Cardiac Sarcomere Signaling in Health and Disease
Abstract
1. Introduction
1.1. The Sarcomere as the Functional Unit of the Muscle Cell
1.2. Cardiac Muscle Contraction
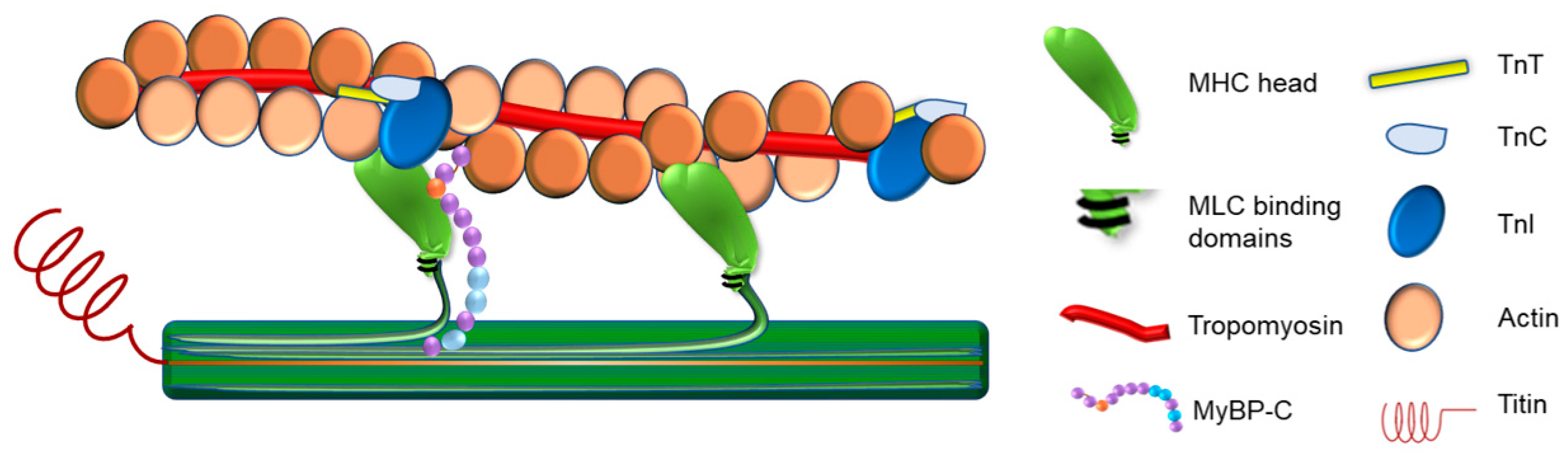
2. Regulation of Sarcomere Contraction
2.1. Thin Filament Regulation of Sarcomere Contraction
2.1.1. TnC-Calcium
2.1.2. TnC-TnI
2.1.3. TnT-Tm
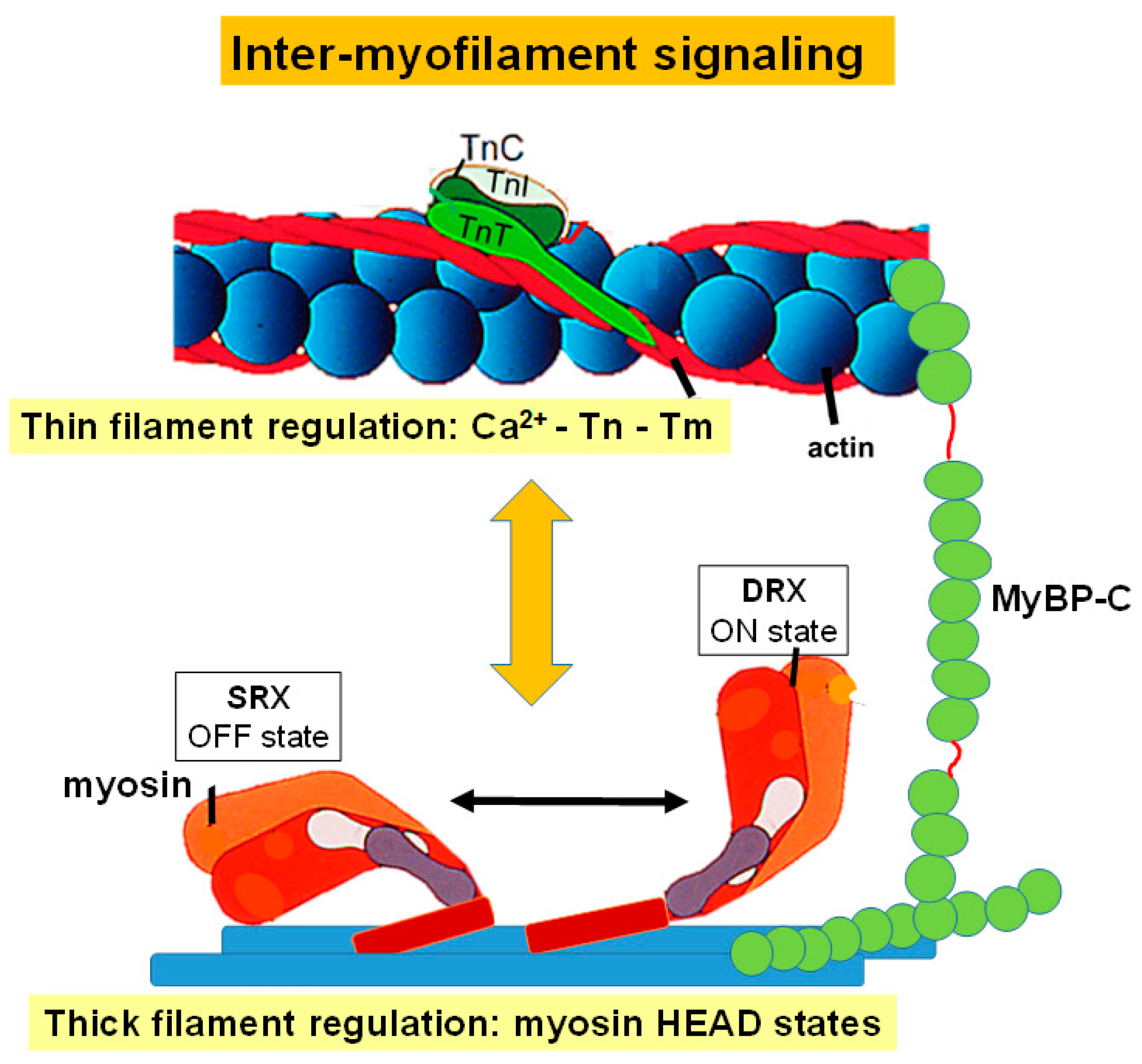
2.2. Thick Filament Regulation of Sarcomere Contraction
2.3. Inter-Myofilament Signaling in the Sarcomere
2.3.1. Myosin-Binding Protein C
2.3.2. Titin
2.4. Post-Translational Modifications in Sarcomere Signaling
2.4.1. Troponin
2.4.2. MyBP-C
2.4.3. Titin
3. Biosensing in the Sarcomere
3.1. Cardiac TnI A164H Histidine Button
3.2. Sarcometer
4. Mechanosensing and Sarcomere Activation
5. Sarcomere Signaling in Disease and Current Therapeutics
5.1. Sarcomeric Cardiomyopathies
5.1.1. Hypertrophic Cardiomyopathy
5.1.2. Restrictive Cardiomyopathy
5.1.3. Dilated Cardiomyopathy
5.2. The Sarcomere as a Therapeutic Target
5.2.1. Mavacamten and Aficamten
5.2.2. Omecamtiv Mercarbil and Danicamtiv
6. Summary and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gordon, A.M.; Homsher, E.; Regnier, M. Regulation of Contraction in Striated Muscle. Physiol. Rev. 2000, 80, 853–924. [Google Scholar] [CrossRef] [PubMed]
- Tobacman, L.S. Thin Filament-Mediated Regulation of Cardiac Contraction. Annu Rev. Physiol. 1996, 58, 447–481. [Google Scholar] [CrossRef] [PubMed]
- Granzier, H.; Labeit, S. Structure-function relations of the giant elastic protein titin in striated and smooth muscle cells. Muscle Nerve 2007, 36, 740–755. [Google Scholar] [CrossRef] [PubMed]
- Heling, L.W.H.J.; Geeves, M.A.; Kad, N.M. MyBP-C: One protein to govern them all. J. Muscle Res. Cell Motil. 2020, 41, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.; Westfall, M.V.; Townsend, D.; Blankinship, M.; Herron, T.J.; Guerrero-Serna, G.; Wang, W.; Devaney, E.; Metzger, J.M. Designing heart performance by gene transfer. Physiol. Rev. 2008, 88, 1567–1651. [Google Scholar] [CrossRef] [PubMed]
- Metzger, J.M.; Westfall, M.V. Covalent and Noncovalent Modification of Thin Filament Action: The Essential Role of Troponin in Cardiac Muscle Regulation. Circ. Res. 2004, 94, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Seidman, C. Genetic causes of inherited cardiac hypertrophy: Robert L. Frye lecture. Mayo Clin. Proc. 2002, 77, 1315–1319. [Google Scholar] [CrossRef]
- Bers, D.M. Cardiac excitation-contraction coupling. Nature 2002, 415, 198–205. [Google Scholar] [CrossRef]
- Eisner, D.A.; Caldwell, J.L.; Kistamás, K.; Trafford, A.W. Calcium and Excitation-Contraction Coupling in the Heart. Circ. Res. 2017, 121, 181–195. [Google Scholar] [CrossRef]
- Badr, M.A.; Pinto, J.R.; Davidson, M.W.; Chase, P.B. Fluorescent Protein-Based Ca2+ Sensor Reveals Global, Divalent Cation-Dependent Conformational Changes in Cardiac Troponin C. PLoS ONE 2016, 11, e0164222. [Google Scholar] [CrossRef]
- Gordon, A.M.; Regnier, M.; Homsher, E. Skeletal and cardiac muscle contractile activation: Tropomyosin “rocks and rolls”. News Physiol. Sci. 2001, 16, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Risi, C.M.; Pepper, I.; Belknap, B.; Landim-Vieira, M.; White, H.D.; Dryden, K.; Pinto, J.R.; Chase, P.B.; Galkin, V.E. The structure of the native cardiac thin filament at systolic Ca2+ levels. Proc. Natl. Acad. Sci. USA 2021, 118, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Brittsan, A.G.; Kranias, E.G. Phospholamban and cardiac contractile function. J. Mol. Cell. Cardiol. 2000, 32, 2131–2139. [Google Scholar] [CrossRef] [PubMed]
- MacLennan, D.H.; Abu-Abed, M.; Kang, C.H. Structure-function relationships in Ca2+ cycling proteins. J. Mol. Cell. Cardiol. 2002, 34, 897–918. [Google Scholar] [CrossRef] [PubMed]
- Kranias, E.G.; Hajjar, R.J. Modulation of cardiac contractility by the phopholamban/SERCA2a regulatome. Circ. Res. 2012, 110, 1646–1660. [Google Scholar] [CrossRef]
- MacLennan, D.H.; Kranias, E.G. Phospholamban: A crucial regulator of cardiac contractility. Nat. Rev. Mol. Cell Biol. 2003, 4, 566–577. [Google Scholar] [CrossRef]
- Farah, C.S.; Reinach, F.C. The troponin complex and regulation of muscle contraction. FASEB J. 1995, 9, 755–767. [Google Scholar] [CrossRef]
- Szikora, S.; Görög, P.; Mihály, J. The Mechanisms of Thin Filament Assembly and Length Regulation in Muscles. Int. J. Mol. Sci. 2022, 23, 5306. [Google Scholar] [CrossRef]
- Yamada, Y.; Namba, K.; Fujii, T. Cardiac muscle thin filament structures reveal calcium regulatory mechanism. Nat. Commun. 2020, 11, 153. [Google Scholar] [CrossRef]
- Yin, Z.; Ren, J.; Guo, W. Sarcomeric protein isoform transitions in cardiac muscle: A journey to heart failure. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2015, 1852, 47–52. [Google Scholar] [CrossRef]
- Herzberg, O.; James, M.N.G. Structure of the calcium regulatory muscle protein troponin-C at 2.8 Å resolution. Nature 1985, 313, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Satyshur, K.A.; Rao, S.T.; Pyzalska, D.; Drendel, W.; Greaser, M.; Sundaralingam, M. Refined structure of chicken skeletal muscle troponin C in the two-calcium state at 2-A resolution. J. Biol. Chem. 1988, 263, 1628–1647. [Google Scholar] [CrossRef] [PubMed]
- Tobacman, L.S. Troponin Revealed: Uncovering the Structure of the Thin Filament On-Off Switch in Striated Muscle. Biophys. J. 2021, 120, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.; Feng, H.Z.; Jin, J.P. The evolutionarily conserved C-terminal peptide of troponin I is an independently configured regulatory structure to function as a myofilament Ca2+-desensitizer. J. Mol. Cell. Cardiol. 2019, 136, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Elliott, K.; Watkins, H.; Redwood, C.S. Altered regulatory properties of human cardiac troponin I mutants that cause hypertrophic cardiomyopathy. J. Biol. Chem. 2000, 275, 22069–22074. [Google Scholar] [CrossRef]
- Gomes, A.V.; Venkatraman, G.; Potter, J.D. The miscommunicative cardiac cell: When good proteins go bad. Ann. N. Y. Acad. Sci. 2005, 1047, 30–37. [Google Scholar] [CrossRef]
- Westfall, M.V.; Albayya, F.P.; Turner, I.I.; Metzger, J.M. Chimera Analysis of Troponin I Domains That Influence Ca2+-Activated Myofilament Tension in Adult Cardiac Myocytes. Circ. Res. 2000, 86, 470–477. [Google Scholar] [CrossRef]
- Anderson, P.A.; Malouf, N.N.; Oakeley, A.E.; Pagani, E.D.; Allen, P.D. Troponin T isoform expression in humans. A comparison among normal and failing adult heart, fetal heart, and adult and fetal skeletal muscle. Circ. Res. 1991, 69, 1226–1233. [Google Scholar] [CrossRef]
- Kobayashi, T.; Solaro, R.J. Calcium, thin filaments, and the integrative biology of cardiac contractility. Annu. Rev. Physiol. 2005, 67, 39–67. [Google Scholar] [CrossRef]
- Butters, C.A.; Willadsen, K.A.; Tobacman, L.S. Cooperative interactions between adjacent troponin-tropomyosin complexes may be transmitted through the actin filament. J. Biol. Chem. 1993, 268, 15565–15570. [Google Scholar] [CrossRef]
- Cooper, J.A. Actin dynamics: Tropomyosin provides stability. Curr. Biol. 2002, 12, R523–R525. [Google Scholar] [CrossRef] [PubMed]
- Orzechowski, M.; Li, X.E.; Fischer, S.; Lehman, W. An atomic model of the tropomyosin cable on F-actin. Biophys. J. 2014, 107, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Tobacman, L.S.; Butters, C.A. A new model of cooperative myosin-thin filament binding. J. Biol. Chem. 2000, 275, 27587–27593. [Google Scholar] [CrossRef] [PubMed]
- Huxley, A.F.; Niedergerke, R. Structural changes in muscle during contraction: Interference microscopy of living muscle fibres. Nature 1954, 173, 971–973. [Google Scholar] [CrossRef] [PubMed]
- Huxley, H.; Hanson, J. Changes in the Cross-Striations of Muscle during Contraction and Stretch and their Structural Interpretation. Nature 1954, 173, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Hooijman, P.; Stewart, M.A.; Cooke, R. A new state of cardiac myosin with very slow ATP turnover: A potential cardioprotective mechanism in the heart. Biophys. J. 2011, 100, 1969–1976. [Google Scholar] [CrossRef]
- McNamara, J.W.; Li, A.; Dos Remedios, C.G.; Cooke, R. The role of super-relaxed myosin in skeletal and cardiac muscle. Biophys. Rev. 2015, 7, 5–14. [Google Scholar] [CrossRef]
- Stewart, M.A.; Franks-Skiba, K.; Chen, S.; Cooke, R. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc. Natl. Acad. Sci. USA 2010, 107, 430–435. [Google Scholar] [CrossRef]
- Harrington, W.F.; Rodgers, M.E. Myosin. Annu. Rev. Biochem. 1984, 53, 35–73. [Google Scholar] [CrossRef]
- Warrick, H.M.; Spudich, J.A. Myosin structure and function in cell motility. Annu. Rev. Cell Biol. 1987, 3, 379–421. [Google Scholar] [CrossRef]
- England, J.; Loughna, S. Heavy and light roles: Myosin in the morphogenesis of the heart. Cell. Mol. Life Sci. 2013, 70, 1221–1239. [Google Scholar] [CrossRef] [PubMed]
- Mornet, D.; Pantel, P.; Audemard, E.; Kassab, R. The limited tryptic cleavage of chymotryptic S-1: An approach to the characterization of the actin site in myosin heads. Biochem. Biophys. Res. Commun. 1979, 89, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Rayment, I.; Rypniewski, W.R.; Schmidt-Bäse, K.; Smith, R.; Tomchick, D.R.; Benning, M.M.; Winkelmann, D.A.; Wesenberg, G.; Holden, H.M. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science 1993, 261, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Harrington, W.F. On the origin of the contractile force in skeletal muscle. Proc. Natl. Acad. Sci. USA 1979, 76, 5066–5070. [Google Scholar] [CrossRef] [PubMed]
- Irving, M. Regulation of Contraction by the Thick Filaments in Skeletal Muscle. Biophys. J. 2017, 113, 2579–2594. [Google Scholar] [CrossRef]
- Naber, N.; Cooke, R.; Pate, E. Slow myosin ATP turnover in the super-relaxed state in tarantula muscle. J. Mol. Biol. 2011, 411, 943–950. [Google Scholar] [CrossRef]
- Scruggs, S.B.; Hinken, A.C.; Thawornkaiwong, A.; Robbins, J.; Walker, L.A.; de Tombe, P.P.; Geenen, D.L.; Buttrick, P.M.; Solaro, R.J. Ablation of ventricular myosin regulatory light chain phosphorylation in mice causes cardiac dysfunction in situ and affects neighboring myofilament protein phosphorylation. J. Biol. Chem. 2009, 284, 5097–5106. [Google Scholar] [CrossRef]
- Trivedi, D.V.; Adhikari, A.S.; Sarkar, S.S.; Ruppel, K.M.; Spudich, J.A. Hypertrophic cardiomyopathy and the myosin mesa: Viewing an old disease in a new light. Biophys. Rev. 2018, 10, 27–48. [Google Scholar] [CrossRef]
- Metzger, J.M.; Greaser, M.L.; Moss, R.L. Variations in cross-bridge attachment rate and tension with phosphorylation of myosin in mammalian skinned skeletal muscle fibers. Implications for twitch potentiation in intact muscle. J. Gen. Physiol. 1989, 93, 855–883. [Google Scholar] [CrossRef]
- Metzger, J.M.; Moss, R.L. Myosin light chain 2 modulates calcium-sensitive cross-bridge transitions in vertebrate skeletal muscle. Biophys. J. 1992, 63, 460–468. [Google Scholar] [CrossRef]
- Szczesna, D. Regulatory light chains of striated muscle myosin. Structure, function and malfunction. Curr. Drug Targets Cardiovasc. Haematol. Disord. 2003, 3, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Ranke, J. Tetanus: Eine Physiologische Studie. Grundwerk; Engelmann: Leipzig, Germany, 1865; Volume 1. [Google Scholar]
- Perrie, W.T.; Smillie, L.B.; Perry, S.V. A phosphorylated light-chain component of myosin. Biochem. J. 1972, 128, 105P–106P. [Google Scholar] [CrossRef] [PubMed]
- Zhi, G.; Ryder, J.W.; Huang, J.; Ding, P.; Chen, Y.; Zhao, Y.; Kamm, K.E.; Stull, J.T. Myosin light chain kinase and myosin phosphorylation effect frequency-dependent potentiation of skeletal muscle contraction. Proc. Natl. Acad. Sci. USA 2005, 102, 17519–17524. [Google Scholar] [CrossRef]
- Stull, J.T.; Kamm, K.E.; Vandenboom, R. Myosin light chain kinase and the role of myosin light chain phosphorylation in skeletal muscle. Arch. Biochem. Biophys. 2011, 510, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.L.; Stull, J.T. Myosin light chain phosphorylation in fast and slow skeletal muscles in situ. Am. J. Physiol. 1984, 247, C462–C471. [Google Scholar] [CrossRef]
- Ryder, J.W.; Lau, K.S.; Kamm, K.E.; Stull, J.T. Enhanced skeletal muscle contraction with myosin light chain phosphorylation by a calmodulin-sensing kinase. J. Biol. Chem. 2007, 282, 20447–20454. [Google Scholar] [CrossRef]
- Kampourakis, T.; Sun, Y.B.; Irving, M. Myosin light chain phosphorylation enhances contraction of heart muscle via structural changes in both thick and thin filaments. Proc. Natl. Acad. Sci. USA 2016, 113, E3039–E3047. [Google Scholar] [CrossRef]
- Persechini, A.; Stull, J.T.; Cooke, R. The effect of myosin phosphorylation on the contractile properties of skinned rabbit skeletal muscle fibers. J. Biol. Chem. 1985, 260, 7951–7954. [Google Scholar] [CrossRef]
- Herring, B.P.; England, P.J. The turnover of phosphate bound to myosin light chain-2 in perfused rat heart. Biochem. J. 1986, 240, 205–214. [Google Scholar] [CrossRef]
- Manning, D.R.; Stull, J.T. Myosin light chain phosphorylation-dephosphorylation in mammalian skeletal muscle. Am. J. Physiol. 1982, 242, C234–C241. [Google Scholar] [CrossRef]
- Chang, A.N.; Mahajan, P.; Knapp, S.; Barton, H.; Sweeney, H.L.; Kamm, K.E.; Stull, J.T. Cardiac myosin light chain is phosphorylated by Ca2+/calmodulin-dependent and -independent kinase activities. Proc. Natl. Acad. Sci. USA 2016, 113, E3824–E3833. [Google Scholar] [CrossRef] [PubMed]
- Alamo, L.; Pinto, A.; Sulbaran, G.; Mavarez, J.; Padron, R. Lessons from a tarantula: New insights into myosin interacting-heads motif evolution and its implications on disease. Biophys. Rev. 2018, 10, 1465–1477. [Google Scholar] [CrossRef] [PubMed]
- Alamo, L.; Qi, D.; Wriggers, W.; Pinto, A.; Zhu, J.; Bilbao, A.; Gillilan, R.E.; Hu, S.; Padron, R. Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis. J. Mol. Biol. 2016, 428, 1142–1164. [Google Scholar] [CrossRef] [PubMed]
- Kampourakis, T.; Irving, M. Phosphorylation of myosin regulatory light chain controls myosin head conformation in cardiac muscle. J. Mol. Cell. Cardiol. 2015, 85, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Van der Velden, J.; Papp, Z.; Boontje, N.M.; Zaremba, R.; de Jong, J.W.; Janssen, P.M.; Hasenfuss, G.; Stienen, G.J. The effect of myosin light chain 2 dephosphorylation on Ca2+ -sensitivity of force is enhanced in failing human hearts. Cardiovasc. Res. 2003, 57, 505–514. [Google Scholar] [CrossRef]
- Van der Velden, J.; Papp, Z.; Zaremba, R.; Boontje, N.M.; de Jong, J.W.; Owen, V.J.; Burton, P.B.; Goldmann, P.; Jaquet, K.; Stienen, G.J. Increased Ca2+-sensitivity of the contractile apparatus in end-stage human heart failure results from altered phosphorylation of contractile proteins. Cardiovasc. Res. 2003, 57, 37–47. [Google Scholar] [CrossRef]
- Abraham, T.P.; Jones, M.; Kazmierczak, K.; Liang, H.Y.; Pinheiro, A.C.; Wagg, C.S.; Lopaschuk, G.D.; Szczesna-Cordary, D. Diastolic dysfunction in familial hypertrophic cardiomyopathy transgenic model mice. Cardiovasc. Res. 2009, 82, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Kerrick, W.G.; Kazmierczak, K.; Xu, Y.; Wang, Y.; Szczesna-Cordary, D. Malignant familial hypertrophic cardiomyopathy D166V mutation in the ventricular myosin regulatory light chain causes profound effects in skinned and intact papillary muscle fibers from transgenic mice. FASEB J. 2009, 23, 855–865. [Google Scholar] [CrossRef]
- Sheikh, F.; Ouyang, K.; Campbell, S.G.; Lyon, R.C.; Chuang, J.; Fitzsimons, D.; Tangney, J.; Hidalgo, C.G.; Chung, C.S.; Cheng, H.; et al. Mouse and computational models link Mlc2v dephosphorylation to altered myosin kinetics in early cardiac disease. J. Clin. Investig. 2012, 122, 1209–1221. [Google Scholar] [CrossRef]
- Yuan, C.C.; Muthu, P.; Kazmierczak, K.; Liang, J.; Huang, W.; Irving, T.C.; Kanashiro-Takeuchi, R.M.; Hare, J.M.; Szczesna-Cordary, D. Constitutive phosphorylation of cardiac myosin regulatory light chain prevents development of hypertrophic cardiomyopathy in mice. Proc. Natl. Acad. Sci. USA 2015, 112, E4138–E4146. [Google Scholar] [CrossRef]
- Hartzell, H.C.; Sale, W.S. Structure of C protein purified from cardiac muscle. J. Cell Biol. 1985, 100, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Starr, R.; Offer, G. Polypeptide chains of intermediate molecular weight in myosin preparations. FEBS Lett. 1971, 15, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Sequeira, V.; Witjas-Paalberends, E.R.; Kuster, D.W.D.; Van Der Velden, J. Cardiac myosin-binding protein C: Hypertrophic cardiomyopathy mutations and structure-function relationships. Pflug. Arch. Eur. J. Physiol. 2014, 466, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Flashman, E.; Redwood, C.; Moolman-Smook, J.; Watkins, H. Cardiac myosin binding protein C: Its role in physiology and disease. Circ. Res. 2004, 94, 1279–1289. [Google Scholar] [CrossRef] [PubMed]
- Palmer, B.M.; Noguchi, T.; Wang, Y.; Heim, J.R.; Alpert, N.R.; Burgon, P.G.; Seidman, C.E.; Seidman, J.G.; Maughan, D.W.; LeWinter, M.M. Effect of cardiac myosin binding protein-C on mechanoenergetics in mouse myocardium. Circ. Res. 2004, 94, 1615–1622. [Google Scholar] [CrossRef]
- Jiang, J.; Burgon, P.G.; Wakimoto, H.; Onoue, K.; Gorham, J.M.; O’Meara, C.C.; Fomovsky, G.; McConnell, B.K.; Lee, R.T.; Seidman, J.G.; et al. Cardiac myosin binding protein C regulates postnatal myocyte cytokinesis. Proc. Natl. Acad. Sci. USA 2015, 112, 9046–9051. [Google Scholar] [CrossRef]
- Previs, M.J.; Michalek, A.J.; Warshaw, D.M. Molecular modulation of actomyosin function by cardiac myosin-binding protein C. Pflugers Arch.-Eur. J. Physiol. 2014, 466, 439–444. [Google Scholar] [CrossRef]
- Lin, B.L.; Song, T.; Sadayappan, S. Myofilaments: Movers and Rulers of the Sarcomere. Compr. Physiol. 2017, 2, 675–692. [Google Scholar]
- Nag, S.; Trivedi, D.V.; Sarkar, S.S.; Adhikari, A.S.; Sunitha, M.S.; Sutton, S.; Ruppel, K.M.; Spudich, J.A. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat. Struct. Mol. Biol. 2017, 24, 525–533. [Google Scholar] [CrossRef]
- Harris, S.P. Making waves: A proposed new role for myosin-binding protein C in regulating oscillatory contractions in vertebrate striated muscle. J. Gen. Physiol. 2020, 153, e202012729. [Google Scholar] [CrossRef]
- McNamara, J.W.; Singh, R.R.; Sadayappan, S. Cardiac myosin binding protein-C phosphorylation regulates the super-relaxed state of myosin. Proc. Natl. Acad. Sci. USA 2019, 116, 11731–11736. [Google Scholar] [CrossRef]
- Spudich, J.A. Three perspectives on the molecular basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Pflug. Arch. Eur. J. Physiol. 2019, 471, 701–717. [Google Scholar] [CrossRef]
- Colson, B.A.; Locher, M.R.; Bekyarova, T.; Patel, J.R.; Fitzsimons, D.P.; Irving, T.C.; Moss, R.L. Differential roles of regulatory light chain and myosin binding protein-C phosphorylations in the modulation of cardiac force development. J. Physiol. 2010, 588, 981–993. [Google Scholar] [CrossRef]
- Moos, C.; Mason, C.M.; Besterman, J.M.; Feng, I.N.M.; Dubin, J.H. The binding of skeletal muscle C-protein to F-actin, and its relation to the interaction of actin with myosin subfragment-1. J. Mol. Biol. 1978, 124, 571–586. [Google Scholar] [CrossRef]
- Mun, J.Y.; Previs, M.J.; Yu, H.Y.; Gulick, J.; Tobacman, L.S.; Previs, S.B.; Robbins, J.; Warshaw, D.M.; Craig, R. Myosin-binding protein C displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc. Natl. Acad. Sci. USA 2014, 111, 2170–2175. [Google Scholar] [CrossRef]
- Granzier, H.L.; Labeit, S. The giant protein titin: A major player in myocardial mechanics, signaling, and disease. Circ. Res. 2004, 94, 284–295. [Google Scholar] [CrossRef]
- Wu, Y.; Bell, S.P.; Trombitas, K.; Witt, C.C.; Labeit, S.; LeWinter, M.M.; Granzier, H. Changes in titin isoform expression in pacing-induced cardiac failure give rise to increased passive muscle stiffness. Circulation 2002, 106, 1384–1389. [Google Scholar] [CrossRef]
- Krüger, M.; Linke, W.A. The giant protein titin: A regulatory node that integrates myocyte signaling pathways. J. Biol. Chem. 2011, 286, 9905–9912. [Google Scholar] [CrossRef]
- Eckels, E.C.; Tapia-Rojo, R.; Rivas-Pardo, J.A.; Fernández, J.M. The Work of Titin Protein Folding as a Major Driver in Muscle Contraction. Annu. Rev. Physiol. 2018, 80, 327–351. [Google Scholar] [CrossRef]
- Herzog, W.; Duvall, M.; Leonard, T.R. Molecular mechanisms of muscle force regulation: A role for titin? Exerc. Sport Sci. Rev. 2012, 40, 50–57. [Google Scholar] [CrossRef]
- Herzog, W. The multiple roles of titin in muscle contraction and force production. Biophys. Rev. 2018, 10, 1187–1199. [Google Scholar] [CrossRef]
- Granzier, H.; Labeit, D.; Wu, Y.; Labeit, S. Titin as a modular spring: Emerging mechanisms for elasticity control by titin in cardiac physiology and pathophysiology. J. Muscle Res. Cell Motil. 2002, 23, 457–471. [Google Scholar] [CrossRef]
- Wang, K.; McClure, J.; Tu, A. Titin: Major myofibrillar components of striated muscle. Proc. Natl. Acad. Sci. USA 1979, 76, 3698–3702. [Google Scholar] [CrossRef]
- Linke, W.A. Titin Gene and Protein Functions in Passive and Active Muscle. Annu. Rev. Physiol. 2018, 80, 389–411. [Google Scholar] [CrossRef]
- Russell, B.; Solís, C. Mechanosignaling pathways alter muscle structure and function by post-translational modification of existing sarcomeric proteins to optimize energy usage. J. Muscle Res. Cell Motil. 2021, 42, 367–380. [Google Scholar] [CrossRef]
- Reinoso, T.R.; Landim-Vieira, M.; Shi, Y.; Johnston, J.R.; Chase, P.B.; Parvatiyar, M.S.; Landstrom, A.P.; Pinto, J.R.; Tadros, H.J. A comprehensive guide to genetic variants and post-translational modifications of cardiac troponin C. J. Muscle Res. Cell Motil. 2021, 42, 323–342. [Google Scholar] [CrossRef]
- Grabarek, Z.; Mabuchi, Y.; Gergely, J. Properties of Troponin C Acetylated at Lysine Residues. Biochemistry 1995, 34, 11872–11881. [Google Scholar] [CrossRef]
- Janssens, J.V.; Ma, B.; Brimble, M.A.; Van Eyk, J.E.; Delbridge, L.M.D.; Mellor, K.M. Cardiac troponins may be irreversibly modified by glycation: Novel potential mechanisms of cardiac performance modulation. Sci. Rep. 2018, 8, 16084. [Google Scholar] [CrossRef]
- Figueiredo-Freitas, C.; Dulce, R.A.; Foster, M.W.; Liang, J.; Yamashita, A.M.S.; Lima-Rosa, F.L.; Thompson, J.W.; Moseley, M.A.; Hare, J.M.; Nogueira, L.; et al. S-Nitrosylation of Sarcomeric Proteins Depresses Myofilament Ca2+ Sensitivity in Intact Cardiomyocytes. Antioxid. Redox Signal. 2015, 23, 1017–1034. [Google Scholar] [CrossRef]
- Irie, T.; Sips, P.Y.; Kai, S.; Kida, K.; Ikeda, K.; Hirai, S.; Moazzami, K.; Jiramongkolchai, P.; Bloch, D.B.; Doulias, P.T.; et al. S-nitrosylation of calcium-handling proteins in cardiac adrenergic signaling and hypertrophy. Circ. Res. 2015, 117, 793–803. [Google Scholar] [CrossRef]
- Burkart, E.M.; Sumandea, M.P.; Tomoyoshi, K.; Nili, M.; Martin, A.F.; Homsher, E.; Solaro, R.J. Phosphorylation or glutamic acid substitution at protein kinase C sites on cardiac troponin I differentially depress myofilament tension and shortening velocity. J. Biol. Chem. 2003, 278, 11265–11272. [Google Scholar] [CrossRef] [PubMed]
- Vahebi, S.; Kobayashi, T.; Warren, C.M.; De Tombe, P.P.; Solaro, R.J. Functional effects of Rho-kinase-dependent phosphorylation of specific sites on cardiac troponin. Circ. Res. 2005, 96, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Solaro, R.J. Multiplex kinase signaling modifies cardiac function at the level of sarcomeric proteins. J. Biol. Chem. 2008, 283, 26829–26833. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, S.I.; Coutu, P.; Sadayappan, S.; Robbins, J.; Metzger, J.M. Cardiac transgenic and gene transfer strategies converge to support an important role for troponin I in regulating relaxation in cardiac myocytes. Circ. Res. 2007, 101, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Main, A.; Fuller, W.; Baillie, G.S. Post-translational regulation of cardiac myosin binding protein-C: A graphical review. Cell. Signal. 2020, 76, 109788. [Google Scholar] [CrossRef] [PubMed]
- Sadayappan, S.; Gulick, J.; Osinska, H.; Martin, L.A.; Hahn, H.S.; Dorn, G.W.; Klevitsky, R.; Seidman, C.E.; Seidman, J.G.; Robbins, J. Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circ. Res. 2005, 97, 1156–1163. [Google Scholar] [CrossRef]
- Tong, C.W.; Stelzer, J.E.; Greaser, M.L.; Powers, P.A.; Moss, R.L. Acceleration of crossbridge kinetics by protein kinase a phosphorylation of cardiac myosin binding protein c modulates cardiac function. Circ. Res. 2008, 103, 974–982. [Google Scholar] [CrossRef]
- Stelzer, J.E.; Patel, J.R.; Walker, J.W.; Moss, R.L. Differential roles of cardiac myosin-binding protein C and cardiac troponin I in the myofibrillar force responses to protein kinase A phosphorylation. Circ. Res. 2007, 101, 503–511. [Google Scholar] [CrossRef]
- Aryal, B.; Jeong, J.; Rao, V.A. Doxorubicin-induced carbonylation and degradation of cardiac myosin binding protein C promote cardiotoxicity. Proc. Natl. Acad. Sci. USA 2014, 111, 2011–2016. [Google Scholar] [CrossRef]
- Stathopoulou, K.; Wittig, I.; Heidler, J.; Piasecki, A.; Richter, F.; Diering, S.; Der Van Velden, J.; Buck, F.; Donzelli, S.; Schröder, E.; et al. S-glutathiolation impairs phosphoregulation and function of cardiac myosin-binding protein C in human heart failure. FASEB J. 2016, 30, 1849–1864. [Google Scholar] [CrossRef]
- Govindan, S.; McElligott, A.; Muthusamy, S.; Nair, N.; Barefield, D.; Martin, J.L.; Gongora, E.; Greis, K.D.; Luther, P.K.; Winegrad, S.; et al. Cardiac myosin binding protein-C is a potential diagnostic biomarker for myocardial infarction. J. Mol. Cell. Cardiol. 2012, 52, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Hamdani, N.; Herwig, M.; Linke, W.A. Tampering with springs: Phosphorylation of titin affecting the mechanical function of cardiomyocytes. Biophys. Rev. 2017, 9, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, R.; Berri, M.; Wu, Y.; Trombitás, K.; McNabb, M.; Kellermayer, M.S.Z.; Witt, C.; Labeit, D.; Labeit, S.; Greaser, M.; et al. Titin-actin interaction in mouse myocardium: Passive tension modulation and its regulation by calcium/S100A1. Biophys. J. 2001, 81, 2297–2313. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, N.; Granzier, H.L. Titin/connectin-based modulation of the Frank-Starling mechanism of the heart. J. Muscle Res. Cell Motil. 2005, 26, 319–323. [Google Scholar] [CrossRef]
- Hidalgo, C.; Hudson, B.; Bogomolovas, J.; Zhu, Y.; Anderson, B.; Greaser, M.; Labeit, S.; Granzier, H. PKC phosphorylation of titin’s PEVK element: A novel and conserved pathway for modulating myocardial stiffness. Circ. Res. 2009, 105, 631–638. [Google Scholar] [CrossRef]
- Geeves, M.A.; Holmes, K.C. The molecular mechanism of muscle contraction. Adv. Protein Chem. 2005, 71, 161–193. [Google Scholar]
- Day, S.M.; Westfall, M.V.; Fomicheva, E.V.; Hoyer, K.; Yasuda, S.; La Cross, N.C.; D’Alecy, L.G.; Ingwall, J.S.; Metzger, J.M. Histidine button engineered into cardiac troponin I protects the ischemic and failing heart. Nat. Med. 2006, 12, 181–189. [Google Scholar] [CrossRef]
- Westfall, M.V.; Rust, E.M.; Metzger, J.M. Slow skeletal troponin I gene transfer, expression, and myofilament incorporation enhances adult cardiac myocyte contractile function. Proc. Natl. Acad. Sci. USA 1997, 94, 5444–5449. [Google Scholar] [CrossRef]
- Takeda, S.; Yamashita, A.; Maeda, K.; Maeda, Y. Structure of the core domain of human cardiac troponin in the Ca2+-saturated form. Nature 2003, 424, 35–41. [Google Scholar] [CrossRef]
- Palpant, N.J.; Houang, E.M.; Sham, Y.Y.; Metzger, J.M. PH-responsive titratable inotropic performance of histidine-modified cardiac troponin i. Biophys. J. 2012, 102, 1570–1579. [Google Scholar] [CrossRef]
- Thompson, B.R.; Houang, E.M.; Sham, Y.Y.; Metzger, J.M. Molecular determinants of cardiac myocyte performance as conferred by isoform-specific tni residues. Biophys. J. 2014, 106, 2105–2114. [Google Scholar] [CrossRef] [PubMed]
- Vetter, A.D.; Martin, A.A.; Thompson, B.R.; Thomas, D.D.; Metzger, J.M. Sarcomere integrated biosensor detects myofilament-activating ligands in real time during twitch contractions in live cardiac muscle. J. Mol. Cell. Cardiol. 2020, 147, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.J.; Arpaǧ, G.; Tüzel, E.; Ostap, E.M. A Perspective on the Role of Myosins as Mechanosensors. Biophys. J. 2016, 110, 2568–2576. [Google Scholar] [CrossRef] [PubMed]
- Piazzesi, G.; Caremani, M.; Linari, M.; Reconditi, M.; Lombardi, V. Thick Filament Mechano-sensing in skeletal and cardiac muscles: A common mechanism able to adapt the energetic cost of the contraction to the task. Front. Physiol. 2018, 9, 736. [Google Scholar] [CrossRef] [PubMed]
- Reconditi, M.; Caremani, M.; Pinzauti, F.; Powers, J.D.; Narayanan, T.; Stienen, G.J.M.; Linari, M.; Lombardi, V.; Piazzesi, G. Myosin filament activation in the heart is tuned to the mechanical task. Proc. Natl. Acad. Sci. USA 2017, 114, 3240–3245. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, F.; Martyn, D.A. Length-dependent Ca2+ activation in cardiac muscle: Some remaining questions. J. Muscle Res. Cell Motil. 2005, 26, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.G.; Kentish, J.C. The cellular basis of the length-tension relation in cardiac muscle. J. Mol. Cell. Cardiol. 1985, 17, 821–840. [Google Scholar] [CrossRef]
- Cazorla, O.; Lacampagne, A. Regional variation in myofilament length-dependent activation. Pflug. Arch. Eur. J. Physiol. 2011, 462, 15–28. [Google Scholar] [CrossRef]
- Gautel, M. The sarcomeric cytoskeleton: Who picks up the strain? Curr. Opin. Cell Biol. 2011, 23, 39–46. [Google Scholar] [CrossRef]
- Puchner, E.M.; Alexandrovich, A.; Ay, L.K.; Hensen, U.; Schäfer, L.V.; Brandmeier, B.; Gräter, F.; Grubmüller, H.; Gaub, H.E.; Gautel, M. Mechanoenzymatics of titin kinase. Proc. Natl. Acad. Sci. USA 2008, 105, 13385–13390. [Google Scholar] [CrossRef]
- Agarkova, I.; Perriard, J.C. The M-band: An elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005, 15, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Fürst, D.O.; Gautel, M. The anatomy of a molecular giant: How the sarcomere cytoskeleton is assembled from immunoglobulin superfamily molecules. J. Mol. Cell. Cardiol. 1995, 27, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Gautel, M.; Djinović-Carugo, K. The sarcomeric cytoskeleton: From molecules to motion. J. Exp. Biol. 2016, 219, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Henderson, C.A.; Gomez, C.G.; Novak, S.M.; Mi-Mi, L.; Gregorio, C.C. Overview of the muscle cytoskeleton. Compr. Physiol. 2017, 7, 891–944. [Google Scholar] [PubMed]
- Sjöblom, B.; Salmazo, A.; Djinović-Carugo, K. α-Actinin structure and regulation. Cell. Mol. Life Sci. 2008, 65, 2688–2701. [Google Scholar] [CrossRef]
- Robison, P.; Prosser, B.L. Microtubule mechanics in the working myocyte. J. Physiol. 2017, 595, 3931–3937. [Google Scholar] [CrossRef]
- Luther, P.K. The vertebrate muscle Z-disc: Sarcomere anchor for structure and signalling. J. Muscle Res. Cell Motil. 2009, 30, 171–185. [Google Scholar] [CrossRef]
- Knöll, R.; Buyandelger, B. Z-disc Transcriptional Coupling, Sarcomeroptosis and Mechanopoptosis. Cell Biochem. Biophys. 2013, 66, 65–71. [Google Scholar] [CrossRef][Green Version]
- Peter, A.K.; Cheng, H.; Ross, R.S.; Knowlton, K.U.; Chen, J. The costamere bridges sarcomeres to the sarcolemma in striated muscle. Prog. Pediatr. Cardiol. 2011, 31, 83–88. [Google Scholar] [CrossRef]
- Van Der Velden, J.; Ho, C.Y.; Tardiff, J.C.; Olivotto, I.; Knollmann, B.C.; Carrier, L. Research priorities in sarcomeric cardiomyopathies. Cardiovasc. Res. 2015, 105, 449–456. [Google Scholar] [CrossRef]
- Spudich, J.A. Hypertrophic and dilated cardiomyopathy: Four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys. J. 2014, 106, 1236–1249. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.J. New Perspectives on the Prevalence of Hypertrophic Cardiomyopathy. J. Am. Coll. Cardiol. 2015, 65, 1249–1254. [Google Scholar]
- Seidman, J.G.; Seidman, C. The Genetic Basis for Cardiomyopathy: From Mutation Identification to Mechanistic Paradigms. Cell 2001, 104, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Geisterfer-Lowrance, A.A.; Kass, S.; Tanigawa, G.; Vosberg, H.P.; McKenna, W.; Seidman, C.E.; Seidman, J.G. A molecular basis for familial hypertrophic cardiomyopathy: A beta cardiac myosin heavy chain gene missense mutation. Cell 1990, 62, 999–1006. [Google Scholar] [CrossRef]
- Bos, J.M.; Towbin, J.A.; Ackerman, M.J. Diagnostic, prognostic, and therapeutic implications of genetic testing for hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2009, 54, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.J.; Maron, M.S. Hypertrophic cardiomyopathy. Lancet 2013, 381, 242–255. [Google Scholar] [CrossRef]
- Ho, C.Y. Genetic considerations in hypertrophic cardiomyopathy. Prog. Cardiovasc. Dis. 2012, 54, 456–460. [Google Scholar] [CrossRef]
- Mogensen, J.; Kubo, T.; Duque, M.; Uribe, W.; Shaw, A.; Murphy, R.; Gimeno, J.R.; Elliott, P.; McKenna, W.J. Idiopathic restrictive cardiomyopathy is part of the clinical expression of cardiac troponin I mutations. J. Clin. Investig. 2003, 111, 209–216. [Google Scholar] [CrossRef]
- Blagova, O.; Pavlenko, E.; Sedov, V.; Kogan, E.; Polyak, M.; Zaklyazminskaya, E.; Lutokhina, Y. Different Phenotypes of Sarcomeric MyBPC3-Cardiomyopathy in the Same Family: Hypertrophic, Left Ventricular Noncompaction and Restrictive Phenotypes (in Association with Sarcoidosis). Genes 2022, 13, 1344. [Google Scholar] [CrossRef]
- Garfinkel, A.C.; Seidman, J.G.; Seidman, C.E. Genetic Pathogenesis of Hypertrophic and Dilated Cardiomyopathy. Heart Fail. Clin. 2018, 14, 139–146. [Google Scholar] [CrossRef]
- Hassoun, R.; Budde, H.; Mügge, A.; Hamdani, N. Cardiomyocyte dysfunction in inherited cardiomyopathies. Int. J. Mol. Sci. 2021, 22, 11154. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.J.; Maron, M.S.; Semsarian, C. Genetics of hypertrophic cardiomyopathy after 20 years: Clinical perspectives. J. Am. Coll. Cardiol. 2012, 60, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Willott, R.H.; Gomes, A.V.; Chang, A.N.; Parvatiyar, M.S.; Pinto, J.R.; Potter, J.D. Mutations in Troponin that cause HCM, DCM AND RCM: What can we learn about thin filament function? J. Mol. Cell. Cardiol. 2010, 48, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Pinto, J.R.; Gomes, A.V.; Xu, Y.; Wang, Y.; Potter, J.D.; Kerrick, W.G. Functional consequences of the human cardiac troponin I hypertrophic cardiomyopathy mutation R145G in transgenic mice. J. Biol. Chem. 2008, 283, 20484–20494. [Google Scholar] [CrossRef]
- Tardiff, J.C.; Hewett, T.E.; Palmer, B.M.; Olsson, C.; Factor, S.M.; Moore, R.L.; Robbins, J.; Leinwand, L.A. Cardiac troponin T mutations result in allele-specific phenotypes in a mouse model for hypertrophic cardiomyopathy. J. Clin. Investig. 1999, 104, 469–481. [Google Scholar] [CrossRef]
- Haim, T.E.; Dowell, C.; Diamanti, T.; Scheuer, J.; Tardiff, J.C. Independent FHC-related cardiac troponin T mutations exhibit specific alterations in myocellular contractility and calcium kinetics. J. Mol. Cell. Cardiol. 2007, 42, 1098–1110. [Google Scholar] [CrossRef]
- Michele, D.E.; Albayya, F.P.; Metzger, J.M. Direct, convergent hypersensitivity of calcium-activated force generation produced by hypertrophic cardiomyopathy mutant alpha-tropomyosins in adult cardiac myocytes. Nat. Med. 1999, 5, 1413–1417. [Google Scholar] [CrossRef]
- Michele, D.E.; Metzger, J.M. Physiological consequences of tropomyosin mutations associated with cardiac and skeletal myopathies. J. Mol. Med. 2000, 78, 543–553. [Google Scholar] [CrossRef]
- Davis, J.; Wen, H.; Edwards, T.; Metzger, J.M. Allele and species dependent contractile defects by restrictive and hypertrophic cardiomyopathy-linked troponin I mutants. J. Mol. Cell. Cardiol. 2008, 44, 891–904. [Google Scholar] [CrossRef][Green Version]
- Chandra, M.; Rundell, V.L.M.; Tardiff, J.C.; Leinwand, L.A.; de Tombe, P.P.; Solaro, R.J. Ca2+ activation of myofilaments from transgenic mouse hearts expressing R92Q mutant cardiac troponin T. Am. J. Physiol. Heart Circ. Physiol. 2001, 280, H705–H713. [Google Scholar] [CrossRef]
- Baudenbacher, F.; Schober, T.; Pinto, J.R.; Sidorov, V.Y.; Hilliard, F.; Solaro, R.J.; Potter, J.D.; Knollmann, B. Myofilament Ca2+ sensitization causes susceptibility to cardiac arrhythmia in mice. J. Clin. Investig. 2008, 118, 3893–3903. [Google Scholar] [CrossRef] [PubMed]
- Huke, S.; Knollmann, B.C. Increased myofilament Ca2+-sensitivity and arrhythmia susceptibility. J. Mol. Cell. Cardiol. 2010, 48, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Tikunova, S.B.; Kline, K.P.; Siddiqui, J.K.; Davis, J.P. Disease-Related Cardiac Troponins Alter Thin Filament Ca2+ Association and Dissociation Rates. PLoS ONE 2012, 7, e38259. [Google Scholar] [CrossRef] [PubMed]
- Duncker, D.J.; Bakkers, J.; Brundel, B.J.; Robbins, J.; Tardiff, J.C.; Carrier, L. Animal and in silico models for the study of sarcomeric cardiomyopathies. Cardiovasc. Res. 2015, 105, 439–448. [Google Scholar] [CrossRef]
- Anderson, R.L.; Trivedi, D.V.; Sarkar, S.S.; Henze, M.; Ma, W.; Gong, H.; Rogers, C.S.; Gorham, J.M.; Wong, F.L.; Morck, M.M.; et al. Deciphering the super relaxed state of human β-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers. Proc. Natl. Acad. Sci. USA 2018, 115, E8143–E8152. [Google Scholar] [CrossRef]
- Toepfer, C.N.; Wakimoto, H.; Garfinkel, A.C.; McDonough, B.; Liao, D.; Jiang, J.; Tai, A.C.; Gorham, J.M.; Lunde, I.G.; Lun, M.; et al. Hypertrophic cardiomyopathy mutations in MYBPC3 dysregulate myosin. Sci. Transl. Med. 2019, 11, eaat1199. [Google Scholar] [CrossRef]
- Chintanaphol, M.; Orgil, B.O.; Alberson, N.R.; Towbin, J.A.; Purevjav, E. Restrictive cardiomyopathy: From genetics and clinical overview to animal modeling. Rev. Cardiovasc. Med. 2022, 23, 108. [Google Scholar] [CrossRef]
- Van den Wijngaard, A.; Volders, P.; Van Tintelen, J.P.; Jongbloed, J.D.; van den Berg, M.P.; Lekanne Deprez, R.H.; Mannens, M.M.; Hofmann, N.; Slegtenhorst, M.; Dooijes, D.; et al. Recurrent and founder mutations in the Netherlands: Cardiac Troponin I (TNNI3) gene mutations as a cause of severe forms of hypertrophic and restrictive cardiomyopathy. Neth. Heart J. 2011, 19, 344–351. [Google Scholar] [CrossRef]
- Gomes, A.V.; Liang, J.; Potter, J.D. Mutations in Human Cardiac Troponin I That Are Associated with Restrictive Cardiomyopathy Affect Basal ATPase Activity and the Calcium Sensitivity of Force Development. J. Biol. Chem. 2005, 280, 30909–30915. [Google Scholar] [CrossRef]
- Wen, Y.; Xu, Y.; Wang, Y.; Pinto, J.R.; Potter, J.D.; Kerrick, W.G. Functional effects of a restrictive-cardiomyopathy-linked cardiac troponin I mutation (R145W) in transgenic mice. J. Mol. Biol. 2009, 392, 1158–1167. [Google Scholar] [CrossRef]
- Davis, J.; Yasuda, S.; Palpant, N.J.; Martindale, J.; Stevenson, T.; Converso, K.; Metzger, J.M. Diastolic dysfunction and thin filament dysregulation resulting from excitation-contraction uncoupling in a mouse model of restrictive cardiomyopathy. J. Mol. Cell. Cardiol. 2012, 53, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.; Wen, H.; Edwards, T.; Metzger, J.M. Thin filament disinhibition by restrictive cardiomyopathy mutant R193H troponin I induces Ca2+-independent mechanical tone and acute myocyte remodeling. Circ. Res. 2007, 100, 1494–1502. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Liu, J.; Feng, H.-Z.; Hossain, M.M.; Gobara, N.; Zhang, C.; Li, Y.; Jean-Charles, P.-Y.; Jin, J.-P.; Huang, X.-P. Impaired relaxation is the main manifestation in transgenic mice expressing a restrictive cardiomyopathy mutation, R193H, in cardiac TnI. Am. J. Physiol. Heart Circ. Physiol. 2008, 294, H2604–H2613. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.R.; Martindale, J.; Metzger, J.M. Sarcomere neutralization in inherited cardiomyopathy: Small-molecule proof-of-concept to correct hyper-Ca2+-sensitive myofilaments. Am. J. Physiol. Heart Circ. Physiol. 2016, 311, 13. [Google Scholar] [CrossRef]
- Towbin, J.A.; Lowe, A.M.; Colan, S.D.; Sleeper, L.A.; Orav, E.J.; Clunie, S.; Messere, J.; Cox, G.F.; Hsu, D.; Canter, C.; et al. Incidence, Causes, and Outcomes of Dilated Cardiomyopathy in Children. J. Am. Med. Assoc. 2017, 296, 1867–1876. [Google Scholar] [CrossRef]
- Hershberger, R.E.; Cowan, J.; Jordan, E.; Kinnamon, D.D. The Complex and Diverse Genetic Architecture of Dilated Cardiomyopathy. Circ. Res. 2021, 128, 1514–1532. [Google Scholar] [CrossRef]
- Mestroni, L.; Brun, F.; Spezzacatene, A.; Sinagra, G.; Taylor, M.R. Genetic Causes of Dilated Cardiomyopathy. Prog. Pediatr. Cardiol. 2014, 37, 13–18. [Google Scholar] [CrossRef]
- Tharp, C.A.; Haywood, M.E.; Sbaizero, O.; Taylor, M.R.G.; Mestroni, L. The Giant Protein Titin’s Role in Cardiomyopathy: Genetic, Transcriptional, and Post-translational Modifications of TTN and Their Contribution to Cardiac Disease. Front. Physiol. 2019, 10, 1436. [Google Scholar] [CrossRef]
- McNally, E.M. Genetics: Broken giant linked to heart failure. Nature 2012, 483, 281–282. [Google Scholar] [CrossRef]
- McNally, E.M.; Golbus, J.R.; Puckelwartz, M.J. Genetic mutations and mechanisms in dilated cardiomyopathy. J. Clin. Investig. 2013, 123, 19–26. [Google Scholar] [CrossRef]
- Hinson, J.T.; Chopra, A.; Nafissi, N.; Polacheck, W.J.; Benson, C.C.; Swist, S.; Gorham, J.; Yang, L.; Schafer, S.; Sheng, C.C.; et al. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science 2015, 349, 982–987. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, J.P.; Debold, E.P.; Ahmad, F.; Armstrong, A.; Frederico, A.; Conner, D.A.; Mende, U.; Lohse, M.J.; Warshaw, D.; Seidman, C.E.; et al. Cardiac myosin missense mutations cause dilated cardiomyopathy in mouse models and depress molecular motor function. Proc. Natl. Acad. Sci. USA 2006, 103, 14525–14530. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.; Metzger, J.M. Combinatorial Effects of Double Cardiomyopathy Mutant Alleles in Rodent Myocytes: A Predictive Cellular Model of Myofilament Dysregulation in Disease. PLoS ONE 2010, 5, e9140. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; McCully, M.E.; Luo, Z.; McMichael, J.; Tu, A.Y.; Daggett, V.; Regnier, M. Structural and functional consequences of cardiac troponin C L57Q and I61Q Ca(2+)-desensitizing variants. Arch. Biochem. Biophys. 2013, 535, 68–75. [Google Scholar] [CrossRef]
- Du, C.K.; Zhan, D.Y.; Morimoto, S. In vivo effects of propyl gallate, a novel Ca(2+) sensitizer, in a mouse model of dilated cardiomyopathy caused by cardiac troponin T mutation. Life Sci. 2014, 109, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Du, C.K.; Morimoto, S.; Nishii, K.; Minakami, R.; Ohta, M.; Tadano, N.; Lu, Q.W.; Wang, Y.Y.; Zhan, D.Y.; Mochizuki, M.; et al. Knock-in mouse model of dilated cardiomyopathy caused by troponin mutation. Circ. Res. 2007, 101, 185–194. [Google Scholar] [CrossRef]
- Gollapudi, S.K.; Tardiff, J.C.; Chandra, M. The functional effect of dilated cardiomyopathy mutation (R144W) in mouse cardiac troponin T is differently affected by alpha- and beta-myosin heavy chain isoforms. Am. J. Physiol. Heart Circ. Physiol. 2015, 308, H884–H893. [Google Scholar] [CrossRef]
- Green, E.M.; Wakimoto, H.; Anderson, R.L.; Evanchik, M.J.; Gorham, J.M.; Harrison, B.C.; Henze, M.; Kawas, R.; Oslob, J.D.; Rodriguez, H.M.; et al. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science 2016, 351, 617–621. [Google Scholar] [CrossRef]
- Rohde, J.A.; Roopnarine, O.; Thomas, D.D.; Muretta, J.M. Mavacamten stabilizes an autoinhibited state of two-headed cardiac myosin. Proc. Natl. Acad. Sci. USA 2018, 115, E7486–E7494. [Google Scholar] [CrossRef]
- Sparrow, A.J.; Watkins, H.; Daniels, M.J.; Redwood, C.; Robinson, P. Mavacamten rescues increased myofilament calcium sensitivity and dysregulation of Ca 2 flux caused by thin filament hypertrophic cardiomyopathy mutations. Am. J. Physiol. Heart Circ. Physiol. 2020, 318, 715–722. [Google Scholar] [CrossRef]
- Mamidi, R.; Li, J.; Chang, B.; Doh, Y.; Sujeet Verma, B.; Stelzer, J.E. Impact of the Myosin Modulator Mavacamten on Force Generation and Cross-Bridge Behavior in a Murine Model of Hypercontractility. J. Am. Heart Assoc. 2018, 7, e009627. [Google Scholar] [CrossRef] [PubMed]
- Dalo, J.D.; Weisman, N.D.; White, C.M. Mavacamten, a First-in-Class Cardiac Myosin Inhibitor for Obstructive Hypertrophic Cardiomyopathy. Ann. Pharmacother. 2022. [Google Scholar] [CrossRef] [PubMed]
- Chuang, C.; Collibee, S.; Ashcraft, L.; Wang, W.; Vander Wal, M.; Wang, X.; Hwee, D.T.; Wu, Y.; Wang, J.; Chin, E.R.; et al. Discovery of Aficamten (CK-274), a Next-Generation Cardiac Myosin Inhibitor for the Treatment of Hypertrophic Cardiomyopathy. J. Med. Chem. 2021, 64, 14142–14152. [Google Scholar] [CrossRef]
- Morelli, C.; Ingrasciotta, G.; Jacoby, D.; Masri, A.; Olivotto, I. Sarcomere protein modulation: The new frontier in cardiovascular medicine and beyond. Eur. J. Intern. Med. 2022, 102, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Malik, F.I.; Hartman, J.J.; Elias, K.A.; Morgan, B.P.; Hector, R.; Brejc, K.; Anderson, R.L.; Sueoka, S.H.; Lee, K.H.; Finer, J.T.; et al. Cardiac Mosin Activation: A Potential Therapeutic Approach for Systolic Heart Failure. Science 2011, 331, 1439–1443. [Google Scholar] [CrossRef]
- Nagy, L.; Kovács, Á.; Bódi, B.; Pásztor, E.T.; Fülöp, G.Á.; Tóth, A.; Édes, I.; Papp, Z. The novel cardiac myosin activator omecamtiv mecarbil increases the calcium sensitivity of force production in isolated cardiomyocytes and skeletal muscle fibres of the rat. Br. J. Pharmacol. 2015, 172, 4506–4518. [Google Scholar] [CrossRef]
- Woody, M.S.; Greenberg, M.J.; Barua, B.; Winkelmann, D.A.; Goldman, Y.E.; Ostap, E.M. Positive cardiac inotrope omecamtiv mecarbil activates muscle despite suppressing the myosin working stroke. Nat. Commun. 2018, 9, 3838. [Google Scholar] [CrossRef]
- Rohde, J.A.; Thomas, D.D.; Muretta, J.M. Heart failure drug changes the mechanoenzymology of the cardiac myosin powerstroke. Proc. Natl. Acad. Sci. USA 2017, 114, E1796–E1804. [Google Scholar] [CrossRef]
- Nanasi, P.; Komaromi, I.; Gaburjakova, M.; Almassy, J. Omecamtiv Mecarbil: A Myosin Motor Activator Agent with Promising Clinical Performance and New in vitro Results. Curr. Med. Chem. 2017, 25, 1720–1728. [Google Scholar] [CrossRef]
- Utter, M.S.; Ryba, D.M.; Li, B.H.; Wolska, B.M.; Solaro, R.J. Omecamtiv mecarbil, a cardiac myosin activator, increases Ca2+ sensitivity in myofilaments with a dilated cardiomyopathy mutant tropomyosin E54K. J. Cardiovasc. Pharmacol. 2015, 66, 347–353. [Google Scholar] [CrossRef]
- Day, S.M.; Tardiff, J.C.; Michael Ostap, E. Myosin modulators: Emerging approaches for the treatment of cardiomyopathies and heart failure. J. Clin. Investig. 2022, 132, e148557. [Google Scholar] [CrossRef] [PubMed]
- Voors, A.A.; Tamby, J.F.; Cleland, J.G.; Koren, M.; Forgosh, L.B.; Gupta, D.; Lund, L.H.; Camacho, A.; Karra, R.; Swart, H.P.; et al. Effects of danicamtiv, a novel cardiac myosin activator, in heart failure with reduced ejection fraction: Experimental data and clinical results from a phase 2a trial. Eur. J. Heart Fail. 2020, 22, 1649–1658. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Sewanan, L.R.; Jacoby, D.L.; Campbell, S.G. Danicamtiv enhances systolic function and frank-starling behavior at minimal diastolic cost in engineered human myocardium. J. Am. Heart Assoc. 2021, 10, e020860. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin, A.A.; Thompson, B.R.; Hahn, D.; Angulski, A.B.B.; Hosny, N.; Cohen, H.; Metzger, J.M. Cardiac Sarcomere Signaling in Health and Disease. Int. J. Mol. Sci. 2022, 23, 16223. https://doi.org/10.3390/ijms232416223
Martin AA, Thompson BR, Hahn D, Angulski ABB, Hosny N, Cohen H, Metzger JM. Cardiac Sarcomere Signaling in Health and Disease. International Journal of Molecular Sciences. 2022; 23(24):16223. https://doi.org/10.3390/ijms232416223
Chicago/Turabian StyleMartin, Ashley A., Brian R. Thompson, Dongwoo Hahn, Addeli Bez Batti Angulski, Nora Hosny, Houda Cohen, and Joseph M. Metzger. 2022. "Cardiac Sarcomere Signaling in Health and Disease" International Journal of Molecular Sciences 23, no. 24: 16223. https://doi.org/10.3390/ijms232416223
APA StyleMartin, A. A., Thompson, B. R., Hahn, D., Angulski, A. B. B., Hosny, N., Cohen, H., & Metzger, J. M. (2022). Cardiac Sarcomere Signaling in Health and Disease. International Journal of Molecular Sciences, 23(24), 16223. https://doi.org/10.3390/ijms232416223





