Relationship between Epithelial-to-Mesenchymal Transition and Tumor-Associated Macrophages in Colorectal Liver Metastases
Abstract
:1. Background
2. The Role of Epithelial-to-Mesenchymal Transition (EMT) in CRC
3. The Molecular Mechanisms of EMT in CRC
4. The Relationship between EMT and TGF-β Signaling Pathway in CRC
5. The Role of Tumor-Associated Macrophages (TAMs) in CRC Progression
6. Mechanisms Exploited by TAMs to Regulate EMT in Colorectal Liver Metastasis (CLMs): A Focus on TGF-β Signaling Pathway
7. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landreau, P.; Drouillard, A.; Launoy, G.; Ortega-Deballon, P.; Jooste, V.; Lepage, C.; Faivre, J.; Facy, O.; Bouvier, A.-M. Incidence and survival in late liver metastases of colorectal cancer: Liver colorectal cancer metastases survival. J. Gastroenterol. Hepatol. 2015, 30, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Mlecnik, B.; Van den Eynde, M.; Bindea, G.; Church, S.E.; Vasaturo, A.; Fredriksen, T.; Lafontaine, L.; Haicheur, N.; Marliot, F.; Debetancourt, D.; et al. Comprehensive Intrametastatic Immune Quantification and Major Impact of Immunoscore on Survival. J. Natl. Cancer Inst. 2018, 110, 97–108. [Google Scholar] [CrossRef] [Green Version]
- Hackl, C.; Neumann, P.; Gerken, M.; Loss, M.; Klinkhammer-Schalke, M.; Schlitt, H.J. Treatment of colorectal liver metastases in Germany: A ten-year population-based analysis of 5772 cases of primary colorectal adenocarcinoma. BMC Cancer 2014, 14, 810. [Google Scholar] [CrossRef] [Green Version]
- Lin, A.; Zhang, J.; Luo, P. Crosstalk Between the MSI Status and Tumor Microenvironment in Colorectal Cancer. Front. Immunol. 2020, 11, 2039. [Google Scholar] [CrossRef]
- Mei, Y.; Xiao, W.; Hu, H.; Lu, G.; Chen, L.; Sun, Z.; Lü, M.; Ma, W.; Jiang, T.; Gao, Y.; et al. Single-cell analyses reveal suppressive tumor microenvironment of human colorectal cancer. Clin. Transl. Med. 2021, 11, e422. [Google Scholar] [CrossRef]
- Mantovani, A.; Marchesi, F.; Malesci, A.; Laghi, L.; Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017, 14, 399–416. [Google Scholar] [CrossRef]
- Cortese, N.; Donadon, M.; Rigamonti, A.; Marchesi, F. Macrophages at the crossroads of anticancer strategies. Front. Biosci. 2019, 24, 1271–1283. [Google Scholar] [CrossRef]
- Cao, H.; Xu, E.; Liu, H.; Wan, L.; Lai, M. Epithelial–mesenchymal transition in colorectal cancer metastasis: A system review. Pathol. Res. Pract. 2015, 211, 557–569. [Google Scholar] [CrossRef]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.-C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.C.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hugo, W.; Zaretsky, J.M.; Sun, L.; Song, C.; Moreno, B.H.; Hu-Lieskovan, S.; Berent-Maoz, B.; Pang, J.; Chmielowski, B.; Cherry, G.; et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016, 165, 35–44. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.; Yelle, N.; Venugopal, C.; Singh, S.K. EMT: Mechanisms and therapeutic implications. Pharmacol. Ther. 2018, 182, 80–94. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Carmona, M.; Lesage, J.; Cataldo, D.; Gilles, C. EMT and inflammation: Inseparable actors of cancer progression. Mol. Oncol. 2017, 11, 805–823. [Google Scholar] [CrossRef]
- Tsilimigras, D.I.; Brodt, P.; Clavien, P.-A.; Muschel, R.J.; D’Angelica, M.I.; Endo, I.; Parks, R.W.; Doyle, M.; de Santibañes, E.; Pawlik, T.M. Liver metastases. Nat. Rev. Dis. Primer 2021, 7, 27. [Google Scholar] [CrossRef]
- Nguyen, D.X.; Bos, P.D.; Massagué, J. Metastasis: From dissemination to organ-specific colonization. Nat. Rev. Cancer 2009, 9, 274–284. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, C. The role of liver sinusoidal endothelial cells in cancer liver metastasis. Am. J. Cancer Res. 2021, 11, 1845–1860. [Google Scholar] [CrossRef] [Green Version]
- Genna, A.; Vanwynsberghe, A.M.; Villard, A.V.; Pottier, C.; Ancel, J.; Polette, M.; Gilles, C. EMT-Associated Heterogeneity in Circulating Tumor Cells: Sticky Friends on the Road to Metastasis. Cancers 2020, 12, 1632. [Google Scholar] [CrossRef]
- Terry, S.; Savagner, P.; Ortiz-Cuaran, S.; Mahjoubi, L.; Saintigny, P.; Thiery, J.-P.; Chouaib, S. New insights into the role of EMT in tumor immune escape. Mol. Oncol. 2017, 11, 824–846. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polyak, K.; Weinberg, R.A. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat. Rev. Cancer 2009, 9, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Jie, X.-X.; Zhang, X.-Y.; Xu, C.-J. Epithelial-to-mesenchymal transition, circulating tumor cells and cancer metastasis: Mechanisms and clinical applications. Oncotarget 2017, 8, 81558–81571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barillari, G. The Impact of Matrix Metalloproteinase-9 on the Sequential Steps of the Metastatic Process. Int. J. Mol. Sci. 2020, 21, 4526. [Google Scholar] [CrossRef]
- Alba, J.; Barcia, R.; Gutiérrez-Berzal, J.; Ramos-Martínez, J.I. Could inhibition of metalloproteinases be used to block the process of metastasis? Cell Biochem. Funct. 2022, 40, 600–607. [Google Scholar] [CrossRef]
- Ding, C.; Luo, J.; Li, L.; Li, S.; Yang, L.; Pan, H.; Liu, Q.; Qin, H.; Chen, C.; Feng, J. Gab2 facilitates epithelial-to-mesenchymal transition via the MEK/ERK/MMP signaling in colorectal cancer. J. Exp. Clin. Cancer Res. 2016, 35, 5. [Google Scholar] [CrossRef] [Green Version]
- Buttacavoli, M.; Di Cara, G.; Roz, E.; Pucci-Minafra, I.; Feo, S.; Cancemi, P. Integrated Multi-Omics Investigations of Metalloproteinases in Colon Cancer: Focus on MMP2 and MMP9. Int. J. Mol. Sci. 2021, 22, 12389. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [Green Version]
- Kozuka, M.; Battaglin, F.; Jayachandran, P.; Wang, J.; Arai, H.; Soni, S.; Zhang, W.; Hirai, M.; Matsusaka, S.; Lenz, H.-J. Clinical Significance of Circulating Tumor Cell Induced Epithelial-Mesenchymal Transition in Patients with Metastatic Colorectal Cancer by Single-Cell RNA-Sequencing. Cancers 2021, 13, 4862. [Google Scholar] [CrossRef]
- Wal, G.; Gouw, A.; Kamps, J.; Moorlag, H.; Bulthuis, M.; Molema, G.; de Jong, K. Angiogenesis in Synchronous and Metachronous Colorectal Liver Metastases the Liver as a Permissive Soil. Ann. Surg. 2011, 255, 86–94. [Google Scholar] [CrossRef]
- Kuczynski, E.A.; Vermeulen, P.B.; Pezzella, F.; Kerbel, R.S.; Reynolds, A.R. Vessel co-option in cancer. Nat. Rev. Clin. Oncol. 2019, 16, 469–493. [Google Scholar] [CrossRef] [PubMed]
- Rada, M.; Lazaris, A.; Kapelanski-Lamoureux, A.; Mayer, T.Z.; Metrakos, P. Tumor microenvironment conditions that favor vessel co-option in colorectal cancer liver metastases: A theoretical model. Semin. Cancer Biol. 2021, 71, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Rada, M.; Kapelanski-Lamoureux, A.; Petrillo, S.; Tabariès, S.; Siegel, P.; Reynolds, A.R.; Lazaris, A.; Metrakos, P. Runt related transcription factor-1 plays a central role in vessel co-option of colorectal cancer liver metastases. Commun. Biol. 2021, 4, 950. [Google Scholar] [CrossRef] [PubMed]
- Rada, M.; Tsamchoe, M.; Kapelanski-Lamoureux, A.; Hassan, N.; Bloom, J.; Petrillo, S.; Kim, D.H.; Lazaris, A.; Metrakos, P. Cancer Cells Promote Phenotypic Alterations in Hepatocytes at the Edge of Cancer Cell Nests to Facilitate Vessel Co-Option Establishment in Colorectal Cancer Liver Metastases. Cancers 2022, 14, 1318. [Google Scholar] [CrossRef] [PubMed]
- Reymond, N.; d’Água, B.B.; Ridley, A.J. Crossing the endothelial barrier during metastasis. Nat. Rev. Cancer 2013, 13, 858–870. [Google Scholar] [CrossRef]
- Paschos, K.A. Natural history of hepatic metastases from colorectal cancer—Pathobiological pathways with clinical significance. World J. Gastroenterol. 2014, 20, 3719–3737. [Google Scholar] [CrossRef]
- Rahimi, N. Defenders and Challengers of Endothelial Barrier Function. Front. Immunol. 2017, 8, 1847. [Google Scholar] [CrossRef] [Green Version]
- Giannotta, M.; Trani, M.; Dejana, E. VE-Cadherin and Endothelial Adherens Junctions: Active Guardians of Vascular Integrity. Dev. Cell 2013, 26, 441–454. [Google Scholar] [CrossRef] [Green Version]
- Dou, R.; Liu, K.; Yang, C.; Zheng, J.; Shi, D.; Lin, X.; Wei, C.; Zhang, C.; Fang, Y.; Huang, S.; et al. EMT-cancer cells-derived exosomal miR-27b-3p promotes circulating tumour cells-mediated metastasis by modulating vascular permeability in colorectal cancer. Clin. Transl. Med. 2021, 11, e595. [Google Scholar] [CrossRef]
- Heeke, S.; Mograbi, B.; Alix-Panabières, C.; Hofman, P. Never Travel Alone: The Crosstalk of Circulating Tumor Cells and the Blood Microenvironment. Cells 2019, 8, 714. [Google Scholar] [CrossRef]
- Chambers, A.F.; Groom, A.C.; MacDonald, I.C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2002, 2, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Aceto, N.; Toner, M.; Maheswaran, S.; Haber, D.A. En Route to Metastasis: Circulating Tumor Cell Clusters and Epithelial-to-Mesenchymal Transition. Trends Cancer 2015, 1, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Haier, J.; Nicolson, G.L. Tumor cell adhesion under hydrodynamic conditions of fluid flow. APMIS 2001, 109, 241–262. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, H.; Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef] [Green Version]
- Bakir, B.; Chiarella, A.M.; Pitarresi, J.R.; Rustgi, A.K. EMT, MET, Plasticity, and Tumor Metastasis. Trends Cell Biol. 2020, 30, 764–776. [Google Scholar] [CrossRef]
- Haerinck, J.; Berx, G. Partial EMT takes the lead in cancer metastasis. Dev. Cell 2021, 56, 3174–3176. [Google Scholar] [CrossRef]
- Pavlič, A.; Urh, K.; Štajer, K.; Boštjančič, E.; Zidar, N. Epithelial-Mesenchymal Transition in Colorectal Carcinoma: Comparison Between Primary Tumor, Lymph Node and Liver Metastases. Front. Oncol. 2021, 11, 662806. [Google Scholar] [CrossRef]
- Cao, Z.-Q.; Wang, Z.; Leng, P. Aberrant N-cadherin expression in cancer. Biomed. Pharmacother. 2019, 118, 109320. [Google Scholar] [CrossRef]
- Paulin, D.; Lilienbaum, A.; Kardjian, S.; Agbulut, O.; Li, Z. Vimentin: Regulation and pathogenesis. Biochimie 2022, 197, 96–112. [Google Scholar] [CrossRef]
- Wang, Q.; Zhu, G.; Lin, C.; Lin, P.; Chen, H.; He, R.; Huang, Y.; Yang, S.; Ye, J. Vimentin affects colorectal cancer proliferation, invasion, and migration via regulated by activator protein 1. J. Cell Physiol. 2021, 236, 7591–7604. [Google Scholar] [CrossRef]
- Niknami, Z.; Muhammadnejad, A.; Ebrahimi, A.; Harsani, Z.; Shirkoohi, R. Significance of E-cadherin and Vimentin as epithelial-mesenchymal transition markers in colorectal carcinoma prognosis. EXCLI J. 2020, 19, 917–926. [Google Scholar] [CrossRef] [PubMed]
- Patten, J.; Wang, K. Fibronectin in development and wound healing. Adv. Drug Deliv. Rev. 2021, 170, 353–368. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Liu, C.; Zhang, T.; Zhu, Y.; Dong, X.; Xue, P. Down-regulation of FN1 inhibits colorectal carcinogenesis by suppressing proliferation, migration, and invasion. J. Cell Biochem. 2018, 119, 4717–4728. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Zhang, R.; Feng, H. Fibronectin promotes tumor cells growth and drugs resistance through a CDC42-YAP-dependent signaling pathway in colorectal cancer. Cell Biol. Int. 2020, 44, 1840–1849. [Google Scholar] [CrossRef] [PubMed]
- Daulagala, A.C.; Bridges, M.C.; Kourtidis, A. E-cadherin Beyond Structure: A Signaling Hub in Colon Homeostasis and Disease. Int. J. Mol. Sci. 2019, 20, 2756. [Google Scholar] [CrossRef] [Green Version]
- Yun, J.-A.; Kim, S.-H.; Hong, H.K.; Yun, S.H.; Kim, H.C.; Chun, H.-K.; Cho, Y.B.; Lee, W.Y. Loss of E-Cadherin Expression Is Associated with a Poor Prognosis in Stage III Colorectal Cancer. Oncology 2014, 86, 318–328. [Google Scholar] [CrossRef]
- Cho, H.-J.; Oh, N.; Park, J.-H.; Kim, K.-S.; Kim, H.-K.; Lee, E.; Hwang, S.; Kim, S.-J.; Park, K.-S. ZEB1 Collaborates with ELK3 to Repress E-Cadherin Expression in Triple-Negative Breast Cancer Cells. Mol. Cancer Res. 2019, 17, 2257–2266. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.; Hu, Y.; Guo, D.; Zhang, A.; Gu, Y.; Zhang, S.; Zhao, C.; Gong, P.; Shen, X.; Li, Y.; et al. DNA methyltransferase 3A isoform b contributes to repressing E-cadherin through cooperation of DNA methylation and H3K27/H3K9 methylation in EMT-related metastasis of gastric cancer. Oncogene 2018, 37, 4358–4371. [Google Scholar] [CrossRef]
- Venhuizen, J.-H.; Jacobs, F.J.C.; Span, P.N.; Zegers, M.M. P120 and E-cadherin: Double-edged swords in tumor metastasis. Semin. Cancer Biol. 2020, 60, 107–120. [Google Scholar] [CrossRef]
- Huang, Y.; Hong, W.; Wei, X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J. Hematol Oncol 2022, 15, 129. [Google Scholar] [CrossRef]
- Villarejo, A.; Cortés-Cabrera, Á.; Molina-Ortíz, P.; Portillo, F.; Cano, A. Differential Role of Snail1 and Snail2 Zinc Fingers in E-cadherin Repression and Epithelial to Mesenchymal Transition. J. Biol. Chem. 2014, 289, 930–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brzozowa, M.; Michalski, M.; Wyrobiec, G.; Piecuch, A.; Dittfeld, A.; Harabin-Słowińska, M.; Boroń, D.; Wojnicz, R. The role of Snail1 transcription factor in colorectal cancer progression and metastasis. Contemp. Oncol. 2015, 19, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Francí, C.; Gallén, M.; Alameda, F.; Baró, T.; Iglesias, M.; Virtanen, I.; García de Herreros, A. Snail1 Protein in the Stroma as a New Putative Prognosis Marker for Colon Tumours. PLoS ONE 2009, 4, e5595. [Google Scholar] [CrossRef] [PubMed]
- Francı, C.; Takkunen, M.; Dave, N.; Alameda, F. Expression of Snail protein in tumor–stroma interface. Oncogene 2006, 25, 5134–5144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, J.E.; Nathan, V.; Osborne, J.K.; Farrow, R.K.; Deb, D.; Sullivan, J.P.; Dospoy, P.D.; Augustyn, A.; Hight, S.K.; Sato, M.; et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J. Clin. Investig. 2016, 126, 3219–3235. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Ng, A.S.; Cai, S.; Li, Q.; Yang, L.; Kerr, D. Novel therapeutic strategies: Targeting epithelial–mesenchymal transition in colorectal cancer. Lancet Oncol. 2021, 22, e358–e368. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.W.; Rao, G.; Kim, J.J.; Shim, H.-S.; Park, K.-S.; An, S.S.; Kim, B.; Steeg, P.S.; Sarfaraz, S.; Changwoo Lee, L.; et al. LAMC2 enhances the metastatic potential of lung adenocarcinoma. Cell Death Differ. 2015, 22, 1341–1352. [Google Scholar] [CrossRef] [Green Version]
- Colangelo, T.; Carbone, A.; Mazzarelli, F.; Cuttano, R.; Dama, E.; Nittoli, T.; Albanesi, J.; Barisciano, G.; Forte, N.; Palumbo, O.; et al. Loss of circadian gene Timeless induces EMT and tumor progression in colorectal cancer via Zeb1-dependent mechanism. Cell Death Differ. 2022, 29, 1552–1568. [Google Scholar] [CrossRef]
- Lindner, P.; Paul, S.; Eckstein, M.; Hampel, C.; Muenzner, J.K.; Erlenbach-Wuensch, K.; Ahmed, H.P.; Mahadevan, V.; Brabletz, T.; Hartmann, A.; et al. EMT transcription factor ZEB1 alters the epigenetic landscape of colorectal cancer cells. Cell Death Dis. 2020, 11, 147. [Google Scholar] [CrossRef] [Green Version]
- Franco, H.L.; Casasnovas, J.; Rodriguez-Medina, J.R.; Cadilla, C.L. Redundant or separate entities?--roles of Twist1 and Twist2 as molecular switches during gene transcription. Nucleic Acids Res. 2011, 39, 1177–1186. [Google Scholar] [CrossRef]
- Abdelmaksoud-Dammak, R.; Chamtouri, N.; Triki, M.; Saadallah-Kallel, A.; Ayadi, W.; Charfi, S.; Khabir, A.; Ayadi, L.; Sallemi-Boudawara, T.; Mokdad-Gargouri, R. Overexpression of miR-10b in colorectal cancer patients: Correlation with TWIST-1 and E-cadherin expression. Tumor Biol. 2017, 39, 1010428317695916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, J.-J.; Zhang, W.; Xu, X.-M.; Zhang, F.; Tao, W.-P.; Ye, J.-J.; Ge, W. Twist mediates an aggressive phenotype in human colorectal cancer cells. Int. J. Oncol. 2016, 48, 1117–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.W.; Lee, Y.S.; Yun, N.H.; Shin, C.H.; Hong, H.K.; Kim, H.H.; Cho, Y.B. MicroRNA-17-5p regulates EMT by targeting vimentin in colorectal cancer. Br. J. Cancer 2020, 123, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, S.; Yin, J.; Xu, R. MiR-566 mediates cell migration and invasion in colon cancer cells by direct targeting of PSKH1. Cancer Cell Int. 2019, 19, 333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwasaki, H.; Shimura, T.; Kitagawa, M.; Yamada, T.; Nishigaki, R.; Fukusada, S.; Okuda, Y.; Katano, T.; Horike, S.; Kataoka, H. A Novel Urinary miRNA Biomarker for Early Detection of Colorectal Cancer. Cancers 2022, 14, 461. [Google Scholar] [CrossRef]
- Zhao, H.; Wei, J.; Sun, J. Roles of TGF-β signaling pathway in tumor microenvirionment and cancer therapy. Int. Immunopharmacol. 2020, 89, 107101. [Google Scholar] [CrossRef]
- Siegel, P.M.; Massagué, J. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat. Rev. Cancer 2003, 3, 807–820. [Google Scholar] [CrossRef]
- Bach, S.P.; Renehan, A.G.; Potten, C.S. Stem cells: The intestinal stem cell as a paradigm. Carcinogenesis 2000, 21, 469–476. [Google Scholar] [CrossRef] [Green Version]
- Pellatt, A.J.; Mullany, L.E.; Herrick, J.S.; Sakoda, L.C.; Wolff, R.K.; Samowitz, W.S.; Slattery, M.L. The TGFβ-signaling pathway and colorectal cancer: Associations between dysregulated genes and miRNAs. J. Transl. Med. 2018, 16, 191. [Google Scholar] [CrossRef] [Green Version]
- Itatani, Y.; Kawada, K.; Sakai, Y. Transforming Growth Factor-β Signaling Pathway in Colorectal Cancer and Its Tumor Microenvironment. Int. J. Mol. Sci. 2019, 20, 5822. [Google Scholar] [CrossRef]
- Peinado, H.; Olmeda, D.; Cano, A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer 2007, 7, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Moustakas, A.; Heldin, C.-H. Induction of epithelial–mesenchymal transition by transforming growth factor β. Semin. Cancer Biol. 2012, 22, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Baker, D.; ten Dijke, P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int. J. Mol. Sci. 2019, 20, 2767. [Google Scholar] [CrossRef] [Green Version]
- Wendt, M.K.; Allington, T.M.; Schiemann, W.P. Mechanisms of the epithelial–mesenchymal transition by TGF-β. Future Oncol. 2009, 5, 1145–1168. [Google Scholar] [CrossRef] [Green Version]
- Hua, W.; Kostidis, S.; Mayboroda, O.; Giera, M.; Hornsveld, M.; ten Dijke, P. Metabolic Reprogramming of Mammary Epithelial Cells during TGF-β-Induced Epithelial-to-Mesenchymal Transition. Metabolites 2021, 11, 626. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Thorikay, M.; van der Zon, G.; van Dinther, M.; ten Dijke, P. Studying TGF-β; Signaling and TGF-β-induced Epithelial-to-mesenchymal Transition in Breast Cancer and Normal Cells. J. Vis. Exp. 2020, 164, e61830. [Google Scholar] [CrossRef]
- Bierie, B.; Moses, H. TGF-β and cancer. Cytokine Growth Factor Rev. 2006, 17, 29–40. [Google Scholar] [CrossRef]
- Chruścik, A.; Gopalan, V.; Lam, A.K. The clinical and biological roles of transforming growth factor beta in colon cancer stem cells: A systematic review. Eur. J. Cell Biol. 2018, 97, 15–22. [Google Scholar] [CrossRef]
- Mooi, J.K.; Wirapati, P.; Asher, R.; Lee, C.K.; Savas, P.; Price, T.J.; Townsend, A.; Hardingham, J.; Buchanan, D.; Williams, D.; et al. The prognostic impact of consensus molecular subtypes (CMS) and its predictive effects for bevacizumab benefit in metastatic colorectal cancer: Molecular analysis of the AGITG MAX clinical trial. Ann. Oncol. 2018, 29, 2240–2246. [Google Scholar] [CrossRef]
- Pohl, M.; Radacz, Y.; Pawlik, N.; Schoeneck, A.; Baldus, S.E.; Munding, J.; Schmiegel, W.; Schwarte-Waldhoff, I.; Reinacher-Schick, A. SMAD4 Mediates Mesenchymal–Epithelial Reversion in SW480 Colon Carcinoma Cells. Anticancer Res. 2010, 18, 601–608. [Google Scholar] [CrossRef]
- Voorneveld, P.W.; Kodach, L.L.; Jacobs, R.J.; Liv, N.; Zonnevylle, A.C.; Hoogenboom, J.P.; Biemond, I.; Verspaget, H.W.; Hommes, D.W.; de Rooij, K.; et al. Loss of SMAD4 Alters BMP Signaling to Promote Colorectal Cancer Cell Metastasis via Activation of Rho and ROCK. Gastroenterology 2014, 147, 196–208.e13. [Google Scholar] [CrossRef] [PubMed]
- Frey, P.; Devisme, A.; Rose, K.; Schrempp, M.; Freihen, V.; Andrieux, G.; Boerries, M.; Hecht, A. SMAD4 mutations do not preclude epithelial–mesenchymal transition in colorectal cancer. Oncogene 2022, 41, 824–837. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Weigel, K.J.; Zhou, H.; Wang, X.-J. Paradoxical roles of TGF-β signaling in suppressing and promoting squamous cell carcinoma. Acta Biochim. Biophys. Sin. 2018, 50, 98–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seoane, J.; Gomis, R.R. TGF-β Family Signaling in Tumor Suppression and Cancer Progression. Cold Spring Harb. Perspect. Biol. 2017, 9, a022277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.-T.; Wu, K.-J. Epigenetic regulation of epithelial-mesenchymal transition: Focusing on hypoxia and TGF-β signaling. J. Biomed. Sci. 2020, 27, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagati, A.; Kumar, S.; Jiang, P.; Pyrdol, J.; Zou, A.E.; Godicelj, A.; Mathewson, N.D.; Cartwright, A.N.R.; Cejas, P.; Brown, M.; et al. Integrin αvβ6–TGFβ–SOX4 Pathway Drives Immune Evasion in Triple-Negative Breast Cancer. Cancer Cell 2021, 39, 54–67.e9. [Google Scholar] [CrossRef] [PubMed]
- Taieb, J.; Svrcek, M.; Cohen, R.; Basile, D.; Tougeron, D.; Phelip, J.-M. Deficient mismatch repair/microsatellite unstable colorectal cancer: Diagnosis, prognosis and treatment. Eur. J. Cancer 2022, 175, 136–157. [Google Scholar] [CrossRef]
- Pino, M.S.; Kikuchi, H.; Zeng, M.; Herraiz, M.; Sperduti, I.; Berger, D.; Park, D.; Iafrate, A.J.; Zukerberg, L.R.; Chung, D.C. Epithelial to Mesenchymal Transition Is Impaired in Colon Cancer Cells with Microsatellite Instability. Gastroenterology 2010, 138, 1406–1417. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, T.; Cloyd, J.M.; Vicente, D.; Omichi, K.; Chun, Y.S.; Kopetz, S.E.; Maru, D.; Conrad, C.; Tzeng, C.-W.D.; Wei, S.H.; et al. SMAD4 gene mutation predicts poor prognosis in patients undergoing resection for colorectal liver metastases. Eur. J. Surg. Oncol. 2018, 44, 684–692. [Google Scholar] [CrossRef]
- Okita, A.; Takahashi, S.; Ouchi, K.; Inoue, M.; Watanabe, M.; Endo, M.; Honda, H.; Yamada, Y.; Ishioka, C. Consensus molecular subtypes classification of colorectal cancer as a predictive factor for chemotherapeutic efficacy against metastatic colorectal cancer. Oncotarget 2018, 9, 18698–18711. [Google Scholar] [CrossRef]
- Jahchan, N.S.; Mujal, A.M.; Pollack, J.L.; Binnewies, M.; Sriram, V.; Reyno, L.; Krummel, M.F. Tuning the Tumor Myeloid Microenvironment to Fight Cancer. Front. Immunol. 2019, 10, 1611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locati, M.; Curtale, G.; Mantovani, A. Diversity, Mechanisms, and Significance of Macrophage Plasticity. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 123–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coussens, L.M.; Zitvogel, L.; Palucka, A.K. Neutralizing Tumor-Promoting Chronic Inflammation: A Magic Bullet? Science 2013, 339, 286–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrison, C. Immuno-oncologists eye up macrophage targets. Nat. Rev. Drug Discov. 2016, 15, 373–374. [Google Scholar] [CrossRef]
- Qian, B.-Z.; Pollard, J.W. Macrophage Diversity Enhances Tumor Progression and Metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Tamura, R.; Tanaka, T.; Yamamoto, Y.; Akasaki, Y.; Sasaki, H. Dual role of macrophage in tumor immunity. Immunotherapy 2018, 10, 899–909. [Google Scholar] [CrossRef]
- Laoui, D.; Van Overmeire, E.; Di Conza, G.; Aldeni, C.; Keirsse, J.; Morias, Y.; Movahedi, K.; Houbracken, I.; Schouppe, E.; Elkrim, Y.; et al. Tumor Hypoxia Does Not Drive Differentiation of Tumor-Associated Macrophages but Rather Fine-Tunes the M2-like Macrophage Population. Cancer Res. 2014, 74, 24–30. [Google Scholar] [CrossRef] [Green Version]
- Galon, J.; Angell, H.K.; Bedognetti, D.; Marincola, F.M. The Continuum of Cancer Immunosurveillance: Prognostic, Predictive, and Mechanistic Signatures. Immunity 2013, 39, 11–26. [Google Scholar] [CrossRef] [Green Version]
- Donadon, M.; Torzilli, G.; Cortese, N.; Soldani, C.; Di Tommaso, L.; Franceschini, B.; Carriero, R.; Barbagallo, M.; Rigamonti, A.; Anselmo, A.; et al. Macrophage morphology correlates with single-cell diversity and prognosis in colorectal liver metastasis. J. Exp. Med. 2020, 217, e20191847. [Google Scholar] [CrossRef]
- Malesci, A.; Bianchi, P.; Celesti, G.; Basso, G.; Marchesi, F.; Grizzi, F.; Di Caro, G.; Cavalleri, T.; Rimassa, L.; Palmqvist, R.; et al. Tumor-associated macrophages and response to 5-fluorouracil adjuvant therapy in stage III colorectal cancer. OncoImmunology 2017, 6, e1342918. [Google Scholar] [CrossRef]
- Mantovani, A.; Allavena, P.; Marchesi, F.; Garlanda, C. Macrophages as tools and targets in cancer therapy. Nat. Rev. Drug Discov. 2022, 21, 799–820. [Google Scholar] [CrossRef] [PubMed]
- Boutilier, A.J.; Elsawa, S.F. Macrophage Polarization States in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 6995. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Atri, C.; Guerfali, F.; Laouini, D. Role of Human Macrophage Polarization in Inflammation during Infectious Diseases. Int. J. Mol. Sci. 2018, 19, 1801. [Google Scholar] [CrossRef] [Green Version]
- Väyrynen, J.P.; Haruki, K.; Lau, M.C.; Väyrynen, S.A.; Zhong, R.; Dias Costa, A.; Borowsky, J.; Zhao, M.; Fujiyoshi, K.; Arima, K.; et al. The Prognostic Role of Macrophage Polarization in the Colorectal Cancer Microenvironment. Cancer Immunol. Res. 2021, 9, 8–19. [Google Scholar] [CrossRef]
- Macciò, A.; Gramignano, G.; Cherchi, M.C.; Tanca, L.; Melis, L.; Madeddu, C. Role of M1-polarized tumor-associated macrophages in the prognosis of advanced ovarian cancer patients. Sci. Rep. 2020, 10, 6096. [Google Scholar] [CrossRef] [Green Version]
- Yunna, C.; Mengru, H.; Lei, W.; Weidong, C. Macrophage M1/M2 polarization. Eur. J. Pharmacol. 2020, 877, 173090. [Google Scholar] [CrossRef]
- Cavalleri, T.; Greco, L.; Rubbino, F.; Hamada, T.; Quaranta, M.; Grizzi, F.; Sauta, E.; Craviotto, V.; Bossi, P.; Vetrano, S.; et al. Tumor-associated macrophages and risk of recurrence in stage III colorectal cancer. J. Pathol. Clin. Res. 2022, 8, 307–312. [Google Scholar] [CrossRef]
- Herrera, M.; Herrera, A.; Domínguez, G.; Silva, J.; García, V.; García, J.M.; Gómez, I.; Soldevilla, B.; Muñoz, C.; Provencio, M. Cancer-associated fibroblast and M 2 macrophage markers together predict outcome in colorectal cancer patients. Cancer Sci. 2013, 104, 437–444. [Google Scholar] [CrossRef]
- Shibutani, M.; Nakao, S.; Maeda, K.; Nagahara, H.; Kashiwagi, S.; Hirakawa, K.; Ohira, M. The Impact of Tumor-associated Macrophages on Chemoresistance via Angiogenesis in Colorectal Cancer. Anticancer Res. 2021, 41, 4447–4453. [Google Scholar] [CrossRef] [PubMed]
- DeNardo, D.G.; Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 2019, 19, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Sugimura-Nagata, A.; Koshino, A.; Inoue, S.; Matsuo-Nagano, A.; Komura, M.; Riku, M.; Ito, H.; Inoko, A.; Murakami, H.; Ebi, M.; et al. Expression and Prognostic Significance of CD47–SIRPA Macrophage Checkpoint Molecules in Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 2690. [Google Scholar] [CrossRef] [PubMed]
- Cortese, N.; Soldani, C.; Franceschini, B.; Barbagallo, M.; Marchesi, F.; Torzilli, G.; Donadon, M. Macrophages in Colorectal Cancer Liver Metastases. Cancers 2019, 11, 633. [Google Scholar] [CrossRef] [Green Version]
- Guilliams, M.; Scott, C.L. Liver macrophages in health and disease. Immunity 2022, 55, 1515–1529. [Google Scholar] [CrossRef]
- Krenkel, O.; Tacke, F. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 2017, 17, 306–321. [Google Scholar] [CrossRef]
- Wei, C.; Yang, C.; Wang, S.; Shi, D.; Zhang, C.; Lin, X.; Liu, Q.; Dou, R.; Xiong, B. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol. Cancer 2019, 18, 64. [Google Scholar] [CrossRef]
- Heijstek, M.W.; Kranenburg, O.; Borel Rinkes, I.H.M. Mouse Models of Colorectal Cancer and Liver Metastases. Dig. Surg. 2005, 22, 16–25. [Google Scholar] [CrossRef]
- Yang, C.; Dou, R.; Wei, C.; Liu, K.; Shi, D.; Zhang, C.; Liu, Q.; Wang, S.; Xiong, B. Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol. Ther. 2021, 29, 2088–2107. [Google Scholar] [CrossRef]
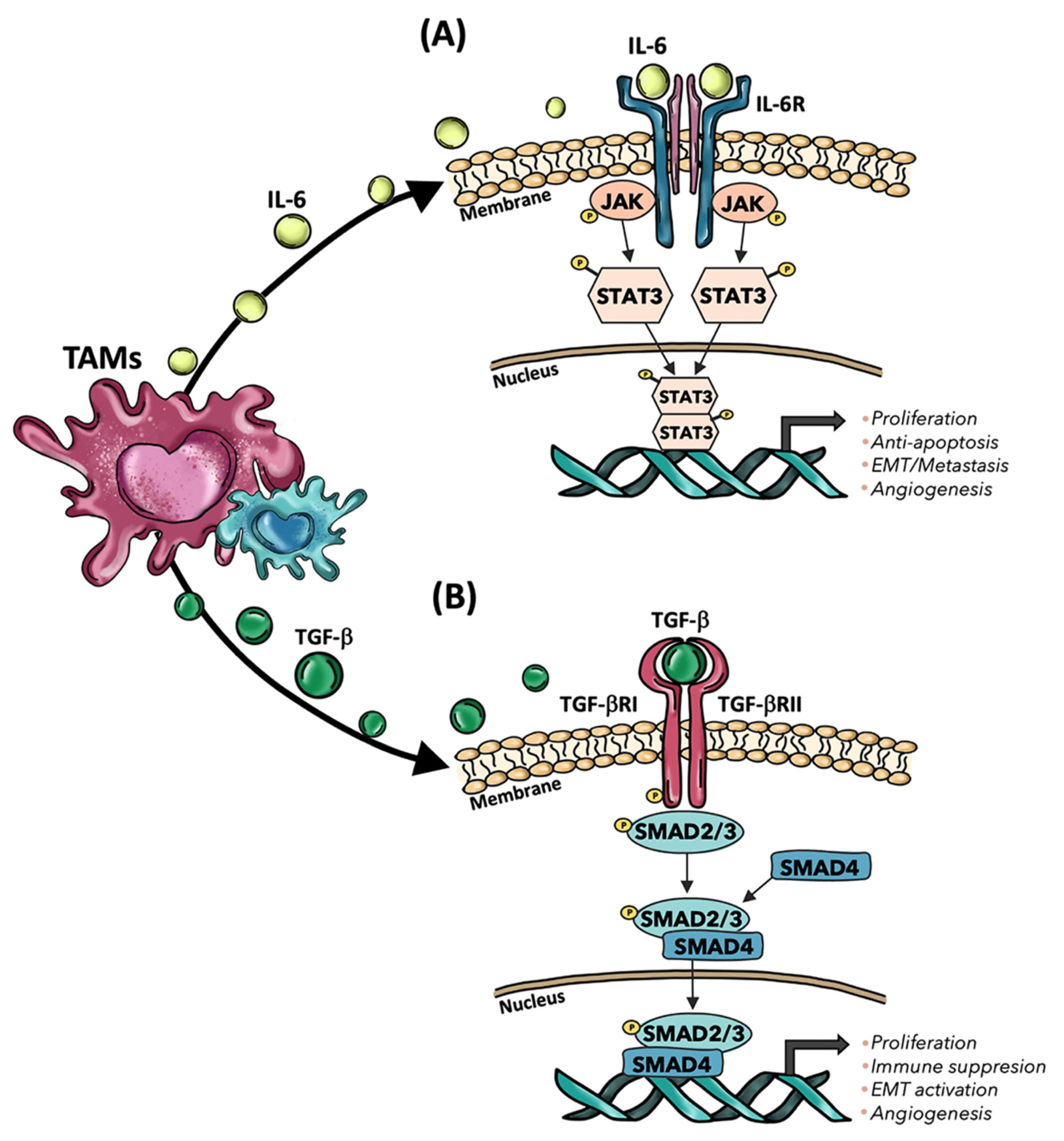
- Yin, Y.; Yao, S.; Hu, Y.; Feng, Y.; Li, M.; Bian, Z.; Zhang, J.; Qin, Y.; Qi, X.; Zhou, L.; et al. The Immune-microenvironment Confers Chemoresistance of Colorectal Cancer through Macrophage-Derived IL6. Clin. Cancer Res. 2017, 23, 7375–7387. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.; Pappan, L.; Galliher-Beckley, A.; Shi, J. IL-1β promotes stemness and invasiveness of colon cancer cells through Zeb1 activation. Mol. Cancer 2012, 11, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Wang, H.-S.; Zhou, B.-H.; Li, C.-L.; Zhang, F.; Wang, X.-F.; Zhang, G.; Bu, X.-Z.; Cai, S.-H.; Du, J. Epithelial–Mesenchymal Transition (EMT) Induced by TNF-α Requires AKT/GSK-3β-Mediated Stabilization of Snail in Colorectal Cancer. PLoS ONE 2013, 8, e56664. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Wang, S.; Sun, M.; Zhang, C.; Wei, C.; Yang, C.; Dou, R.; Liu, Q.; Xiong, B. miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J. Hematol. Oncol. 2019, 12, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Liu, L.; Lai, W.; Zeng, Y.; Xu, H.; Lan, Q.; Su, P.; Chu, Z. Interaction with tumor-associated macrophages promotes PRL-3-induced invasion of colorectal cancer cells via MAPK pathway-induced EMT and NF-κB signaling-induced angiogenesis. Oncol. Rep. 2019, 41, 2790–2802. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Zeng, W.; Zhang, D.; Wang, L.; Deng, X.; Lai, J.; Li, J.; Gong, J.; Xiang, G. SNAIL Induces EMT and Lung Metastasis of Tumours Secreting CXCL2 to Promote the Invasion of M2-Type Immunosuppressed Macrophages in Colorectal Cancer. Int. J. Biol. Sci. 2022, 18, 2867–2881. [Google Scholar] [CrossRef]
- Li, S.; Xu, F.; Zhang, J.; Wang, L.; Zheng, Y.; Wu, X.; Wang, J.; Huang, Q.; Lai, M. Tumor-associated macrophages remodeling EMT and predicting survival in colorectal carcinoma. OncoImmunology 2018, 7, e1380765. [Google Scholar] [CrossRef] [Green Version]
- Cai, J.; Xia, L.; Li, J.; Ni, S.; Song, H.; Wu, X. Tumor-Associated Macrophages Derived TGF-β–Induced Epithelial to Mesenchymal Transition in Colorectal Cancer Cells through Smad2,3-4/Snail Signaling Pathway. Cancer Res. Treat. 2019, 51, 252–266. [Google Scholar] [CrossRef]
- Zhang, X.-L.; Hu, L.-P.; Yang, Q.; Qin, W.-T.; Wang, X.; Xu, C.-J.; Tian, G.-A.; Yang, X.-M.; Yao, L.-L.; Zhu, L.; et al. CTHRC1 promotes liver metastasis by reshaping infiltrated macrophages through physical interactions with TGF-β receptors in colorectal cancer. Oncogene 2021, 40, 3959–3973. [Google Scholar] [CrossRef]
- Ni, S.; Ren, F.; Xu, M.; Tan, C.; Weng, W.; Huang, Z.; Sheng, W.; Huang, D. CTHRC1 overexpression predicts poor survival and enhances epithelial-mesenchymal transition in colorectal cancer. Cancer Med. 2018, 7, 5643–5654. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Li, X.; Zhu, L.; Ming, S.; Wang, H.; Xie, J.; Ren, L.; Huang, J.; Liang, D.; Xiong, L.; et al. CD163+ macrophages suppress T cell response by producing TGF-β in pediatric colorectal polyps. Int. Immunopharmacol. 2021, 96, 107644. [Google Scholar] [CrossRef]
- Ma, X.; Gao, Y.; Chen, Y.; Liu, J.; Yang, C.; Bao, C.; Wang, Y.; Feng, Y.; Song, X.; Qiao, S. M2-Type Macrophages Induce Tregs Generation by Activating the TGF-β/Smad Signalling Pathway to Promote Colorectal Cancer Development. OncoTargets Ther. 2021, 14, 5391–5402. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Qiu, X.; Li, J.; Zheng, S.; Li, L.; Zhao, H. TGF-β secreted by tumor-associated macrophages promotes proliferation and invasion of colorectal cancer via miR-34a-VEGF axis. Cell Cycle 2018, 17, 2766–2778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gulubova, M.; Ananiev, J.; Yovchev, Y.; Julianov, A.; Karashmalakov, A.; Vlaykova, T. The density of macrophages in colorectal cancer is inversely correlated to TGF-β1 expression and patients’ survival. J. Mol. Histol. 2013, 44, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Teuwen, L.-A.; De Rooij, L.P.M.H.; Cuypers, A.; Rohlenova, K.; Dumas, S.J.; García-Caballero, M.; Meta, E.; Amersfoort, J.; Taverna, F.; Becker, L.M.; et al. Tumor vessel co-option probed by single-cell analysis. Cell Rep. 2021, 35, 109253. [Google Scholar] [CrossRef]
- Peng, D.; Fu, M.; Wang, M.; Wei, Y.; Wei, X. Targeting TGF-β signal transduction for fibrosis and cancer therapy. Mol. Cancer 2022, 21, 104. [Google Scholar] [CrossRef]
- Dai, G.; Sun, B.; Gong, T.; Pan, Z.; Meng, Q.; Ju, W. Ginsenoside Rb2 inhibits epithelial-mesenchymal transition of colorectal cancer cells by suppressing TGF-β/Smad signaling. Phytomedicine 2019, 56, 126–135. [Google Scholar] [CrossRef]
- Villalba, M.; Evans, S.R.; Vidal-Vanaclocha, F.; Calvo, A. Role of TGF-β in metastatic colon cancer: It is finally time for targeted therapy. Cell Tissue Res. 2017, 370, 29–39. [Google Scholar] [CrossRef]
- House, M.G.; Ito, H.; Gönen, M.; Fong, Y.; Allen, P.J.; DeMatteo, R.P.; Brennan, M.F.; Blumgart, L.H.; Jarnagin, W.R.; D’Angelica, M.I. Survival after Hepatic Resection for Metastatic Colorectal Cancer: Trends in Outcomes for 1600 Patients during Two Decades at a Single Institution. J. Am. Coll. Surg. 2010, 210, 744–752. [Google Scholar] [CrossRef]
- Hughes, K.; Simon, R.; Songhorabodi, S.; Adson, M.; Ilstrup, D. Resection of the liver for colorectal carcinoma metastases: A multi-institutional study of patterns of recurrence. Surgery 1986, 31, 278–284. [Google Scholar] [CrossRef] [Green Version]
- Hołówko, W.; Grąt, M.; Hinderer, B.; Orlińska, I.; Krawczyk, M. Prediction of Survival in Patients with Unresectable Colorectal Liver Metastases. Pol. J. Surg. 2014, 86, 319–324. [Google Scholar] [CrossRef]
- Wagner, J.; Adson, M.; Van Heerden, J.; Adson, M.; Ilstrup, D. The natural history of hepatic metastases from colorectal cancer. A comparison with resective treatment. Ann. Surg. 1984, 199, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Kopetz, S.; Chang, G.J.; Overman, M.J.; Eng, C.; Sargent, D.J.; Larson, D.W.; Grothey, A.; Vauthey, J.-N.; Nagorney, D.M.; McWilliams, R.R. Improved Survival in Metastatic Colorectal Cancer Is Associated with Adoption of Hepatic Resection and Improved Chemotherapy. J. Clin. Oncol. 2009, 27, 3677–3683. [Google Scholar] [CrossRef] [PubMed]
- Donadon, M.; Cortese, N.; Marchesi, F.; Cimino, M.; Mantovani, A.; Torzilli, G. Hepatobiliary surgeons meet immunologists: The case of colorectal liver metastases patients. HepatoBiliary Surg. Nutr. 2019, 8, 370–377. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Cheng, X.; Zhou, C.; Tang, Y.; Li, F.; Zhang, B.; Huang, T.; Wang, J.; Tu, S. Fruquintinib Enhances the Antitumor Immune Responses of Anti-Programmed Death Receptor-1 in Colorectal Cancer. Front. Oncol. 2022, 12, 841977. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, K.M.; Jiang, X.; Guha, P.; Lausted, C.; Carter, J.A.; Hsu, C.; Labadie, K.P.; Kohli, K.; Kenerson, H.L.; Daniel, S.K.; et al. Blockade of interleukin 10 potentiates antitumour immune function in human colorectal cancer liver metastases. Gut 2022, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, L.; Peng, X.; Zhan, X. Progress of tumor-associated macrophages in the epithelial-mesenchymal transition of tumor. Front. Oncol. 2022, 12, 911410. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, X.; Hao, H. Macrophage phenotype-switching in cancer. Eur. J. Pharmacol. 2022, 931, 175229. [Google Scholar] [CrossRef]
- Xu, W.; Cheng, Y.; Guo, Y.; Yao, W.; Qian, H. Targeting tumor associated macrophages in hepatocellular carcinoma. Biochem. Pharmacol. 2022, 199, 114990. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, Y.; Dai, H.; Han, B. Epithelial–Mesenchymal Transition-Mediated Tumor Therapeutic Resistance. Molecules 2022, 27, 4750. [Google Scholar] [CrossRef]
- Maslankova, J.; Vecurkovska, I.; Rabajdova, M.; Katuchova, J.; Kicka, M.; Gayova, M.; Katuch, V. Regulation of transforming growth factor-β signaling as a therapeutic approach to treating colorectal cancer. World J. Gastroenterol. 2022, 28, 4744–4761. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, Y.Y.; Chen, Y.; Wang, J.; Wang, Q.; Lu, H. TGF-β Signaling and Resistance to Cancer Therapy. Front. Cell Dev. Biol. 2021, 9, 786728. [Google Scholar] [CrossRef] [PubMed]
- Sui, H.; Zhao, J.; Zhou, L.; Wen, H.; Deng, W.; Li, C.; Ji, Q.; Liu, X.; Feng, Y.; Chai, N.; et al. Tanshinone IIA inhibits β-catenin/VEGF-mediated angiogenesis by targeting TGF-β1 in normoxic and HIF-1α in hypoxic microenvironments in human colorectal cancer. Cancer Lett. 2017, 403, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Gunderson, A.J.; Gilchrist, M.; Whiteford, M.; Kiely, M.X.; Hayman, A.; O’Brien, D.; Ahmad, R.; Manchio, J.V.; Fox, N.; et al. Galunisertib plus neoadjuvant chemoradiotherapy in patients with locally advanced rectal cancer: A single-arm, phase 2 trial. Lancet Oncol. 2022, 23, 1189–1200. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Liu, H.; He, P.; An, D.; Guo, X.; Zhang, X.; Feng, M. Inhibition of PCSK9 enhances the antitumor effect of PD-1 inhibitor in colorectal cancer by promoting the infiltration of CD8+ T cells and the exclusion of Treg cells. Front. Immunol. 2022, 13, 947756. [Google Scholar] [CrossRef]
- Chen, T.-W.; Hung, W.-Z.; Chiang, S.-F.; Chen, W.T.-L.; Ke, T.-W.; Liang, J.-A.; Huang, C.-Y.; Yang, P.-C.; Huang, K.C.-Y.; Chao, K.S.C. Dual inhibition of TGFβ signaling and CSF1/CSF1R reprograms tumor-infiltrating macrophages and improves response to chemotherapy via suppressing PD-L1. Cancer Lett. 2022, 543, 215795. [Google Scholar] [CrossRef] [PubMed]

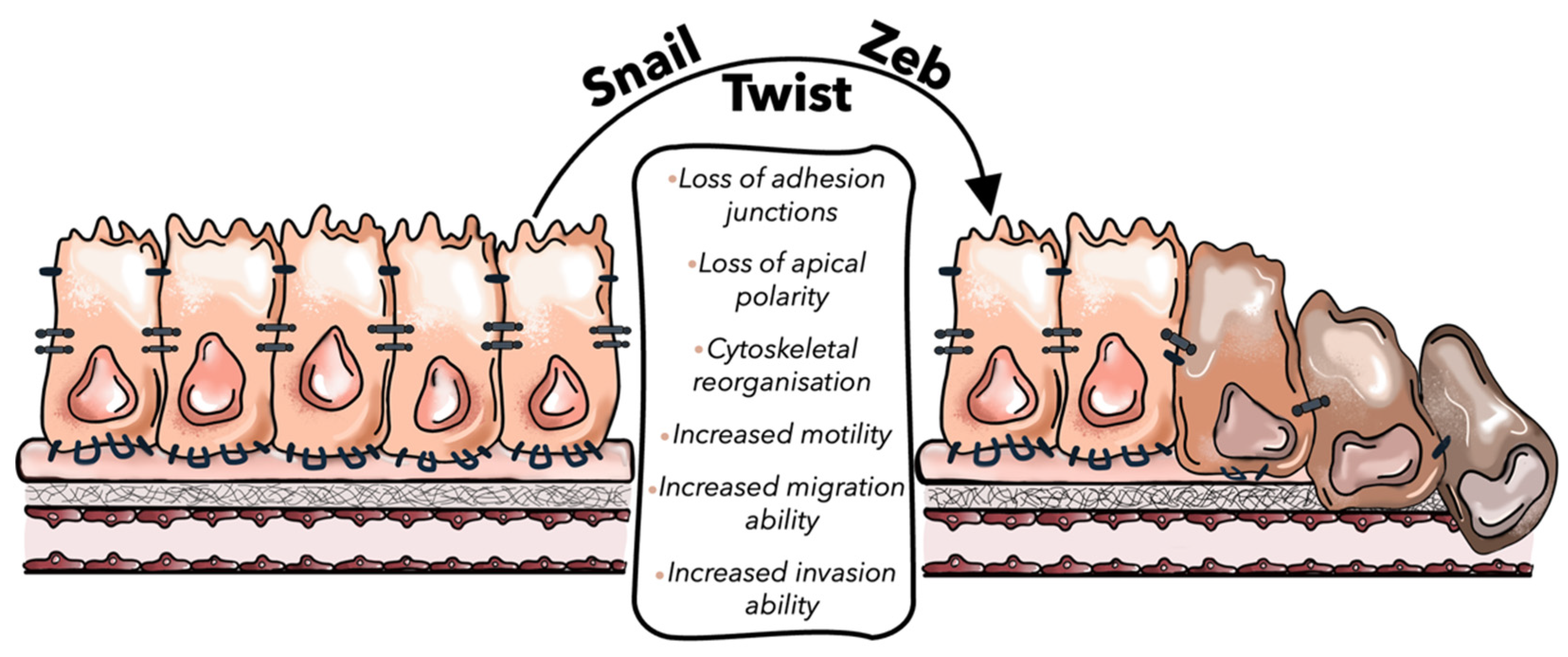
| EMT-TFs | Members of TF Family | Mechanisms in CLM-Related EMT | References |
|---|---|---|---|
| Snail | Snail1, Snail2 (SLUG genes) |
| [60,61] |
| Zeb | Zeb1, Zeb2 |
| [67,68,69] |
| Twist | Twist1, Twist2 |
| [70,71,72] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gazzillo, A.; Polidoro, M.A.; Soldani, C.; Franceschini, B.; Lleo, A.; Donadon, M. Relationship between Epithelial-to-Mesenchymal Transition and Tumor-Associated Macrophages in Colorectal Liver Metastases. Int. J. Mol. Sci. 2022, 23, 16197. https://doi.org/10.3390/ijms232416197
Gazzillo A, Polidoro MA, Soldani C, Franceschini B, Lleo A, Donadon M. Relationship between Epithelial-to-Mesenchymal Transition and Tumor-Associated Macrophages in Colorectal Liver Metastases. International Journal of Molecular Sciences. 2022; 23(24):16197. https://doi.org/10.3390/ijms232416197
Chicago/Turabian StyleGazzillo, Aurora, Michela Anna Polidoro, Cristiana Soldani, Barbara Franceschini, Ana Lleo, and Matteo Donadon. 2022. "Relationship between Epithelial-to-Mesenchymal Transition and Tumor-Associated Macrophages in Colorectal Liver Metastases" International Journal of Molecular Sciences 23, no. 24: 16197. https://doi.org/10.3390/ijms232416197







