CRISPR-Cas Controls Cryptic Prophages
Abstract
:1. Introduction
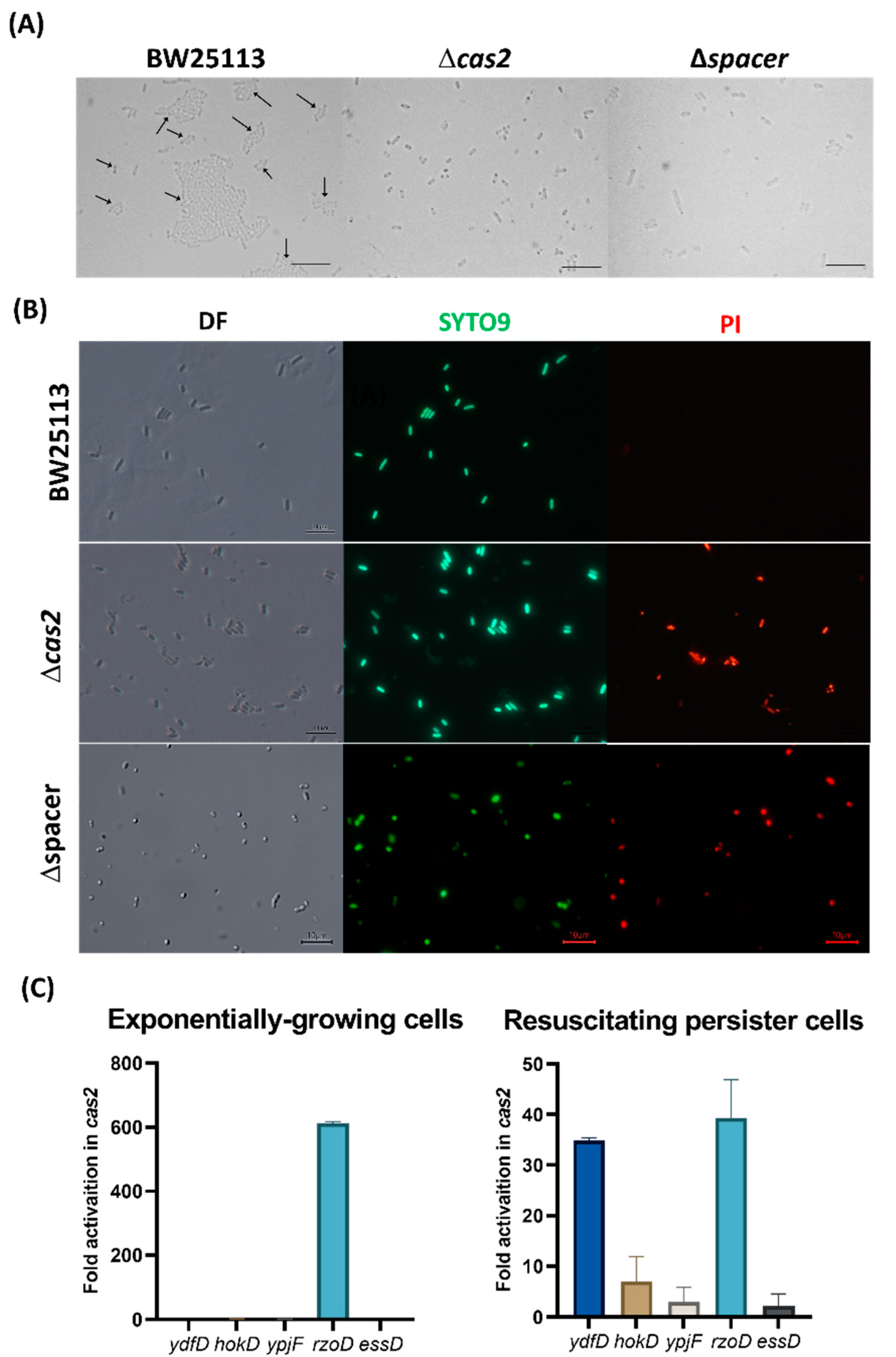
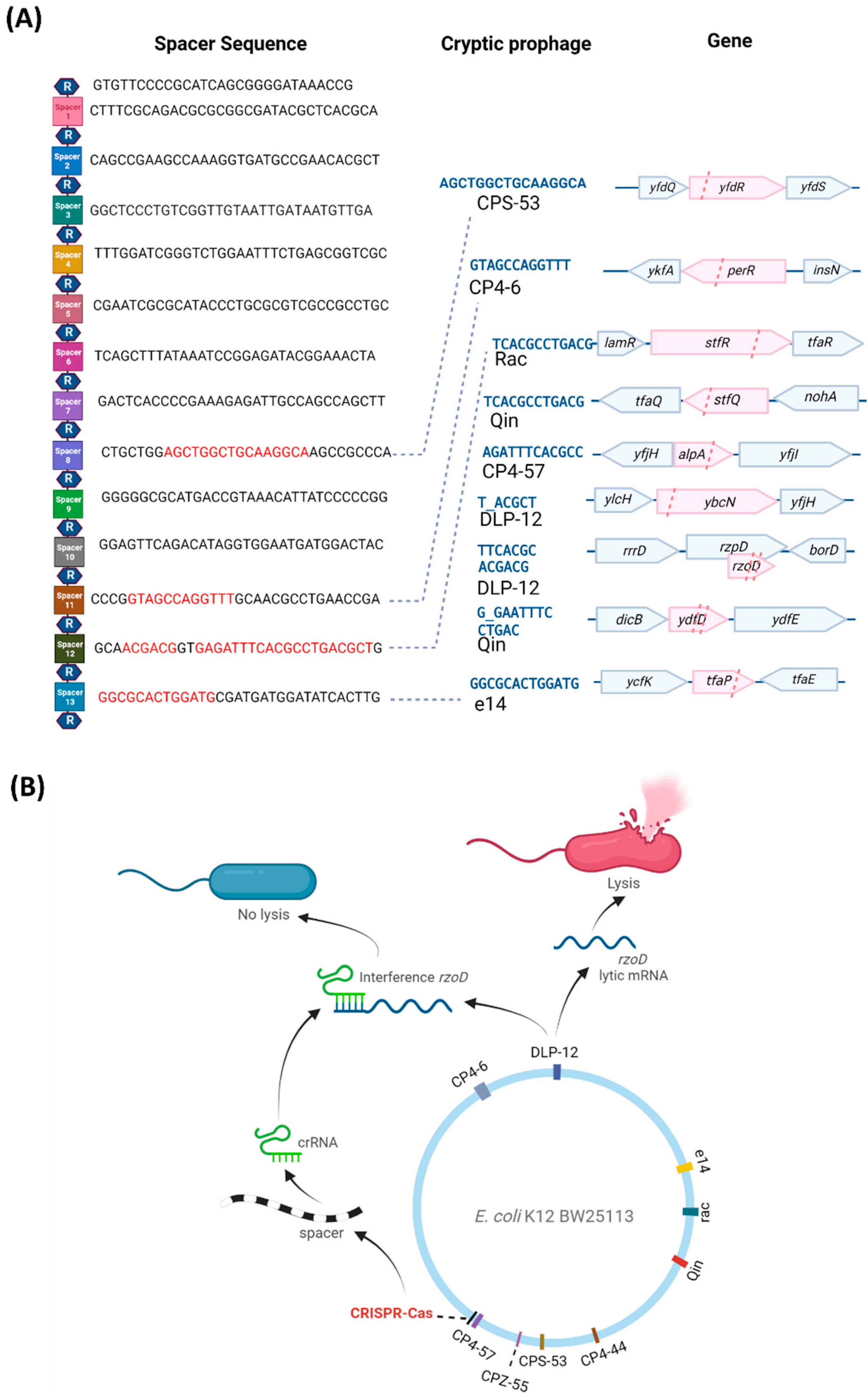
2. Results
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Vasu, K.; Nagaraja, V. Diverse Functions of Restriction-Modification Systems in Addition to Cellular Defense. Microbiol. Mol. Biol. Rev. 2013, 77, 53–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pecota, D.C.; Wood, T.K. Exclusion of T4 Phage by the hok/sok Killer Locus from Plasmid R1. J. Bacteriol. 1996, 178, 2044–2050. [Google Scholar] [CrossRef] [Green Version]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR–Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lord, D.M.; Cheng, H.-Y.; Osbourne, D.O.; Hong, S.H.; Sanchez-Torres, V.; Quiroga, C.; Zheng, K.; Herrmann, T.; Peti, W.; et al. A Novel Type V TA System Where mRNA for Toxin GhoT is Cleaved by Antitoxin GhoS. Nat. Chem. Biol. 2012, 8, 855–861. [Google Scholar] [CrossRef] [Green Version]
- Meeske, A.J.; Nakandakari-Higa, S.; Marraffini, L.A. Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature 2019, 570, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Gong, L.; Cheng, F.; Yu, H.; Zhao, D.; Wang, R.; Wang, T.; Zhang, S.; Zhou, J.; Shmakov, S.A.; et al. Toxin-antitoxin RNA pairs safeguard CRISPR-Cas systems. Science 2021, 372, eabe5601. [Google Scholar] [CrossRef]
- Goldberg, G.W.; McMillan, E.A.; Varble, A.; Modell, J.W.; Samai, P.; Jiang, W.; Marraffini, L.A. Incomplete prophage tolerance by type III-A CRISPR-Cas systems reduces the fitness of lysogenic hosts. Nat. Commun. 2018, 9, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.; Qimron, U. The Escherichia coli CRISPR System Protects from λ Lysogenization, Lysogens, and Prophage Induction. J. Bacteriol. 2010, 192, 6291–6294. [Google Scholar] [CrossRef] [Green Version]
- Touchon, M.; Charpentier, S.; Clermont, O.; Rocha, E.P.C.; Denamur, E.; Branger, C. CRISPR Distribution within the Escherichia coli Species Is Not Suggestive of Immunity-Associated Diversifying Selection. J. Bacteriol. 2011, 193, 2460–2467. [Google Scholar] [CrossRef] [Green Version]
- Shmakov, S.A.; Sitnik, V.; Makarova, K.S.; Wolf, Y.I.; Severinov, K.V.; Koonin, E.V. The CRISPR Spacer Space Is Dominated by Sequences from Species-Specific Mobilomes. mBio 2017, 8, e01397-17. [Google Scholar] [CrossRef]
- Pougach, K.; Semenova, E.; Bogdanova, E.; Datsenko, K.A.; Djordjevic, M.; Wanner, B.L.; Severinov, K. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol. 2010, 77, 1367–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Kim, Y.; Ma, Q.; Hong, S.H.; Pokusaeva, K.; Sturino, J.M.; Wood, T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010, 1, 147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, T.K.; Song, S. Forming and waking dormant cells: The ppGpp ribosome dimerization persister model. Biofilm 2020, 2, 100018. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Kim, J.-S.; Yamasaki, R.; Oh, S.; Benedik, M.J.; Wood, T.K. Escherichia coli cryptic prophages sense nutrients to influence persister cell resuscitation. Environ. Microbiol. 2021, 23, 7245–7254. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-S.; Yamasaki, R.; Song, S.; Zhang, W.; Wood, T.K. Single Cell Observations Show Persister Cells Wake Based on Ribosome Content. Environ. Microbiol. 2018, 20, 2085–2098. [Google Scholar] [CrossRef]
- Yamasaki, R.; Song, S.; Benedik, M.J.; Wood, T.K. Persister Cells Resuscitate Using Membrane Sensors that Activate Chemotaxis, Lower cAMP Levels, and Revive Ribosomes. iScience 2020, 23, 100792. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Wood, T.K. Persister Cells Resuscitate via Ribosome Modification by 23S rRNA Pseudouridine Synthase RluD. Environ. Microbiol. 2020, 22, 850–857. [Google Scholar] [CrossRef]
- Kwan, B.W.; Valenta, J.A.; Benedik, M.J.; Wood, T.K. Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 2013, 57, 1468–1473. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Wood, T.K. Are We Really Studying Persister Cells? Environ. Microbiol. Rep. 2021, 13, 3–7. [Google Scholar] [CrossRef]
- Cheng, H.-Y.; Soo, V.W.C.; Islam, S.; McAnulty, M.J.; Benedik, M.J.; Wood, T.K. Toxin GhoT of the GhoT/GhoS toxin/antitoxin system damages the cell membrane to reduce adenosine triphosphate and to reduce growth under stress. Environ. Microbiol. 2014, 16, 1741–1754. [Google Scholar] [CrossRef] [PubMed]
- Militello, K.T.; Lazatin, J.C. Discovery of Escherichia coli CRISPR sequences in an undergraduate laboratory. Biochem. Molec. Biol. Edu. 2017, 45, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Brouns, S.J.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.H.; Snijders, A.P.L.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; van der Oost, J. Small CRISPR RNAs Guide Antiviral Defense in Prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrangou, R.; Luciano, M.A. CRISPR-Cas Systems: Prokaryotes Upgrade to Adaptive Immunity. Mol. Cell 2014, 54, 234–244. [Google Scholar] [CrossRef] [Green Version]
- García Contreras, R.; Zhang, X.-S.; Kim, Y.; Wood, T.K. Protein Translation and Cell Death: The Role of Rare tRNAs in Biofilm Formation and in Activating Dormant Phage Killer Genes. PLoS ONE 2008, 3, e2394. [Google Scholar] [CrossRef]
- Marimon, O.; Teixeira, J.M.C.; Cordeiro, T.N.; Soo, V.W.C.; Wood, T.L.; Mayzel, M.; Amata, I.; García, J.; Morera, A.; Gay, M.; et al. An oxygen-sensitive toxin–antitoxin system. Nat. Commun. 2016, 7, 13634. [Google Scholar] [CrossRef] [Green Version]
- Masuda, H.; Awano, N.; Inouye, M. ydfD encodes a novel lytic protein in Escherichia coli. FEMS Microbiol. Lett. 2016, 363, fnw039. [Google Scholar] [CrossRef] [Green Version]
- Mohanraju, P.; Saha, C.; van Baarlen, P.; Louwen, R.; Staals, R.H.J.; van der Oost, J. Alternative functions of CRISPR–Cas systems in the evolutionary arms race. Nat. Rev. Microbiol. 2022, 20, 351–364. [Google Scholar] [CrossRef]
- Bozic, B.; Repac, J.; Djordjevic, M. Endogenous Gene Regulation as a Predicted Main Function of Type I-E CRISPR/Cas System in E. coli. Molecules 2019, 24, 784. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Fang, L.; Tan, S.; Yu, M.; Li, X.; He, S.; Wei, Y.; Li, G.; Jiang, J.; Wu, M. Type I CRISPR-Cas targets endogenous genes and regulates virulence to evade mammalian host immunity. Cell Res. 2016, 26, 1273–1287. [Google Scholar] [CrossRef]
- Jore, M.M.; Lundgren, M.; van Duijn, E.; Bultema, J.B.; Westra, E.R.; Waghmare, S.P.; Wiedenheft, B.; Pul, Ü.; Wurm, R.; Wagner, R.; et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011, 18, 529–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rollie, C.; Chevallereau, A.; Watson, B.N.J.; Chyou, T.-y.; Fradet, O.; McLeod, I.; Fineran, P.C.; Brown, C.M.; Gandon, S.; Westra, E.R. Targeting of temperate phages drives loss of type I CRISPR–Cas systems. Nature 2020, 578, 149–153. [Google Scholar] [CrossRef]
- Perna, N.T.; Plunkett, G.; Burland, V.; Mau, B.; Glasner, J.D.; Rose, D.J.; Mayhew, G.F.; Evans, P.S.; Gregor, J.; Kirkpatrick, H.A.; et al. Genome sequence of enterohaemorrhagic Escherichia coli O157: H7. Nature 2001, 409, 529–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savitskaya, E.; Lopatina, A.; Medvedeva, S.; Kapustin, M.; Shmakov, S.; Tikhonov, A.; Artamonova, I.I.; Logacheva, M.; Severinov, K. Dynamics of Escherichia coli type I-E CRISPR spacers over 42 000 years. Mol. Ecol. 2017, 26, 2019–2026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafeezunnisa, M.; Chhakchhuak, P.I.R.; Krishnakumar, J.; Sen, R. Rho-dependent transcription termination regulates the toxin–antitoxin modules of cryptic prophages to silence their expression in Escherichia coli. FEBS Lett. 2021, 595, 2057–2067. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, A.; Shimada, T. Hierarchy of transcription factor network in Escherichia coli K-12: H-NS-mediated silencing and Anti-silencing by global regulators. FEMS Microbiol. Rev. 2021, 45, fuab032. [Google Scholar] [CrossRef]
- Bertani, G. Studies on Lysogenesis.1. The Mode of Phage Liberation by Lysogenic Escherichia-Coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, R.L.; Tait, R.C. Recombinant DNA Techniques: An. Introduction; Benjamin/Cummings Publishing: Menlo Park, CA, USA, 1983. [Google Scholar]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K-12 ORF Archive): Unique Resources for Biological Research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef] [Green Version]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [Green Version]
- Donegan, K.; Matyac, C.; Seidler, R.; Porteous, A. Evaluation of methods for sampling, recovery, and enumeration of bacteria applied to the phylloplane. Appl. Environ. Microbiol. 1991, 57, 51–56. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006–2008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datsenko, K.A.; Pougach, K.; Tikhonov, A.; Wanner, B.L.; Severinov, K.; Semenova, E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012, 3, 945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Kim, Y.; Wood, T.K. Control and Benefits of CP4-57 Prophage Excision in Escherichia coli Biofilms. ISME J. 2009, 3, 1164–1179. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, S.; Semenova, E.; Severinov, K.; Fernández-García, L.; Benedik, M.J.; Maeda, T.; Wood, T.K. CRISPR-Cas Controls Cryptic Prophages. Int. J. Mol. Sci. 2022, 23, 16195. https://doi.org/10.3390/ijms232416195
Song S, Semenova E, Severinov K, Fernández-García L, Benedik MJ, Maeda T, Wood TK. CRISPR-Cas Controls Cryptic Prophages. International Journal of Molecular Sciences. 2022; 23(24):16195. https://doi.org/10.3390/ijms232416195
Chicago/Turabian StyleSong, Sooyeon, Ekaterina Semenova, Konstantin Severinov, Laura Fernández-García, Michael J. Benedik, Toshinari Maeda, and Thomas K. Wood. 2022. "CRISPR-Cas Controls Cryptic Prophages" International Journal of Molecular Sciences 23, no. 24: 16195. https://doi.org/10.3390/ijms232416195






