Mapping PP1c and Its Inhibitor 2 Interactomes Reveals Conserved and Specific Networks in Asexual and Sexual Stages of Plasmodium
Abstract
:1. Introduction
2. Results and Discussion
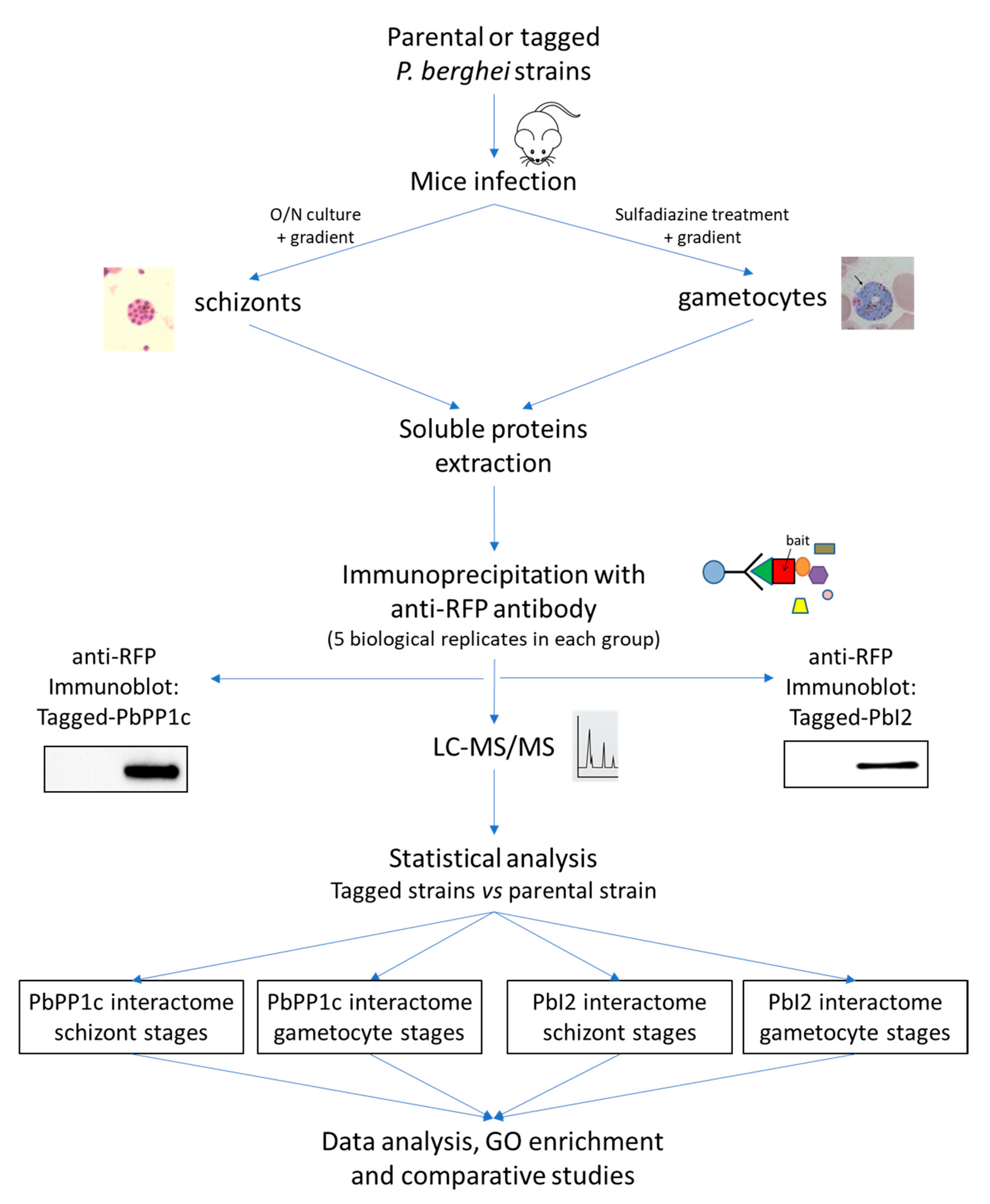
2.1. Transgenic P. berghei Lines Expressing Tagged Baits
2.2. Identification of PbPP1c-Interacting Proteins in Schizonts and Gametocytes
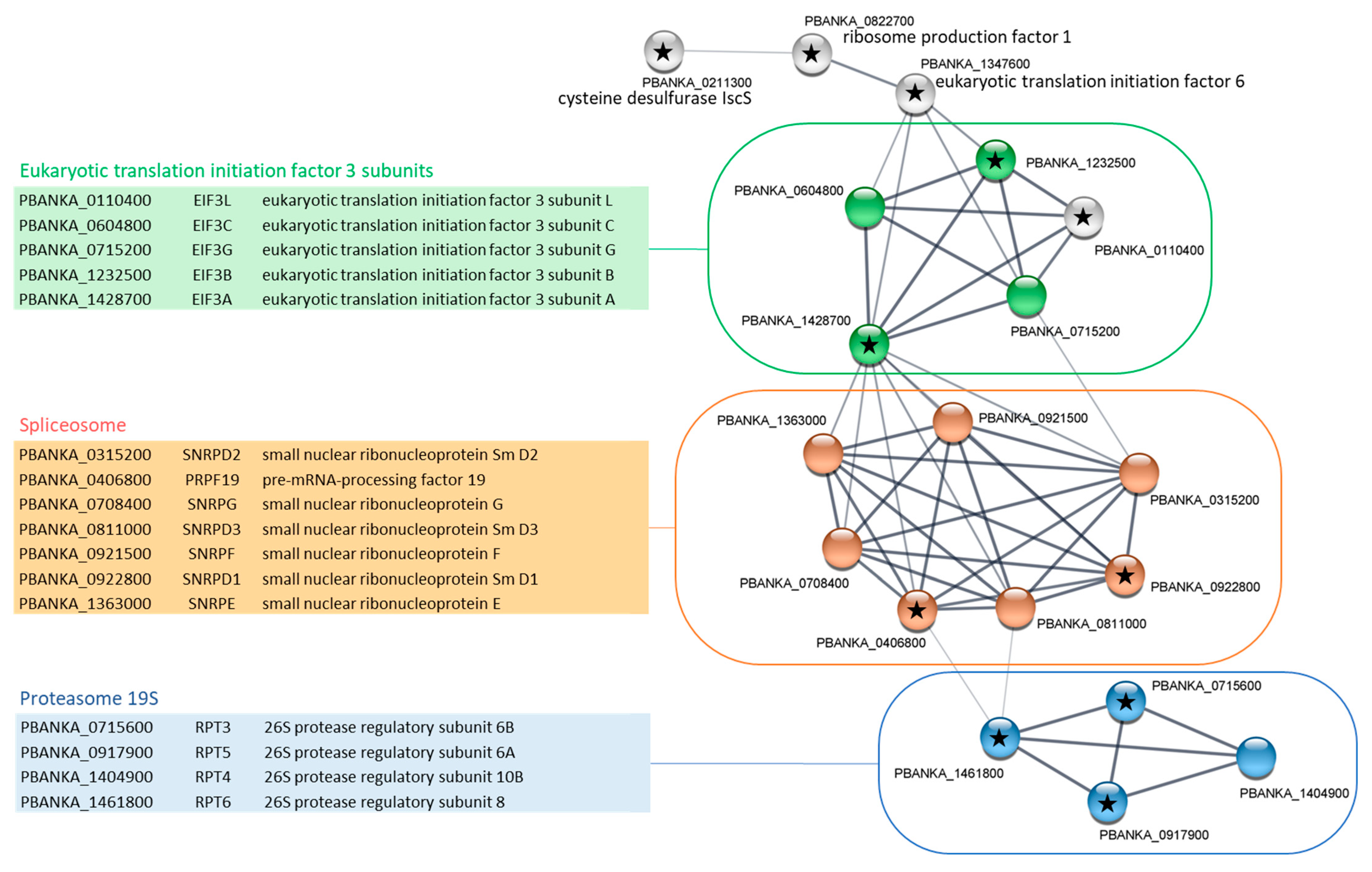
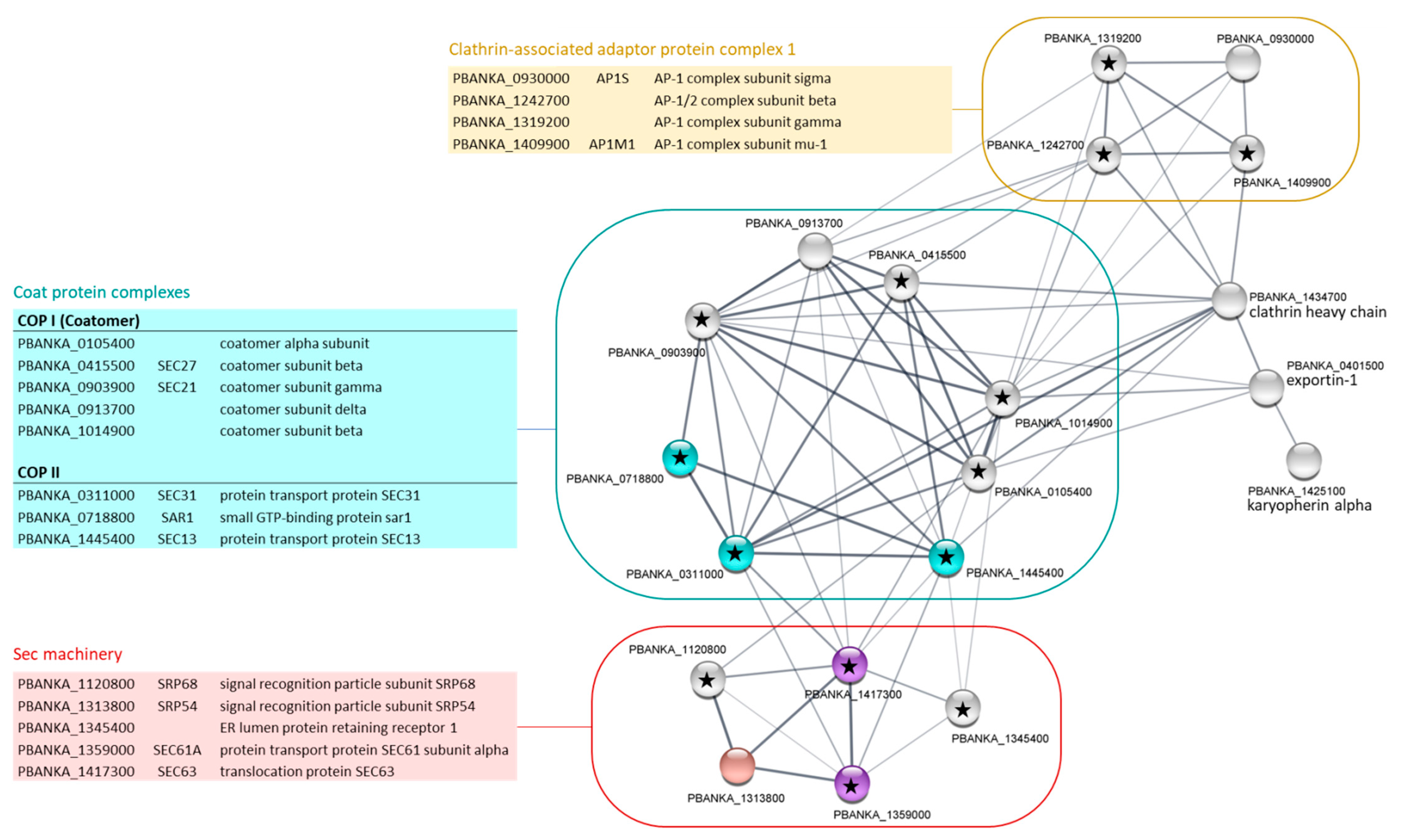
2.3. Gene Ontology Enrichment Analyses of PbPP1c-Interacting Proteins
2.4. Proteomic Identification of PbI2 Interactions in Schizonts and Gametocytes
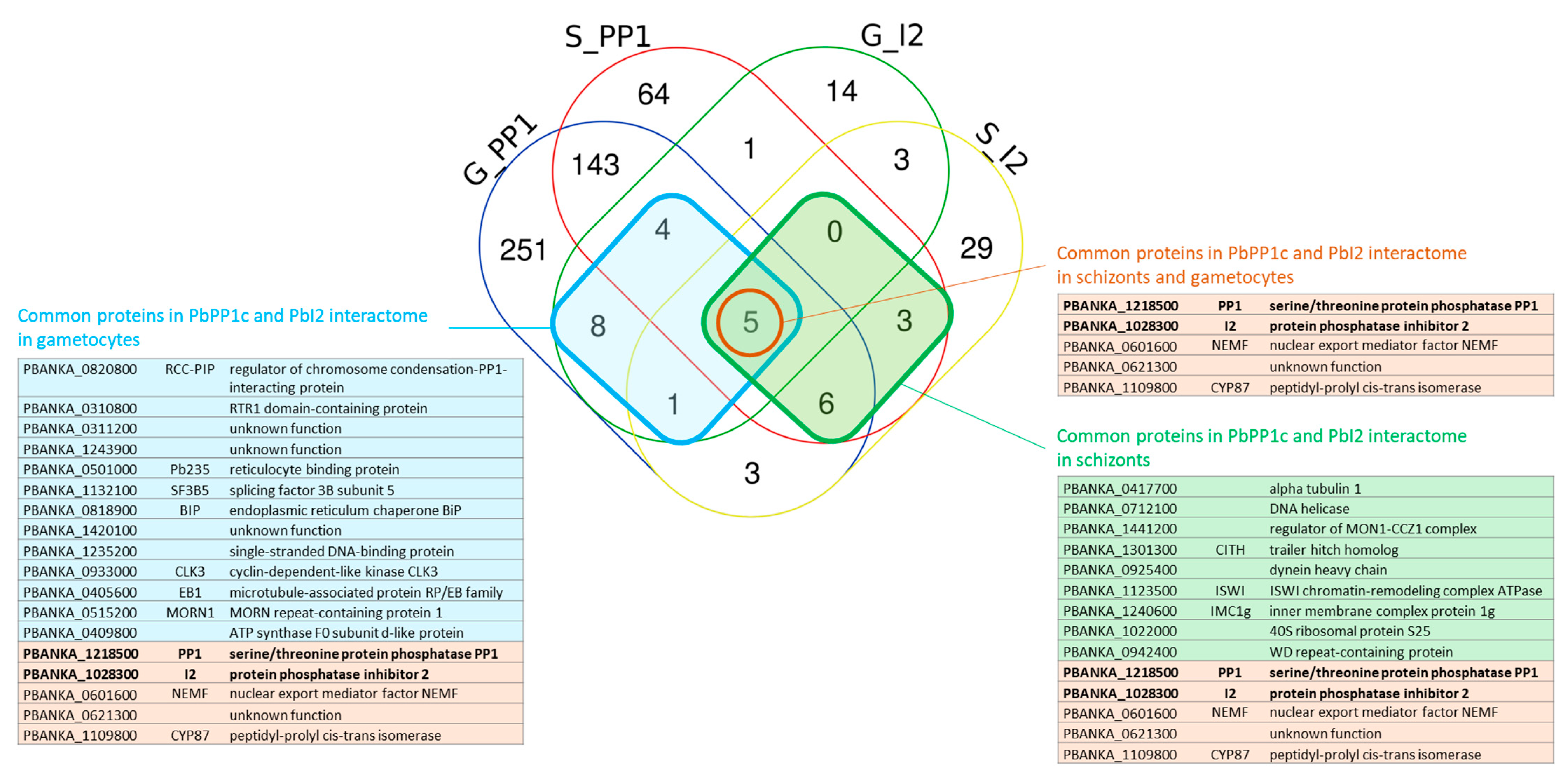
2.5. Identification of Shared Proteins and Interactions of PP1c and I2 in Schizonts and Gametocytes
2.6. Conclusions
3. Materials and Methods
3.1. Ethics Statement
3.2. Materials
3.3. Generation of P. berghei Transgenic Parasites
3.4. Isolation of P. berghei Schizonts and Gametocytes
3.5. Immunoblot Assays
3.6. Sample Preparation and Immunoprecipitation
3.7. Sample Preparation for Interactome Analysis
3.8. NanoLC-MS/MS Protein Identification and Quantification
3.9. Proteome Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Menard, D.; Dondorp, A. Antimalarial Drug Resistance: A Threat to Malaria Elimination. Cold Spring Harb. Perspect. Med. 2017, 7, a025619. [Google Scholar] [CrossRef] [Green Version]
- Casamayor, A.; Ariño, J. Controlling Ser/Thr protein phosphatase PP1 activity and function through interaction with regulatory subunits. Adv. Protein Chem. Struct. Biol. 2020, 122, 231–288. [Google Scholar]
- Barker, H.M.; Brewis, N.D.; Street, A.J.; Spurr, N.K.; Cohen, P.T.W. Three genes for protein phosphatase 1 map to different human chromosomes: Sequence, expression and gene localisation of protein serine/threonine phosphatase 1 beta (PPP1CB). Biochim. Biophys. Acta 1994, 1220, 212–218. [Google Scholar] [CrossRef]
- Bhattacharyya, M.K.; Hong, Z.; Kongkasuriyachai, D.; Kumar, N. Plasmodium falciparum protein phosphatase type 1 functionally complements a glc7 mutant in Saccharomyces cerevisiae. Int. J. Parasitol. 2002, 32, 739–747. [Google Scholar] [CrossRef]
- Yokoyama, D.; Saito-Ito, A.; Asao, N.; Tanabe, K.; Yamamoto, M.; Matsumura, T. Modulation of the growth of Plasmodium falciparum in vitro by protein serine/threonine phosphatase inhibitors. Biochem. Biophys. Res. Commun. 1998, 247, 18–23. [Google Scholar] [CrossRef]
- Paul, A.S.; Miliu, A.; Paulo, J.A.; Goldberg, J.M.; Bonilla, A.M.; Berry, L.; Séveno, M.; Braun-Breton, C.; Kosber, A.L.; Elsworth, B.; et al. Co-option of Plasmodium falciparum PP1 for egress from host erythrocytes. Nat. Commun. 2020, 11, 3532. [Google Scholar] [CrossRef]
- Choy, M.S.; Hieke, M.; Kumar, G.S.; Lewis, G.R.; Gonzalez-DeWhitt, K.R.; Kessler, R.P.; Stein, B.J.; Hessenberger, M.; Nairn, A.C.; Peti, W.; et al. Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code. Proc. Natl. Acad. Sci. USA 2014, 111, 4097–4102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendrickx, A.; Beullens, M.; Ceulemans, H.; Den Abt, T.; Van Eynde, A.; Nicolaescu, E.; Lesage, B.; Bollen, M. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 2009, 16, 365–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heroes, E.; Lesage, B.; Gornemann, J.; Beullens, M.; Van Meervelt, L.; Bollen, M. The PP1 binding code: A molecular-lego strategy that governs specificity. FEBS J. 2013, 280, 584–595. [Google Scholar] [CrossRef]
- Moorhead, G.B.G.; Trinkle-Mulcahy, L.; Nimick, M.; De Wever, V.; Campbell, D.G.; Gourlay, R.; Lam, Y.W.; Lamond, A.I. Displacement affinity chromatography of protein phosphatase one (PP1) complexes. BMC Biochem. 2008, 9, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollin, T.; De Witte, C.; Lenne, A.; Pierrot, C.; Khalife, J. Analysis of the interactome of the Ser/Thr Protein Phosphatase type 1 in Plasmodium falciparum. BMC Genom. 2016, 17, 246. [Google Scholar] [CrossRef] [Green Version]
- Freville, A.; Cailliau-Maggio, K.; Pierrot, C.; Tellier, G.; Kalamou, H.; Lafitte, S.; Martoriati, A.; Pierce, R.J.; Bodart, J.F.; Khalife, J. Plasmodium falciparum encodes a conserved active inhibitor-2 for Protein Phosphatase type 1: Perspectives for novel anti-plasmodial therapy. BMC Biol. 2013, 11, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierrot, C.; Zhang, X.; Zhangi, G.; Freville, A.; Rebollo, A.; Khalife, J. Peptides derived from Plasmodium falciparum leucine-rich repeat 1 bind to serine/threonine phosphatase type 1 and inhibit parasite growth in vitro. Drug Des. Dev. Ther. 2018, 12, 85–88. [Google Scholar] [CrossRef] [Green Version]
- Hollin, T.; De Witte, C.; Freville, A.; Guerrera, I.C.; Chhuon, C.; Saliou, J.M.; Herbert, F.; Pierrot, C.; Khalife, J. Essential role of GEXP15, a specific Protein Phosphatase type 1 partner, in Plasmodium berghei in asexual erythrocytic proliferation and transmission. PLoS Pathog. 2019, 15, e1007973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeeshan, M.; Pandey, R.; Subudhi, A.K.; Ferguson, D.J.P.; Kaur, G.; Rashpa, R.; Nugmanova, R.; Brady, D.; Bottrill, A.R.; Vaughan, S.; et al. Protein phosphatase 1 regulates atypical mitotic and meiotic division in Plasmodium sexual stages. Commun. Biol. 2021, 4, 760. [Google Scholar] [CrossRef] [PubMed]
- Daher, W.; Browaeys, E.; Pierrot, C.; Jouin, H.; Dive, D.; Meurice, E.; Dissous, C.; Capron, M.; Tomavo, S.; Doerig, C.; et al. Regulation of protein phosphatase type 1 and cell cycle progression by PfLRR1, a novel leucine-rich repeat protein of the human malaria parasite Plasmodium falciparum. Mol. Microbiol. 2006, 60, 578–590. [Google Scholar] [CrossRef]
- Freville, A.; Landrieu, I.; Garcia-Gimeno, M.A.; Vicogne, J.; Montbarbon, M.; Bertin, B.; Verger, A.; Kalamou, H.; Sanz, P.; Werkmeister, E.; et al. Plasmodium falciparum inhibitor-3 homolog increases protein phosphatase type 1 activity and is essential for parasitic survival. J. Biol. Chem. 2012, 287, 1306–1321. [Google Scholar] [CrossRef] [Green Version]
- Lenne, A.; De Witte, C.; Tellier, G.; Hollin, T.; Aliouat, E.M.; Martoriati, A.; Cailliau, K.; Saliou, J.-M.; Khalife, J.; Pierrot, C. Characterization of a Protein Phosphatase Type-1 and a Kinase Anchoring Protein in Plasmodium falciparum. Front. Microbiol. 2018, 9, 2617. [Google Scholar] [CrossRef]
- Wakula, P.; Beullens, M.; Ceulemans, H.; Stalmans, W.; Bollen, M. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J. Biol. Chem. 2003, 278, 18817–18823. [Google Scholar] [CrossRef] [Green Version]
- Beullens, M.; Stalmans, W.; Bollen, M. Characterization of a Ribosomal Inhibitory Polypeptide of Protein Phosphatase-1 from Rat Liver. Eur. J. Biochem. FEBS 1996, 239, 183–189. [Google Scholar] [CrossRef]
- Hirano, K.; Ito, M.; Hartshorne, D.J. Interaction of the Ribosomal Protein, L5, with Protein Phosphatase Type 1. J. Biol. Chem. 1995, 270, 19786–19790. [Google Scholar] [CrossRef] [Green Version]
- Hutchinson, J.A.; Shanware, N.P.; Chang, H.; Tibbetts, R.S. Regulation of ribosomal protein S6 phosphorylation by casein kinase 1 and protein phosphatase 1. J. Biol. Chem. 2011, 286, 8688–8696. [Google Scholar] [CrossRef] [Green Version]
- Chamousset, D.; De Wever, V.; Moorhead, G.B.; Chen, Y.; Boisvert, F.-M.; Lamond, A.I.; Trinkle-Mulcahy, L. RRP1B targets PP1 to mammalian cell nucleoli and is associated with Pre-60S ribosomal subunits. Mol. Biol. Cell 2010, 21, 4212–4226. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Hu, Y.; Huang, P.; Toleman, C.A.; Paterson, A.J.; Kudlow, J.E. Proteasome Function Is Regulated by Cyclic AMP-dependent Protein Kinase through Phosphorylation of Rpt6. J. Biol. Chem. 2007, 282, 22460–22471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Reddy, B.; Manley, J.L. PP1/PP2A Phosphatases Are Required for the Second Step of Pre-mRNA Splicing and Target Specific snRNP Proteins. Mol. Cell 2006, 23, 819–829. [Google Scholar] [CrossRef]
- Hillier, C.; Pardo, M.; Yu, L.; Bushell, E.; Sanderson, T.; Metcalf, T.; Herd, C.; Anar, B.; Rayner, J.C.; Billker, O.; et al. Landscape of the Plasmodium Interactome Reveals Both Conserved and Species-Specific Functionality. Cell Rep. 2019, 28, 1635–1647.e1635. [Google Scholar] [CrossRef] [Green Version]
- Choy, M.S.; Yusoff, P.; Lee, I.C.; Newton, J.C.; Goh, C.W.; Page, R.; Shenolikar, S.; Peti, W. Structural and Functional Analysis of the GADD34:PP1 eIF2α Phosphatase. Cell Rep. 2015, 11, 1885–1891. [Google Scholar] [CrossRef] [Green Version]
- Tellier, G.; Lenne, A.; Cailliau-Maggio, K.; Cabezas-Cruz, A.; Valdes, J.J.; Martoriati, A.; Aliouat el, M.; Gosset, P.; Delaire, B.; Freville, A.; et al. Identification of Plasmodium falciparum Translation Initiation eIF2beta Subunit: Direct Interaction with Protein Phosphatase Type 1. Front. Microbiol. 2016, 7, 777. [Google Scholar] [CrossRef] [PubMed]
- Wakula, P.; Beullens, M.; van Eynde, A.; Ceulemans, H.; Stalmans, W.; Bollen, M. The translation initiation factor eIF2beta is an interactor of protein phosphatase-1. Biochem. J. 2006, 400, 377–383. [Google Scholar] [CrossRef]
- Kaderi Kibria, K.M.; Rawat, K.; Klinger, C.M.; Datta, G.; Panchal, M.; Singh, S.; Iyer, G.R.; Kaur, I.; Sharma, V.; Dacks, J.B.; et al. A role for adaptor protein complex 1 in protein targeting to rhoptry organelles in Plasmodium falciparum. Biochim. Biophys. Acta 2015, 1853, 699–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adisa, A.; Albano, F.R.; Reeder, J.; Foley, M.; Tilley, L. Evidence for a role for a Plasmodium falciparum homologue of Sec31p in the export of proteins to the surface of malaria parasite-infected erythrocytes. J. Cell Sci. 2001, 114, 3377–3386. [Google Scholar] [CrossRef] [PubMed]
- Albano, F.R.; Berman, A.; La Greca, N.; Hibbs, A.R.; Wickham, M.; Foley, M.; Tilley, L. A homologue of Sar1p localises to a novel trafficking pathway in malaria-infected erythrocytes. Eur. J. Cell Biol. 1999, 78, 453–462. [Google Scholar] [CrossRef]
- Adisa, A.; Rug, M.; Foley, M.; Tilley, L. Characterisation of a δ-COP homologue in the malaria parasite, Plasmodium falciparum. Mol. Biochem. Parasitol. 2002, 123, 11–21. [Google Scholar] [CrossRef]
- Luo, P.M.; Boyce, M. Directing Traffic: Regulation of COPI Transport by Post-translational Modifications. Front. Cell Dev. Biol. 2019, 7, 190. [Google Scholar] [CrossRef]
- Sheff, D.; Lowe, M.; Kreis, T.E.; Mellman, I. Biochemical Heterogeneity and Phosphorylation of Coatomer Subunits. J. Biol. Chem. 1996, 271, 7230–7236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]


| GO ID | GO Term | Number of Genes Detected 1 | Fold Enrichment | p-Value | Benjamini FDR | Bonferroni Adjusted p-Value |
|---|---|---|---|---|---|---|
| GO:0022607 | Cellular component assembly | 20/108 | 2.6 | 4.63 × 10−5 | 2.55 × 10−4 | 1.53 × 10−3 |
| GO:0065003 | Protein-containing complex assembly | 18/80 | 3.16 | 6.93 × 10−6 | 4.57 × 10−5 | 2.29 × 10−4 |
| GO:0022618 | Ribonucleoprotein complex assembly | 13/43 | 4.25 | 4.39 × 10−6 | 3.62 × 10−5 | 1.45 × 10−4 |
| GO:0009058 | Biosynthetic process | 71/531 | 1.88 | 4.53 × 10−9 | 7.47 × 10−8 | 1.49 × 10−7 |
| GO:0034641 | Cellular nitrogen compound metabolic process | 87/756 | 1.62 | 7.49 × 10−8 | 8.24 × 10−7 | 2.47 × 10−6 |
| GO:0006412 | Translation | 57/232 | 3.45 | 4.86 × 10−19 | 1.60 × 10−17 | 1.60 × 10−17 |
| GO ID | GO Term | Number of Genes Detected 1 | Fold Enrichment | p-Value | Benjamini FDR | Bonferroni Adjusted p-Value |
|---|---|---|---|---|---|---|
| GO:0022607 | Cellular component assembly | 27/108 | 2.18 | 4.48 × 10−5 | 1.04 × 10−3 | 1.88 × 10−3 |
| GO:0065003 | Protein-containing complex assembly | 22/80 | 2.4 | 4.97 × 10−5 | 1.04 × 10−3 | 2.09 × 10−3 |
| GO:0022618 | Ribonucleoprotein complex assembly | 14/43 | 2.84 | 1.75 × 10−4 | 2.45 × 10−3 | 7.35 × 10−3 |
| GO:0015031 | Protein transport | 24/109 | 1.92 | 9.32 × 10−4 | 9.79 × 10−3 | 3.91 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Witte, C.; Aliouat, E.M.; Chhuon, C.; Guerrera, I.C.; Pierrot, C.; Khalife, J. Mapping PP1c and Its Inhibitor 2 Interactomes Reveals Conserved and Specific Networks in Asexual and Sexual Stages of Plasmodium. Int. J. Mol. Sci. 2022, 23, 1069. https://doi.org/10.3390/ijms23031069
De Witte C, Aliouat EM, Chhuon C, Guerrera IC, Pierrot C, Khalife J. Mapping PP1c and Its Inhibitor 2 Interactomes Reveals Conserved and Specific Networks in Asexual and Sexual Stages of Plasmodium. International Journal of Molecular Sciences. 2022; 23(3):1069. https://doi.org/10.3390/ijms23031069
Chicago/Turabian StyleDe Witte, Caroline, El Moukhtar Aliouat, Cerina Chhuon, Ida Chiara Guerrera, Christine Pierrot, and Jamal Khalife. 2022. "Mapping PP1c and Its Inhibitor 2 Interactomes Reveals Conserved and Specific Networks in Asexual and Sexual Stages of Plasmodium" International Journal of Molecular Sciences 23, no. 3: 1069. https://doi.org/10.3390/ijms23031069
APA StyleDe Witte, C., Aliouat, E. M., Chhuon, C., Guerrera, I. C., Pierrot, C., & Khalife, J. (2022). Mapping PP1c and Its Inhibitor 2 Interactomes Reveals Conserved and Specific Networks in Asexual and Sexual Stages of Plasmodium. International Journal of Molecular Sciences, 23(3), 1069. https://doi.org/10.3390/ijms23031069






