 2, 2, 1, 1 | 0, 0, 0, 0 | 0, 1, 1, 0 | 0, 0, 0, 0 | 2, 0, 2, 2 | ||||
| mgll | Monoglyceride lipase | Q7ZWC2_DANRE | 33.66 | S | mSH | 0, 0, 0, 0 | 7, 4, 7, 10 | 0, 0, 0, 0 | 8, 9, 9, 11 | 0, 0, 0, 0 | 6, 7, 6, 10 |
| N/A | 2-arachidonoylglycerol hydrolase ABHD12 | A0A2R8QQD0_DANRE | 40.472 | S | SP | 0, 0, 0, 0 | 4, 4, 4, 4 | 0, 0, 0, 0 | 4, 5, 5, 4 | 0, 0, 0, 0 | 5, 4, 4, 5 |
| nceh1b.1 | Neutral cholesterol ester hydrolase 1b, tandem duplicate 1 | F1Q8P9_DANRE | 45.39 | S | mSH | 0, 0, 0, 0 | 7, 2, 5, 7 | 0, 0, 0, 0 | 5, 7, 8, 7 | 0, 0, 0, 0 | 3, 3, 4, 7 |
| pla2g15 | Phospholipase A2, group XV (Fragment) | A0A0R4IE29_DANRE | 53.521 | S | mSH | 0, 0, 0, 0 | 1, 3, 5, 9 | 0, 0, 0, 0 | 2, 7, 7, 9 | 0, 0, 0, 0 | 2, 5, 4, 8 |
| pla2g7 | Platelet-activating factor acetylhydrolase | Q5RHM0_DANRE | 50.429 | S | mSH | 0, 0, 0, 0 | 1, 2, 4, 8 | 0, 0, 0, 0 | 2, 4, 4, 10 | 0, 0, 0, 0 | 2, 2, 3, 7 |
| ppme1 | Protein phosphatase methylesterase 1 | Q7ZV37_DANRE | 41.64 | S | mSH | 0, 0, 0, 0 | 4, 0, 4, 6 | 0, 0, 0, 0 | 3, 3, 5, 9 | 0, 0, 0, 0 | 4, 1, 5, 8 |
| ppt2b | Novel protein similar to verebrate palmitoyl-protein thioesterase 2 (PPT2) | A3KPZ6_DANRE | 32.366 | S | mSH | 0, 0, 0, 0 | 1, 2, 1, 2 | 0, 0, 0, 0 | 2, 2, 2, 3 | 0, 0, 0, 0 | 2, 2, 1, 3 |
| prcp | Prolylcarboxypeptidase (Angiotensinase c) | Q6DG46_DANRE | 55.132 | S | SP | 0, 0, 0, 0 | 3, 0, 0, 5 | 0, 0, 0, 0 | 0, 2, 3, 4 | 0, 0, 0, 0 | 0, 0, 0, 5 |
| prep | Prolyl endopeptidase | Q503E2_DANRE | 80.336 | S | SP | 0, 0, 0, 1 | 22, 14, 20, 22 | 0,0,0,1 | 20, 20, 25, 22 | 0, 1, 0, 0 | 19, 11, 22, 25 |
| rbbp9 | Retinoblastoma-binding protein 9 | Q1MT41_DANRE | 20.887 | S | mSH | 0, 0, 0, 0 | 6, 3, 4, 4 | 0, 0, 0, 0 | 6, 6, 6, 4 | 0, 0, 0, 0 | 4, 2, 4, 5 |
| si:ch211-117n7.6 | Si:ch211-117n7.6 | A8E7H0_DANRE | 38.7 | S | mSH | 0, 0, 0, 0 | 5, 2, 6, 8 | 0, 0, 0, 0 | 7, 9, 9, 8 | 0, 0, 0, 0 | 7, 3, 5, 8 |
| si:ch211-117n7.7 | Si:ch211-117n7.7 | A8E7G9_DANRE | 39.501 | S | mSH | 0, 0, 0, 0 | 7, 5, 3, 9 | 0, 0, 0, 0 | 7, 6, 8, 9 | 0, 0, 0, 0 | 5, 5, 5, 8 |
| si:ch211-122f10.4 | Carboxypeptidase | F1QYP6_DANRE | 51.525 | S | mSH | 0, 0, 0, 0 | 7, 7, 4, 5 | 0, 0, 0, 0 | 8, 4, 5, 6 | 0, 0, 0, 0 | 6, 7, 6, 5 |
| si:ch211-71n6.4 | Carboxylic ester hydrolase | A0A0R4IYT8_DANRE | 67.565 | S | mSH | 0, 0, 0, 0 | 11, 7, 6, 14 | 0, 0, 0, 0 | 9, 11, 12, 15 | 0, 0, 0, 0 | 13, 3, 8, 14 |
| si:ch73-89b15.3 | Carboxylic ester hydrolase | A0A0R4IPW5_DANRE | 63.523 | S | mSH | 0, 0, 0, 0 | 3, 1, 3, 10 | 0, 0, 0, 0 | 5, 6, 6, 10 | 0, 0, 0, 0 | 5, 1, 4, 11 |
| si:dkey-21e2.16 | Si:dkey-21e2.16 (Fragment) | Q1LUQ6_DANRE | 25.62 | S | SP | 0, 0, 0, 0 | 2, 2, 0, 3 | 0, 0, 0, 0 | 0, 4, 2, 4 | 0, 0, 0, 0 | 0, 2, 0, 3 |
| siae | Sialic acid acetylesterase | Q1LUX8_DANRE | 56.624 | S | SP | 0, 0, 0, 0 | 3, 2, 2, 6 | 0, 0, 0, 0 | 2, 3, 3, 2 | 0, 0, 0, 0 | 0, 1, 2, 4 |
| tpp2 | Tripeptidyl-peptidase 2 | R4GDQ0_DANRE | 139.25 | I | SP | 0, 0, 0, 0 | 8, 8, 7, 7 | 0, 0, 0, 0 | 11, 13, 17, 10 | 0, 0, 0, 0 | 20, 6, 8, 9 |
| zgc:154142 | Zgc:154142 | A5PKM4_DANRE | 118.63 | S | SP | 0, 0, 0, 0 | 1, 1, 1, 2 | 0, 0, 0, 0 | 0, 7, 2, 4 | 0, 0, 0, 0 | 0, 3, 1, 3 |
| Other proteins | |||||||||||
| clic3 | Chloride intracellular channel 3 | E7F4S2_DANRE | 151.36 | S | - | 0, 0, 0, 0 | 1, 0, 0, 6 | 0, 0, 0, 0 | 1, 2, 3, 6 | 0, 0, 0, 0 | 0, 0, 2, 8 |
| a2ml | Alpha-2-macroglobulin-like | A0A0R4IDD1_DANRE | 159.76 | S | - | 0, 0, 0, 0 | 11, 12, 9, 10 | 0, 0, 0, 0 | 5, 14, 11, 11 | 0, 0, 0, 0 | 5, 10, 4, 12 |
| colq | Collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase | F1Q7Y1_DANRE | 47.825 | I | - | 0, 0, 3, 0 | 3, 5, 3, 3 | 0, 0, 0, 0 | 3, 4, 4, 3 | 1, 0, 1, 0 | 4, 3, 2, 3 |
| Genes and [Reference] | Primers (5′–3′) | Amplicon (pb) |
|---|---|---|
| Elongation factor 1 alpha (ef1-α) [64] | Foward: TACAAATGCGGTGGAATCGAC Reverse: GTCAGCCTGAGAAGTACCAGT | 248 |
| Ribosomal protein L 13a (rpl13a) [64] | Foward: TCTGGAGGACTGTAAGAGGTATGC Reverse: AGACGCACAATCTTGAGAGCAG | 149 |
| Vitellogenin (vtg7) [62] | Foward: GCCAAAAAGCTGGGTAAACA Reverse: AGTTCCGTCTGGATTGATGG | 210 |
| Acyl-coenzyme-A-oxidase 1 (acox-1) | Foward: TCCATGAGTCCCACAACAAG Reverse: CCTTTCTTCCCCTTTCTTGC | 75 |
| 17β-hydroxysteroid dehydrogenase (17b-HSD 12a) | Foward: ACCAGACCAACGGCTATTTC Reverse: ATCGCAGTTTTCTCCTCCTG | 141 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yedji, R.S.; Sohm, B.; Salnot, V.; Guillonneau, F.; Cossu-Leguille, C.; Battaglia, E. First Identification of a Large Set of Serine Hydrolases by Activity-Based Protein Profiling in Dibutyl Phthalate-Exposed Zebrafish Larvae. Int. J. Mol. Sci. 2022, 23, 16060. https://doi.org/10.3390/ijms232416060
Yedji RS, Sohm B, Salnot V, Guillonneau F, Cossu-Leguille C, Battaglia E. First Identification of a Large Set of Serine Hydrolases by Activity-Based Protein Profiling in Dibutyl Phthalate-Exposed Zebrafish Larvae. International Journal of Molecular Sciences. 2022; 23(24):16060. https://doi.org/10.3390/ijms232416060
Chicago/Turabian StyleYedji, Rodrigue S., Bénédicte Sohm, Virginie Salnot, François Guillonneau, Carole Cossu-Leguille, and Eric Battaglia. 2022. "First Identification of a Large Set of Serine Hydrolases by Activity-Based Protein Profiling in Dibutyl Phthalate-Exposed Zebrafish Larvae" International Journal of Molecular Sciences 23, no. 24: 16060. https://doi.org/10.3390/ijms232416060
APA StyleYedji, R. S., Sohm, B., Salnot, V., Guillonneau, F., Cossu-Leguille, C., & Battaglia, E. (2022). First Identification of a Large Set of Serine Hydrolases by Activity-Based Protein Profiling in Dibutyl Phthalate-Exposed Zebrafish Larvae. International Journal of Molecular Sciences, 23(24), 16060. https://doi.org/10.3390/ijms232416060







