Acting on the CFTR Membrane-Spanning Domains Interface Rescues Some Misfolded Mutants
Abstract
1. Introduction
2. Results
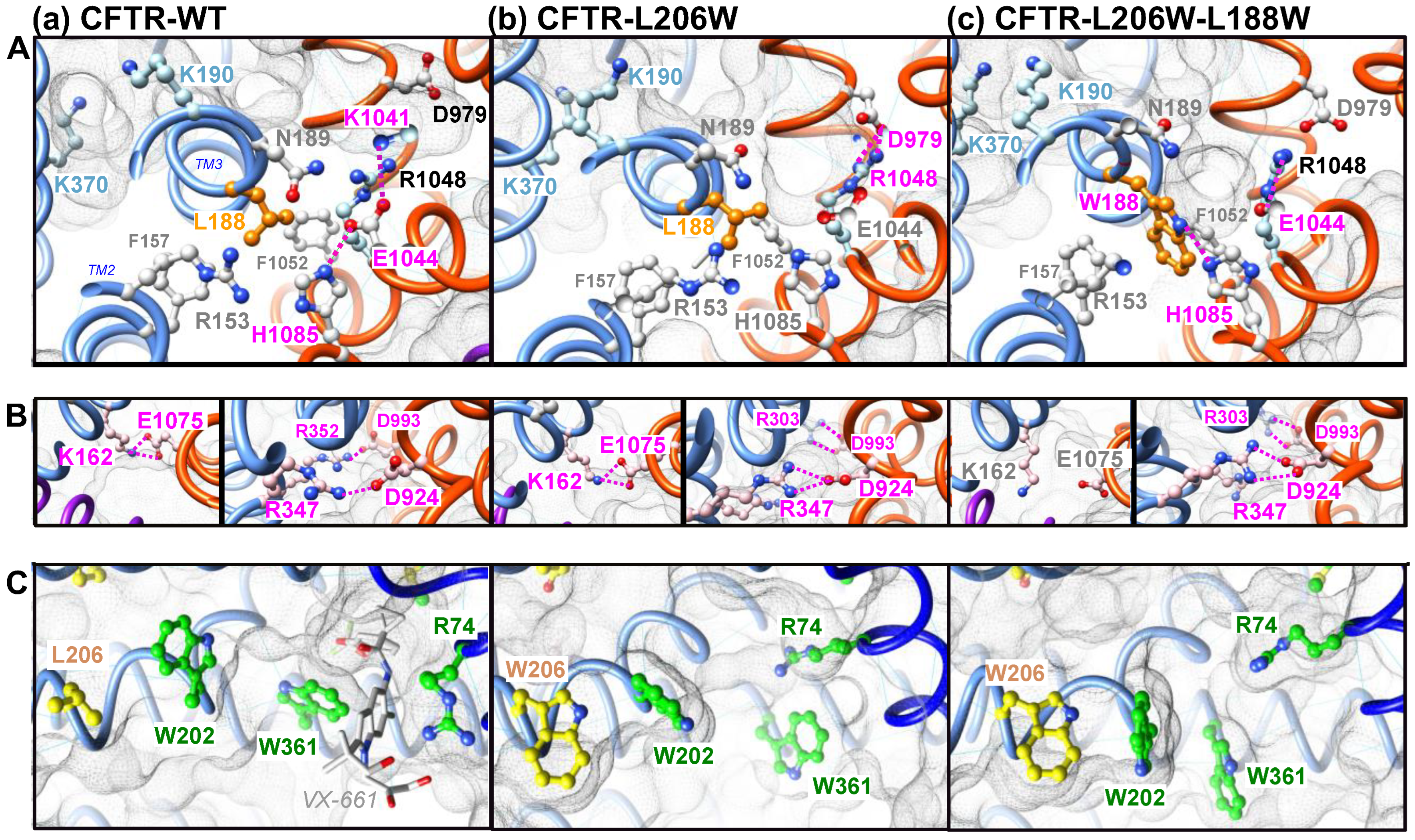
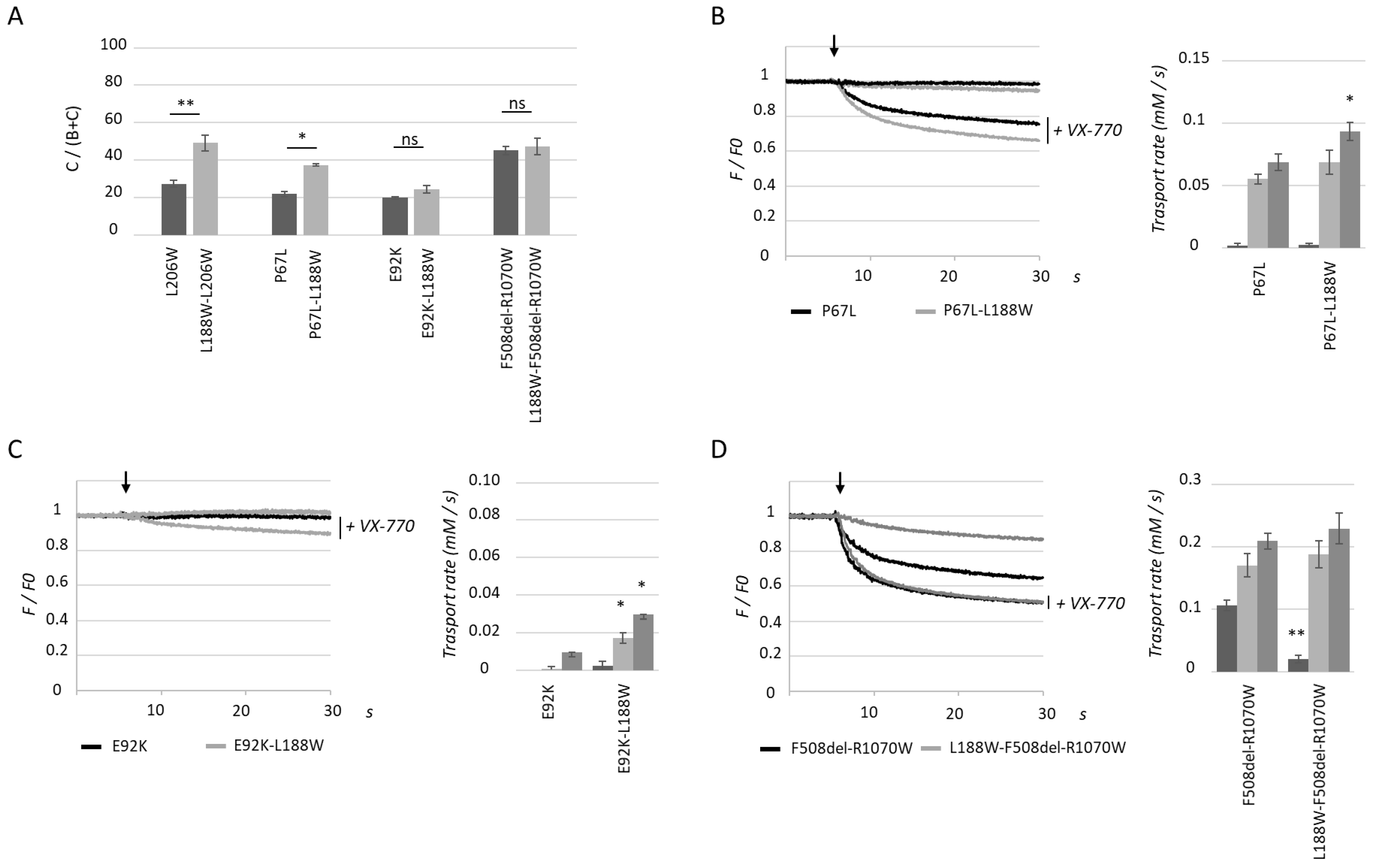
2.1. L188W Rescues MSD1 Class II Mutation L206W
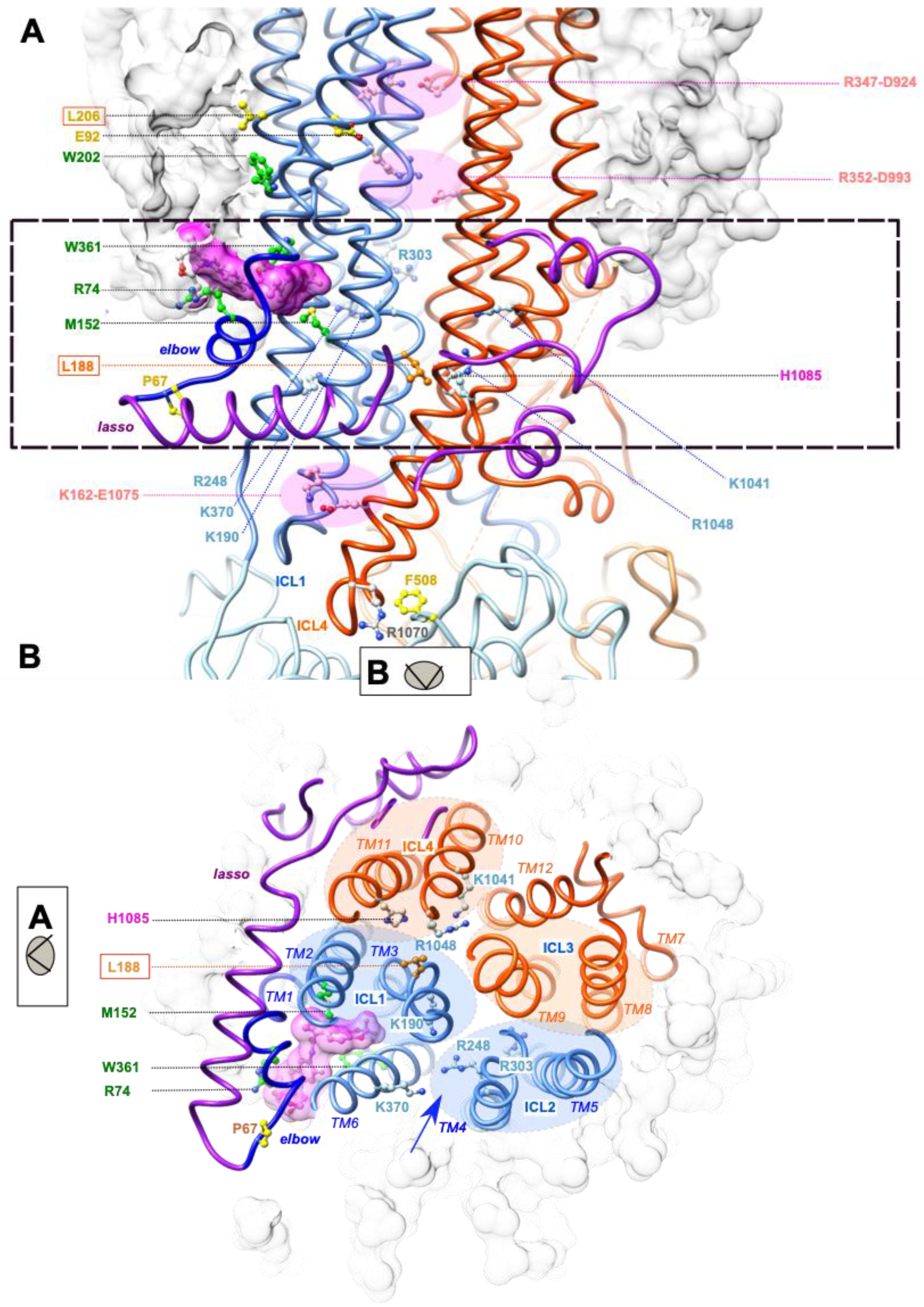
2.2. L188W Acts on the MSD1:MSD2 Interface
2.3. L188W Rescues Other Class II Mutations Localized within the Lasso Motif or MSD1
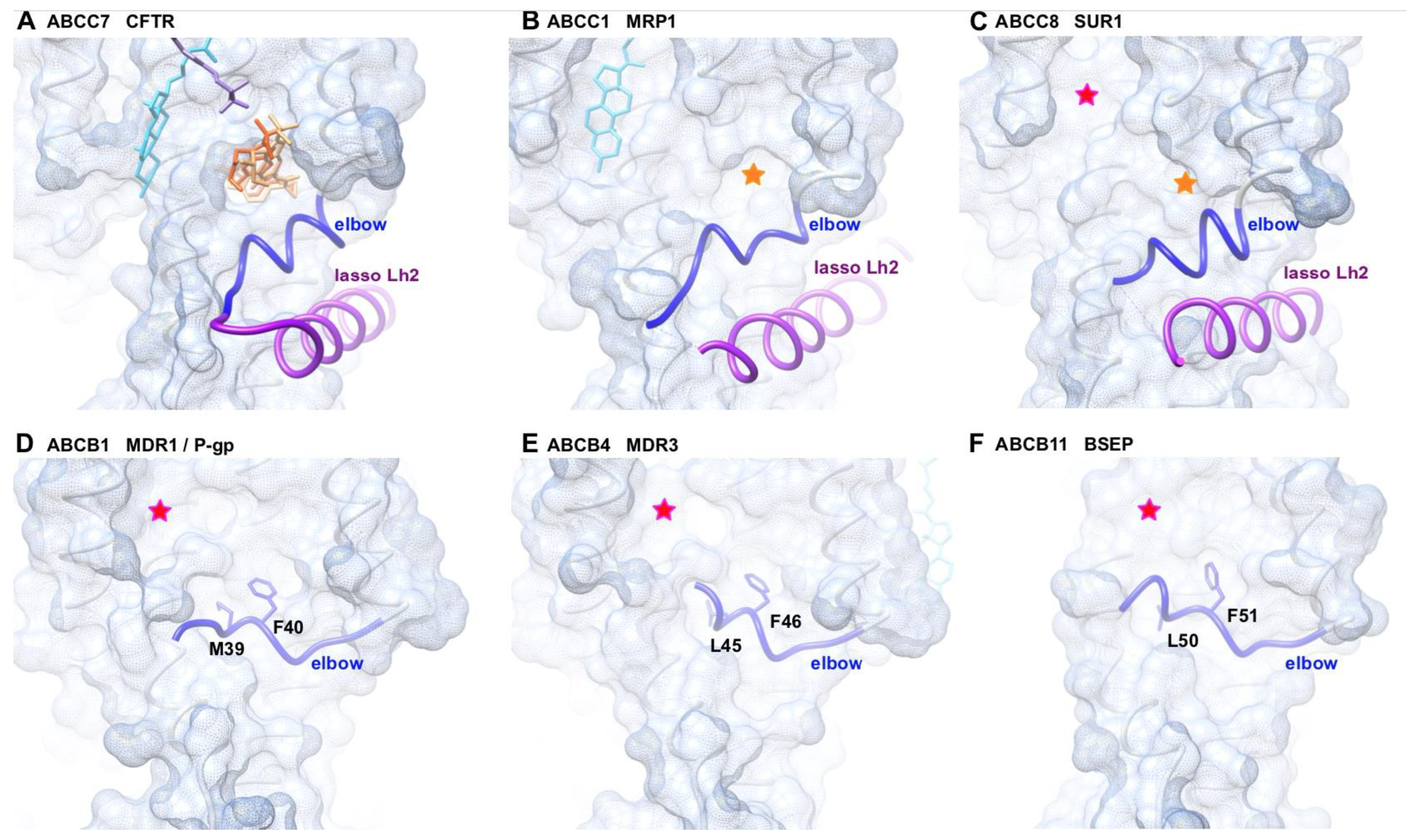
2.4. The MSD1 Instability Is Likely Associated with the Presence of an ABCC-Specific Lasso Motif
3. Discussion
4. Materials and Methods
4.1. Plasmids and Mutagenesis
4.2. Cell Culture and Transfection
4.3. Western Blot Analysis
4.4. Halide Sensitive Functional Assay
4.5. Cell Surface Quantification
4.6. Statistical Analysis
4.7. Molecular Dynamics Simulations/3D Structure Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thomas, C.; Aller, S.G.; Beis, K.; Carpenter, E.P.; Chang, G.; Chen, L.; Dassa, E.; Dean, M.; Duong Van Hoa, F.; Ekiert, D.; et al. Structural and functional diversity calls for a new classification of ABC transporters. FEBS Lett. 2020, 594, 3767–3775. [Google Scholar] [CrossRef] [PubMed]
- Delaunay, J.L.; Durand-Schneider, A.M.; Dossier, C.; Falguières, T.; Gautherot, J.; Davit-Spraul, A.; Aït-Slimane, T.; Housset, C.; Jacquemin, E.; Maurice, M. A functional classification of ABCB4 variations causing progressive familial intrahepatic cholestasis type 3. Hepatology 2016, 63, 1620–1631. [Google Scholar] [CrossRef] [PubMed]
- Soroka, C.J.; Boyer, J.L. Biosynthesis and trafficking of the bile salt export pump, BSEP: Therapeutic implications of BSEP mutations. Mol. Asp. Med. 2014, 37, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Skach, W.R. Mechanisms of CFTR Folding at the Endoplasmic Reticulum. Front. Pharm. 2012, 3, 201. [Google Scholar] [CrossRef]
- Kim, S.J.; Yoon, J.S.; Shishido, H.; Yang, Z.; Rooney, L.A.; Barral, J.M.; Skach, W.R. Protein folding. Translational tuning optimizes nascent protein folding in cells. Science 2015, 348, 444–448. [Google Scholar] [CrossRef]
- Kleizen, B.; van Vlijmen, T.; de Jonge, H.R.; Braakman, I. Folding of CFTR is predominantly cotranslational. Mol. Cell 2005, 20, 277–287. [Google Scholar] [CrossRef]
- Kleizen, B.; van Willigen, M.; Mijnders, M.; Peters, F.; Grudniewska, M.; Hillenaar, T.; Thomas, A.; Kooijman, L.; Peters, K.W.; Frizzell, R.; et al. Co-Translational Folding of the First Transmembrane Domain of ABC-Transporter CFTR is Supported by Assembly with the First Cytosolic Domain. J. Mol. Biol. 2021, 433, 166955. [Google Scholar] [CrossRef] [PubMed]
- Lukacs, G.L.; Verkman, A.S. CFTR: Folding, misfolding and correcting the DeltaF508 conformational defect. Trends Mol. Med. 2012, 18, 81–91. [Google Scholar] [CrossRef]
- Mendoza, J.L.; Schmidt, A.; Li, Q.; Nuvaga, E.; Barrett, T.; Bridges, R.J.; Feranchak, A.P.; Brautigam, C.A.; Thomas, P.J. Requirements for efficient correction of DF508 CFTR revealed by analyses of evolved sequences. Cell 2012, 148, 164–174. [Google Scholar] [CrossRef]
- Mijnders, M.; Kleizen, B.; Braakman, I. Correcting CFTR folding defects by small-molecule correctors to cure cystic fibrosis. Curr. Opin. Pharm. 2017, 34, 83–90. [Google Scholar] [CrossRef]
- Protasevich, I.; Yang, Z.; Wang, C.; Atwell, S.; Zhao, X.; Emtage, S.; Wetmore, D.; Hunt, J.; Brouillette, C.G. Thermal unfolding studies show the disease causing F508del mutation in CFTR thermodynamically destabilizes nucleotide-binding domain 1. Protein Sci. 2010, 19, 1917–1931. [Google Scholar] [CrossRef] [PubMed]
- Colas, J.; Faure, G.; Saussereau, E.; Trudel, S.; Rabeh, W.M.; Bitam, S.; Guerrera, I.C.; Fritsch, J.; Sermet-Gaudelus, I.; Davezac, N.; et al. Disruption of cytokeratin-8 interaction with F508del-CFTR corrects its functional defect. Hum. Mol. Genet 2012, 21, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Thibodeau, P.H.; Richardson, J.M.; Wang, W.; Millen, L.; Watson, J.; Mendoza, J.L.; Du, K.; Fischman, S.; Senderowitz, H.; Lukacs, G.L.; et al. The cystic fibrosis-causing mutation deltaF508 affects multiple steps in cystic fibrosis transmembrane conductance regulator biogenesis. J. Biol. Chem. 2010, 285, 35825–35835. [Google Scholar] [CrossRef]
- Wang, W.; Wu, J.; Bernard, K.; Li, G.; Wang, G.; Bevensee, M.O.; Kirk, K.L. ATP-independent CFTR channel gating and allosteric modulation by phosphorylation. Proc. Natl. Acad. Sci. USA 2010, 107, 3888–3893. [Google Scholar] [CrossRef]
- Loo, T.W.; Barlett, M.C.; Clarke, D.M. The V510D suppressor mutation stablilizes DeltaF508-CFTR at the cell surface. Biochemistry 2010, 49, 6352–6357. [Google Scholar] [CrossRef] [PubMed]
- DeCarvalho, A.C.; Gansheroff, L.J.; Teem, J.L. Mutations in the nucleotide binding domain 1 signature motif region rescue processing and functional defects of cystic fibrosis transmembrane conductance regulator delta f508. J. Biol. Chem. 2002, 277, 35896–35905. [Google Scholar] [CrossRef]
- Teem, J.L.; Carson, M.R.; Welsh, M.J. Mutation of R555 in CFTR-delta F508 enhances function and partially corrects defective processing. Recept. Channels 1996, 4, 63–72. [Google Scholar]
- Pissarra, L.S.; Farinha, C.M.; Xu, Z.; Schmidt, A.; Thibodeau, P.H.; Cai, Z.; Thomas, P.; Sheppard, D.N.; Amaral, M.D. Solubilizing mutations used to crystallize one CFTR domain attenuate the trafficking and channel defects caused by the major cystic fibrosis mutation. Chem. Biol. 2008, 15, 62–69. [Google Scholar] [CrossRef]
- Aleksandrov, A.A.; Kota, P.; Aleksandrov, L.A.; He, L.; Jensen, T.; Cui, L.; Gentzsch, M.; Dokholyan, N.V.; Riordan, J.R. Regulatory insertion removal restores maturation, stability and function of DeltaF508 CFTR. J. Mol. Biol. 2010, 401, 194–210. [Google Scholar] [CrossRef]
- Chang, X.-B.; Cui, L.; Hou, Y.-X.; Jensen, T.J.; Aleksandrov, A.A.; Mengos, A.; Riordan, J.R. Removal of multiple arginine-framed trafficking signals overcomes misprocessing of ΔF508 CFTR present in most patients with cystic fibrosis. Mol. Cell 1999, 4, 137–142. [Google Scholar] [CrossRef]
- Farinha, C.M.; King-Underwood, J.; Sousa, M.; Correia, A.R.; Henriques, B.J.; Roxo-Rosa, M.; Da Paula, A.C.; Williams, J.; Hirst, S.; Gomes, C.M.; et al. Revertants, low temperature, and correctors reveal the mechanism of F508del-CFTR rescue by VX-809 and suggest multiple agents for full correction. Chem. Biol. 2013, 20, 943–955. [Google Scholar] [CrossRef]
- Baatallah, N.; Elbahnsi, A.; Mornon, J.-P.; Chevalier, B.; Pranke, I.; Servel, N.; Zelli, R.; Décout, J.-L.; Edelman, A.; Sermet-Gaudelus, I.; et al. Pharmacological chaperones improve intra-domain stability and inter-domain assembly via distinct binding sites to rescue misfolded CFTR. Cell Mol. Life Sci. 2021, 78, 7813–7829. [Google Scholar] [CrossRef] [PubMed]
- Fiedorczuk, K.; Chen, J. Mechanism of CFTR correction by type I folding correctors. Cell 2022, 185, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Galietta, L.J.; Haggie, P.M.; Verkman, A.S. Green fluorescent protein-based halide indicators with improved chloride and iodide affinities. FEBS Lett. 2001, 499, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Cotten, J.F.; Welsh, M.J. Cystic fibrosis-associated mutations at arginine 347 alter the pore architecture of CFTR. Evidence for disruption of a salt bridge. J. Biol. Chem. 1999, 274, 5429–5435. [Google Scholar] [CrossRef]
- Cui, G.; Freeman, C.S.; Knotts, T.; Prince, C.Z.; Kuang, C.; McCarty, N.A. Two salts bridges differentially contribute to the maintenance of Cystic Fibrosis Transmebrane Conductance Regulator (CFTR) channel function. J. Biol. Chem. 2013, 288, 20758–20767. [Google Scholar] [CrossRef]
- Cui, G.; Zhang, Z.R.; O’Brien, A.R.; Song, B.; McCarty, N.A. Mutation at arginine 352 alters the pore architecture of CFTR. J. Membr. Biol. 2008, 222, 91–106. [Google Scholar] [CrossRef]
- El Hiani, Y.; Linsdell, P. Functional architecture of the cytoplasmic entrance to the Cystic Fibrosis Transmembrane conductance Regulator chloride channel pore. J. Biol. Chem. 2015, 290, 15855–15865. [Google Scholar] [CrossRef] [PubMed]
- El Hiani, Y.; Negoda, A.; Linsdell, P. Cytoplasmic pathway followed by chloride ions to enter the CFTR channel pore. Cell Mol. Life Sci. 2016, 73, 1917–1925. [Google Scholar] [CrossRef]
- Wang, L.; Johnson, Z.L.; Wasserman, M.R.; Levring, J.; Chen, J.; Liu, S. Characterization of the kinetic cycle of an ABC transporter by single-molecule and cryo-EM analyses. Elife 2020, 9, e56451. [Google Scholar] [CrossRef]
- Zhao, C.; MacKinnon, R. Molecular structure of an open human K(ATP) channel. Proc. Natl. Acad. Sci. USA 2021, 118, e2112267118. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Chen, J. Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. Science 2018, 359, 915–919. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.A.; Alam, A.; Kowal, J.; Stieger, B.; Locher, K.P. Structure of the human lipid exporter ABCB4 in a lipid environment. Nat. Struct Mol. Biol. 2020, 27, 62–70. [Google Scholar] [CrossRef]
- Wang, L.; Hou, W.T.; Wang, J.; Xu, D.; Guo, C.; Sun, L.; Ruan, K.; Zhou, C.Z.; Chen, Y. Structures of human bile acid exporter ABCB11 reveal a transport mechanism facilitated by two tandem substrate-binding pockets. Cell Res. 2022, 32, 501–504. [Google Scholar] [CrossRef] [PubMed]
- Aleksandrov, A.A.; Kota, P.; Cui, L.; Jensen, T.; Alekseev, A.E.; Reyes, S.; He, L.; Gentzsch, M.; Aleksandrov, L.A.; Dokholyan, N.V.; et al. Allosteric modulation balances thermodynamic stability and restores function of DF508 CFTR. J. Mol. Biol. 2012, 419, 41–60. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Pissarra, L.S.; Farinha, C.M.; Liu, J.; Cai, Z.; Thibodeau, P.H.; Amaral, M.D.; Sheppard, D.N. Revertant mutants modify, but do not rescue, the gating defect of the cystic fibrosis mutant G551D-CFTR. J. Physiol. 2014, 592, 1931–1947. [Google Scholar] [CrossRef] [PubMed]
- Roxo-Rosa, M.; Xu, Z.; Schmidt, A.; Neto, M.; Cai, Z.; Soares, C.M.; Sheppard, D.N.; Amaral, M.D. Revertant mutants G550E and 4RK rescue cystic fibrosis mutants in the first nucleotide-binding domain of CFTR by different mechanisms. Proc. Natl. Acad. Sci. USA 2006, 103, 17891–17896. [Google Scholar] [CrossRef]
- Fanen, P.; Clain, J.; Labarthe, R.; Hulin, P.; Girodon, E.; Pagesy, P.; Goossens, M.; Edelman, A. Structure-function analysis of a double-mutant cystic fibrosis transmembrane conductance regulator protein occurring in disorders related to cystic fibrosis. FEBS Lett. 1999, 452, 371–374. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- MacKerell, A.J.; Feig, M.; Brooks, C.I. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comp. Chem. 2004, 25, 1400–1415. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511–519. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.; Fraaije, J.G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baatallah, N.; Elbahnsi, A.; Chevalier, B.; Castanier, S.; Mornon, J.-P.; Pranke, I.; Edelman, A.; Sermet-Gaudelus, I.; Callebaut, I.; Hinzpeter, A. Acting on the CFTR Membrane-Spanning Domains Interface Rescues Some Misfolded Mutants. Int. J. Mol. Sci. 2022, 23, 16225. https://doi.org/10.3390/ijms232416225
Baatallah N, Elbahnsi A, Chevalier B, Castanier S, Mornon J-P, Pranke I, Edelman A, Sermet-Gaudelus I, Callebaut I, Hinzpeter A. Acting on the CFTR Membrane-Spanning Domains Interface Rescues Some Misfolded Mutants. International Journal of Molecular Sciences. 2022; 23(24):16225. https://doi.org/10.3390/ijms232416225
Chicago/Turabian StyleBaatallah, Nesrine, Ahmad Elbahnsi, Benoit Chevalier, Solène Castanier, Jean-Paul Mornon, Iwona Pranke, Aleksander Edelman, Isabelle Sermet-Gaudelus, Isabelle Callebaut, and Alexandre Hinzpeter. 2022. "Acting on the CFTR Membrane-Spanning Domains Interface Rescues Some Misfolded Mutants" International Journal of Molecular Sciences 23, no. 24: 16225. https://doi.org/10.3390/ijms232416225
APA StyleBaatallah, N., Elbahnsi, A., Chevalier, B., Castanier, S., Mornon, J.-P., Pranke, I., Edelman, A., Sermet-Gaudelus, I., Callebaut, I., & Hinzpeter, A. (2022). Acting on the CFTR Membrane-Spanning Domains Interface Rescues Some Misfolded Mutants. International Journal of Molecular Sciences, 23(24), 16225. https://doi.org/10.3390/ijms232416225





