The KRAB Domain of ZNF10 Guides the Identification of Specific Amino Acids That Transform the Ancestral KRAB-A-Related Domain Present in Human PRDM9 into a Canonical Modern KRAB-A Domain
Abstract
:1. Introduction
2. Results
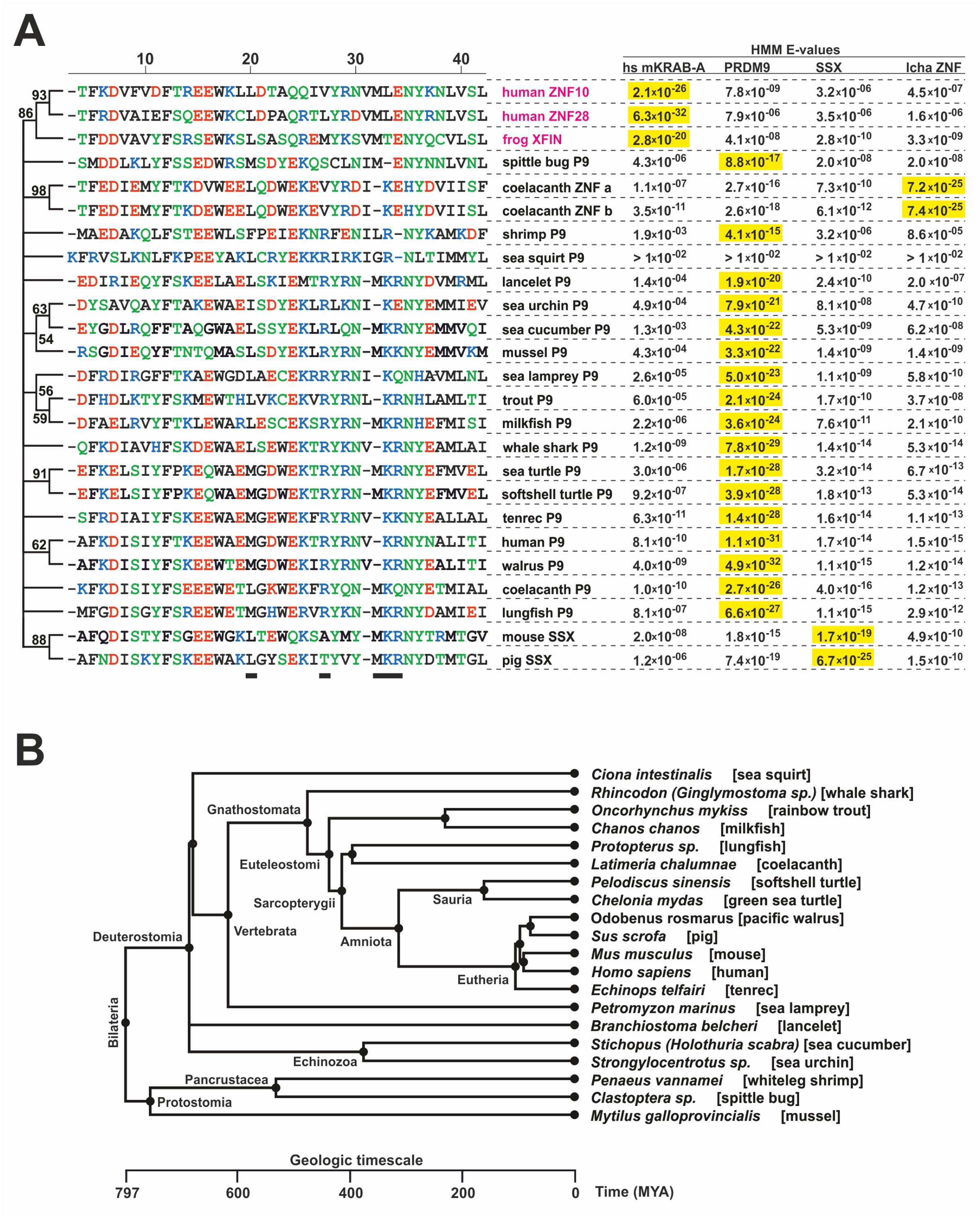
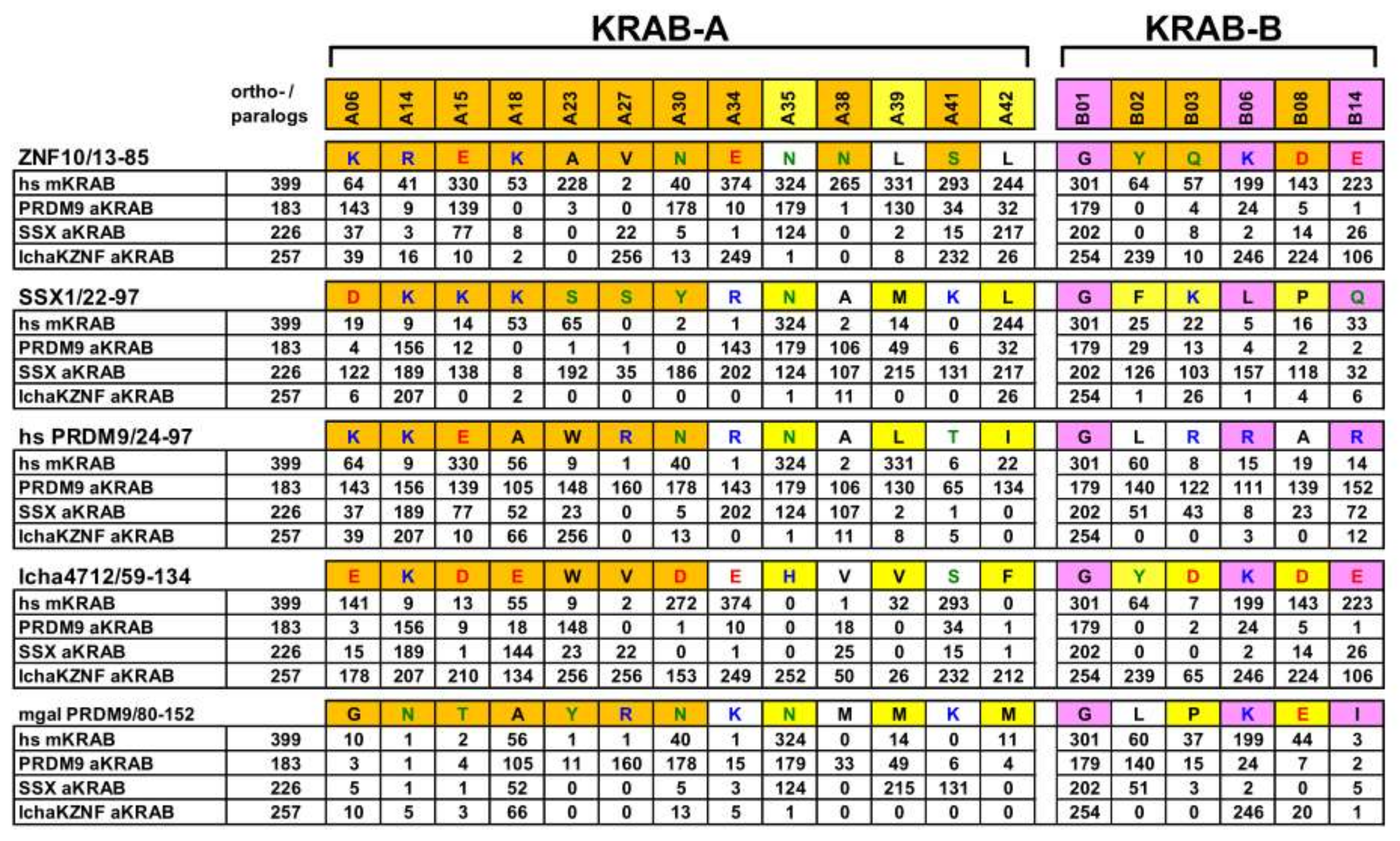
2.1. Consolidation of the Ancestral KRAB Domain (aKRAB)
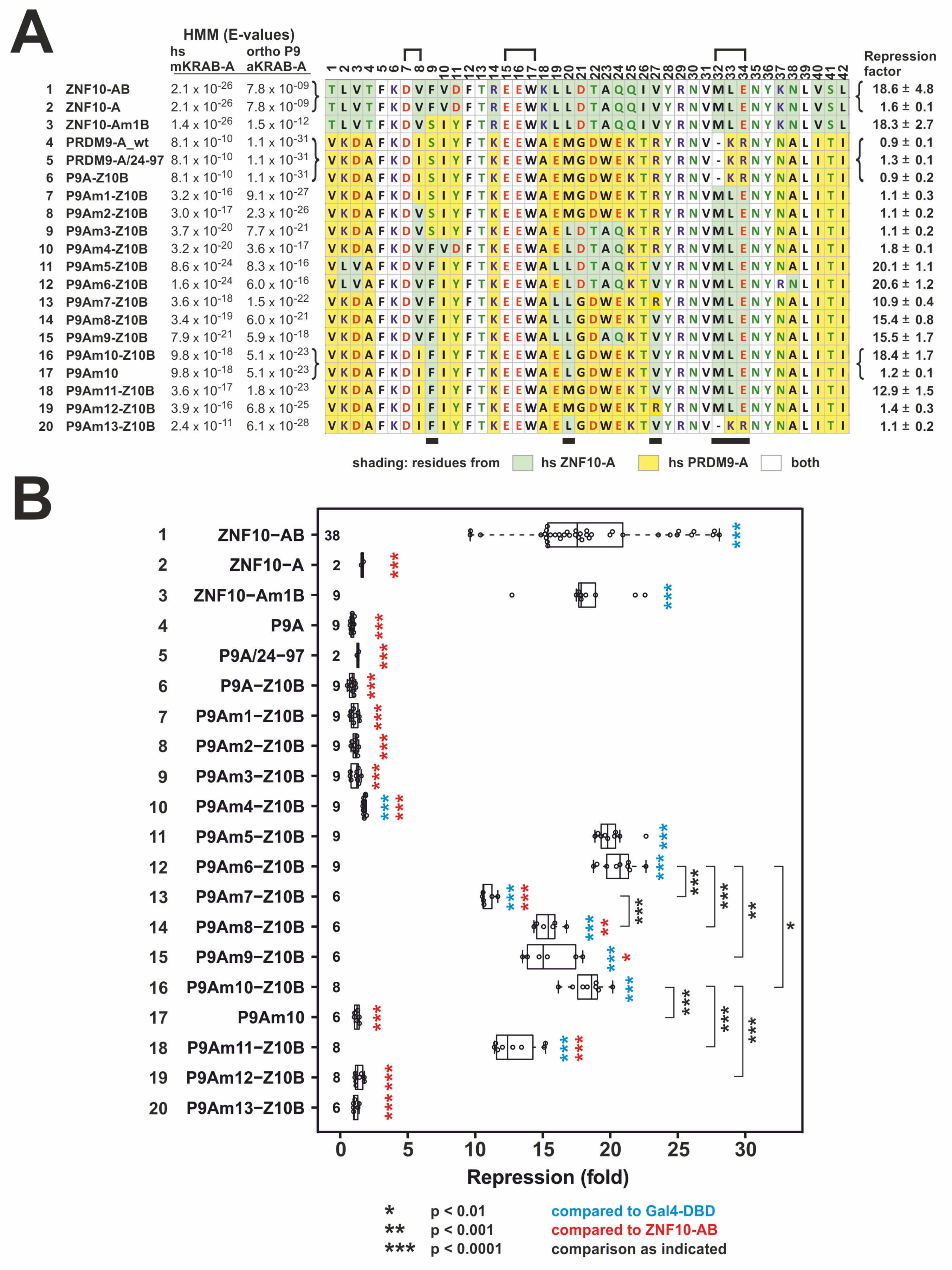
2.2. Transformation of PRDM9 aKRAB-A into a Modern Canonical KRAB
2.3. Heterologous Reporter Assay for Transcriptional Repression
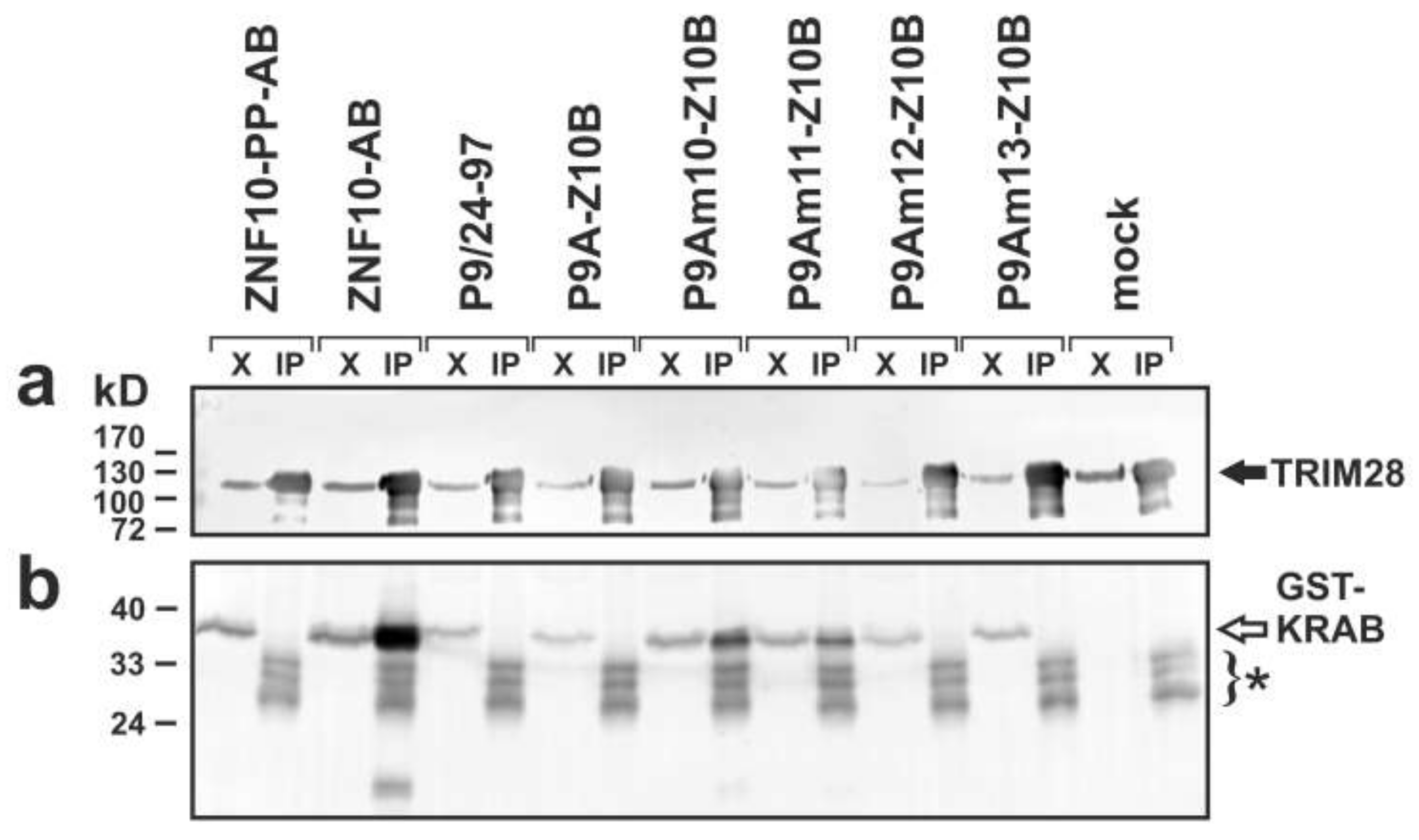
2.4. Complex Formation with TRIM28
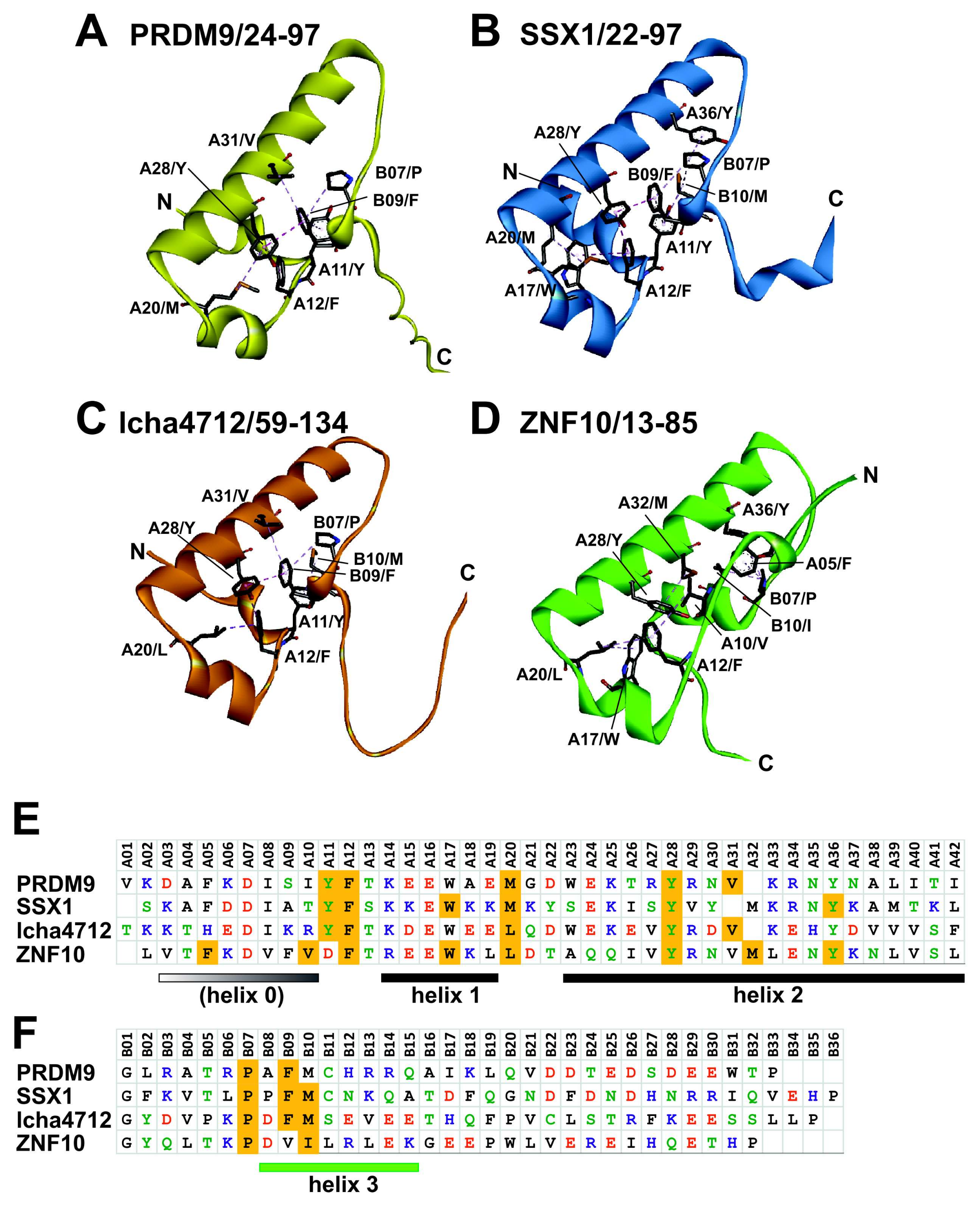
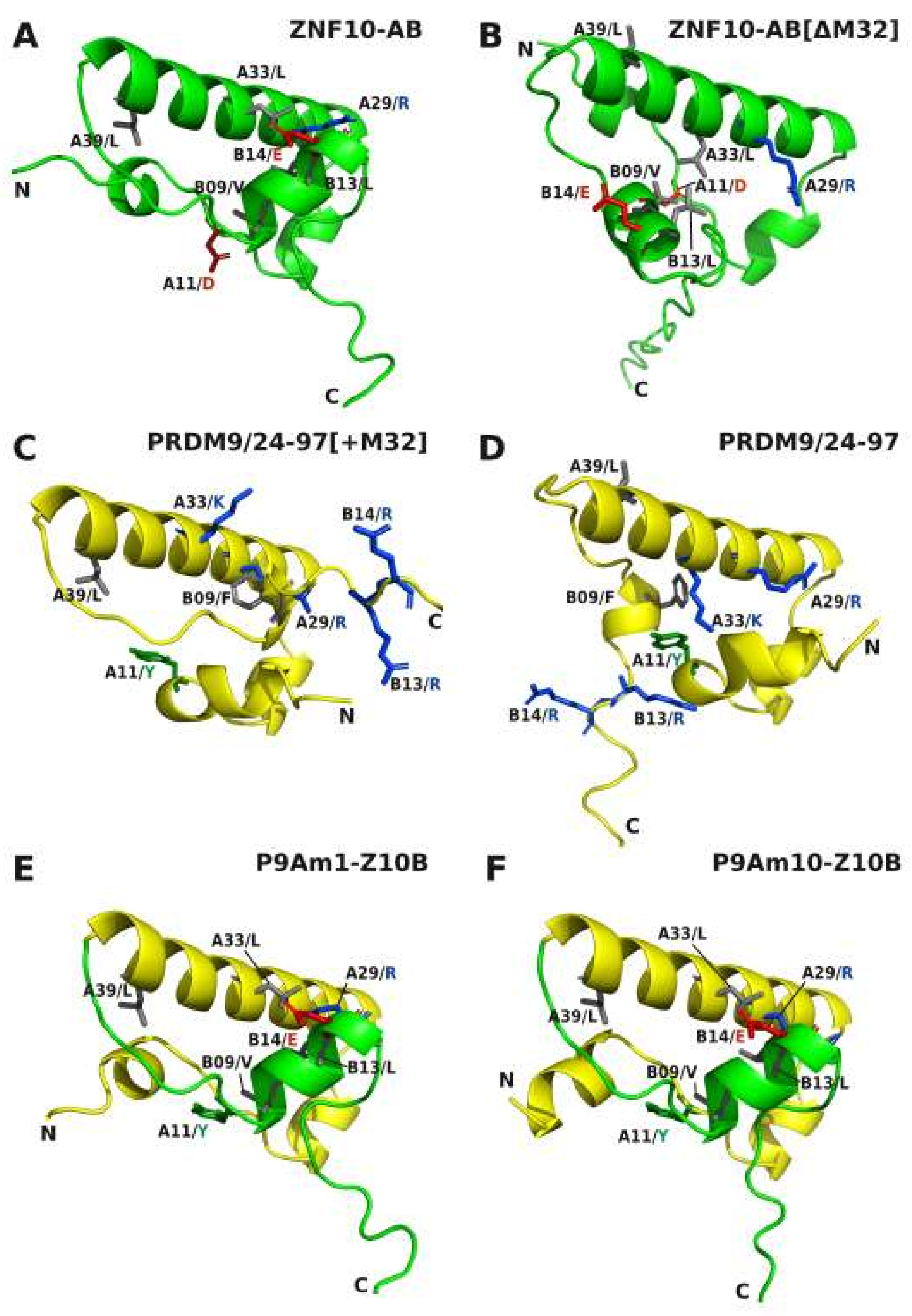
2.5. Comparative Topologies of Ancestral versus Modern KRAB Domains
3. Discussion
4. Methods
4.1. Compilation and Computational Analysis of Sequences
4.2. Sequence Logos and Tree Construction
4.3. Protein 3-D Structure Models
4.4. Cell Culture and Transfection
4.5. Immunoprecipitation and Western Blotting
4.6. Luciferase Reporter Assays
4.7. DNA Constructs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GST | Glutathione S-transferase |
| KO | Knockout |
| KRAB | Krüppel-associated box |
| aKRAB | Ancestral KRAB |
| mKRAB | Modern KRAB |
| KZNF | KRAB zinc finger |
| PRDM | PR/SET Domain |
| SETDB1 | SET domain bifurcated histone lysine methyltransferase 1 |
| SSX | Synovial sarcoma X breakpoint |
| TRIM28 | Tripartite motif containing 28 |
| ZNF | C2H2 zinc finger |
References
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 175, 598–599. [Google Scholar] [CrossRef] [Green Version]
- Vaquerizas, J.M.; Kummerfeld, S.K.; Teichmann, S.A. Luscombe NM. A census of human transcription factors: Function, expression and evolution. Nat. Rev. Genet. 2009, 10, 252–263. [Google Scholar] [CrossRef]
- Ninova, M.; Fejes Toth, K.; Aravin, A.A. The control of gene expression and cell identity by H3K9 trimethylation. Development 2019, 146, dev181180. [Google Scholar] [CrossRef] [Green Version]
- Schlesinger, S.; Meshorer, E. Open Chromatin, Epigenetic Plasticity, and Nuclear Organization in Pluripotency. Dev. Cell 2019, 48, 135–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, T.I.; Young, R.A. Transcriptional regulation and its misregulation in disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segert, J.A.; Gisselbrecht, S.S.; Bulyk, M.L. Transcriptional Silencers: Driving Gene Expression with the Brakes On. Trends in genetics. TIG 2021, 37, 514–527. [Google Scholar] [CrossRef]
- Perissi, V.; Jepsen, K.; Glass, C.K.; Rosenfeld, M.G. Deconstructing repression: Evolving models of co-repressor action. Nat. Rev. Genet. 2010, 11, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Ooi, L.; Wood, I.C. Chromatin crosstalk in development and disease: Lessons from REST. Nat. Rev. Genet. 2007, 8, 544–554. [Google Scholar] [CrossRef]
- Thiesen, H.J. Multiple genes encoding zinc finger domains are expressed in human T cells. New Biol. 1990, 2, 363–374. [Google Scholar]
- Emerson, R.O.; Thomas, J.H. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009, 5, e1000325. [Google Scholar] [CrossRef] [Green Version]
- Imbeault, M.; Helleboid, P.Y.; Trono, D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 2017, 543, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chang, L.H.; Sun, Y.; Lu, X.; Stubbs, L. Deep vertebrate roots for mammalian zinc finger transcription factor subfamilies. Genome Biol. Evol. 2014, 6, 510–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birtle, Z.; Ponting, C.P. Meisetz and the birth of the KRAB motif. Bioinformatics 2006, 22, 2841–2845. [Google Scholar] [CrossRef] [PubMed]
- Baker, Z.; Schumer, M.; Haba, Y.; Bashkirova, L.; Holland, C.; Rosenthal, G.G.; Przeworski, M. Repeated losses of PRDM9-directed recombination despite the conservation of PRDM9 across vertebrates. eLife 2017, 6, e24133. [Google Scholar] [CrossRef]
- Deuschle, U.; Meyer, W.K.; Thiesen, H.J. Tetracycline-reversible silencing of eukaryotic promoters. Mol. Cell Biol. 1995, 15, 1907–1914. [Google Scholar] [CrossRef] [Green Version]
- Ecco, G.; Imbeault, M.; Trono, D. KRAB zinc finger proteins. Development 2017, 144, 2719–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lupo, A.; Cesaro, E.; Montano, G.; Zurlo, D.; Izzo, P.; Costanzo, P. KRAB-Zinc Finger Proteins: A Repressor Family Displaying Multiple Biological Functions. Curr. Genom. 2013, 14, 268–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.T.; Kuo, C.Y.; Ann, D.K. KAPtain in charge of multiple missions: Emerging roles of KAP1. World J. Biol. Chem. 2014, 5, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Iyengar, S.; Farnham, P.J. KAP1 protein: An enigmatic master regulator of the genome. J. Biol. Chem. 2011, 286, 26267–26276. [Google Scholar] [CrossRef] [Green Version]
- Cesaro, E.; Lupo, A.; Rapuano, R.; Pastore, A.; Grosso, M.; Costanzo, P. ZNF224 Protein: Multifaceted Functions Based on Its Molecular Partners. Molecules 2021, 26, 6296. [Google Scholar] [CrossRef]
- Czerwinska, P.; Mazurek, S.; Wiznerowicz, M. The complexity of TRIM28 contribution to cancer. J. Biomed. Sci. 2017, 24, 63. [Google Scholar] [CrossRef] [PubMed]
- Sobocinska, J.; Molenda, S.; Machnik, M.; Oleksiewicz, U. KRAB-ZFP Transcriptional Regulators Acting as Oncogenes and Tumor Suppressors: An Overview. Int. J. Mol. Sci. 2021, 22, 2212. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.H.; Schneider, S. Coevolution of retroelements and tandem zinc finger genes. Genome Res. 2011, 21, 1800–1812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, G.; de Iaco, A.; Sun, M.A.; Bruno, M.; Tinkham, M.; Hoang, D.; Mitra, A.; Ralls, S.; Trono, D.; Macfarlan, T.S. KRAB-zinc finger protein gene expansion in response to active retrotransposons in the murine lineage. eLife 2020, 9, e56337. [Google Scholar] [CrossRef]
- Bruno, M.; Mahgoub, M.; Macfarlan, T.S. The Arms Race Between KRAB-Zinc Finger Proteins and Endogenous Retroelements and Its Impact on Mammals. Annu. Rev. Genet. 2019, 53, 393–416. [Google Scholar] [CrossRef]
- Pontis, J.; Planet, E.; Offner, S.; Turelli, P.; Duc, J.; Coudray, A.; Theunissen, T.W.; Jaenisch, R.; Trono, D. Hominoid-Specific Transposable Elements and KZFPs Facilitate Human Embryonic Genome Activation and Control Transcription in Naive Human ESCs. Cell Stem. Cell 2019, 24, 724–735.e5. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacLeod, R.S.; Cawley, K.M.; Gubrij, I.; Nookaew, I.; Onal, M.; O’Brien, C.A. Effective CRISPR interference of an endogenous gene via a single transgene in mice. Sci. Rep. 2019, 9, 17312. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, E.; Prado Balcazar, J.; Wu, Z.; Xiang, K.; Wang, Y.; Huang, Q.; Negrete, M.; Chen, K.Y.; Li, W.; et al. Chromatin Remodeling of Colorectal Cancer Liver Metastasis is Mediated by an HGF-PU.1-DPP4 Axis. Adv. Sci. 2021, 8, e2004673. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, L.; Turkoz, A.; Chen, P.; Harari-Steinfeld, R.; Bobbin, M.; Stefanson, O.; Choi, H.; Pietrobon, V.; Alphson, B.; et al. Contextual reprogramming of CAR-T cells for treatment of HER2(+) cancers. J. Transl. Med. 2021, 19, 459. [Google Scholar] [CrossRef]
- Das, S.; Chadwick, B.P. CRISPR mediated targeting of DUX4 distal regulatory element represses DUX4 target genes dysregulated in Facioscapulohumeral muscular dystrophy. Sci. Rep. 2021, 11, 12598. [Google Scholar] [CrossRef]
- Olson, A.; Basukala, B.; Lee, S.; Gagne, M.; Wong, W.W.; Henderson, A.J. Targeted Chromatinization and Repression of HIV-1 Provirus Transcription with Repurposed CRISPR/Cas9. Viruses 2020, 12, 1154. [Google Scholar] [CrossRef]
- Born, N.; Thiesen, H.J.; Lorenz, P. The B-subdomain of the Xenopus laevis XFIN KRAB-AB domain is responsible for its weaker transcriptional repressor activity compared to human ZNF10/Kox1. PLoS ONE 2014, 9, e87609. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Gibson, L.C.; Capili, A.D.; Borden, K.L.; Osborne, M.J.; Harper, S.L.; Speicher, D.W.; Zhao, K.; Marmorstein, R.; Rock, T.A.; et al. The structurally disordered KRAB repression domain is incorporated into a protease resistant core upon binding to KAP-1-RBCC domain. J. Mol. Biol. 2007, 370, 269–289. [Google Scholar] [CrossRef]
- Vissing, H.; Meyer, W.K.; Aagaard, L.; Tommerup, N.; Thiesen, H.J. Repression of transcriptional activity by heterologous KRAB domains present in zinc finger proteins. FEBS Lett. 1995, 369, 153–157. [Google Scholar] [CrossRef] [Green Version]
- Huntley, S.; Baggott, D.M.; Hamilton, A.T.; Tran-Gyamfi, M.; Yang, S.; Kim, J.; Gordon, L.; Branscomb, E.; Stubbs, L. A comprehensive catalog of human KRAB-associated zinc finger genes: Insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006, 16, 669–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grey, C.; Baudat, F.; de Massy, B. PRDM9, a driver of the genetic map. PLoS Genet. 2018, 14, e1007479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spruce, C.; Dlamini, S.; Ananda, G.; Bronkema, N.; Tian, H.; Paigen, K.; Carter, G.W.; Baker, C.L. HELLS and PRDM9 form a pioneer complex to open chromatin at meiotic recombination hot spots. Genes Dev. 2020, 34, 398–412. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.L.; Soulez, M.; Koczan, D.; Thiesen, H.J.; Knight, J.C. A KRAB-related domain and a novel transcription repression domain in proteins encoded by SSX genes that are disrupted in human sarcomas. Oncogene 1998, 17, 2013–2018. [Google Scholar] [CrossRef] [Green Version]
- Mihola, O.; Trachtulec, Z.; Vlcek, C.; Schimenti, J.C.; Forejt, J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 2009, 323, 373–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, P.L.; Goodstadt, L.; Bayes, J.J.; Birtle, Z.; Roach, K.C.; Phadnis, N.; Beatson, S.A.; Lunter, G.; Malik, H.S.; Ponting, C.P. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009, 5, e1000753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, J.H.; Emerson, R.O.; Shendure, J. Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS ONE 2009, 4, e8505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, Y.; Baudat, F.; Taillepierre, M.; Stanzione, M.; Toth, A.; de Massy, B. The PRDM9 KRAB domain is required for meiosis and involved in protein interactions. Chromosoma 2017, 126, 681–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, A.; Horton, J.R.; Wilson, G.G.; Zhang, X.; Cheng, X. Structural basis for human PRDM9 action at recombination hot spots. Genes Dev. 2016, 30, 257–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gure, A.O.; Wei, I.J.; Old, L.J.; Chen, Y.T. The SSX gene family: Characterization of 9 complete genes. Int. J. Cancer 2002, 101, 448–453. [Google Scholar] [CrossRef]
- Johansen, S.; Gjerstorff, M.F. Interaction between Polycomb and SSX Proteins in Pericentromeric Heterochromatin Function and Its Implication in Cancer. Cells 2020, 9, 226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, H.A.; McNeel, D.G. The SSX family of cancer-testis antigens as target proteins for tumor therapy. Clin. Dev. Immunol. 2010, 2010, 150591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helleboid, P.Y.; Heusel, M.; Duc, J.; Piot, C.; Thorball, C.W.; Coluccio, A.; Pontis, J.; Imbeault, M.; Turelli, P.; Aebersold, R.; et al. The interactome of KRAB zinc finger proteins reveals the evolutionary history of their functional diversification. EMBO J. 2019, 38, e101220. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef]
- Al Chiblak, M.; Steinbeck, F.; Thiesen, H.J.; Lorenz, P. DUF3669, a “domain of unknown function” within ZNF746 and ZNF777, oligomerizes and contributes to transcriptional repression. BMC Mol. Cell Biol. 2019, 20, 60. [Google Scholar] [CrossRef]
- Margolin, J.F.; Friedman, J.R.; Meyer, W.K.; Vissing, H.; Thiesen, H.J.; Rauscher, F.J., 3rd. Kruppel-associated boxes are potent transcriptional repression domains. Proc. Natl. Acad. Sci. USA 1994, 91, 4509–4513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, H.; Ivanov, A.V.; Oh, H.J.; Lau, Y.F.; Rauscher, F.J., 3rd. Epigenetic gene silencing by the SRY protein is mediated by a KRAB-O protein that recruits the KAP1 co-repressor machinery. J. Biol. Chem. 2009, 284, 35670–35680. [Google Scholar] [CrossRef] [Green Version]
- Witzgall, R.; O’Leary, E.; Leaf, A.; Onaldi, D.; Bonventre, J.V. The Kruppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc. Natl. Acad. Sci. USA 1994, 91, 4514–4518. [Google Scholar] [CrossRef] [Green Version]
- Thiesen, H.J. From repression domains to designer zinc finger proteins: A novel strategy of intracellular immunization against HIV. Gene Expr. 1996, 5, 229–243. [Google Scholar] [PubMed]
- Friedman, J.R.; Fredericks, W.J.; Jensen, D.E.; Speicher, D.W.; Huang, X.P.; Neilson, E.G.; Rauscher, F.J., 3rd. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996, 10, 2067–2078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moosmann, P.; Georgiev, O.; Le Douarin, B.; Bourquin, J.P.; Schaffner, W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res. 1996, 24, 4859–4867. [Google Scholar] [CrossRef] [Green Version]
- Schultz, D.C.; Ayyanathan, K.; Negorev, D.; Maul, G.G.; Rauscher, F.J., 3rd. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002, 16, 919–932. [Google Scholar] [CrossRef] [Green Version]
- Sripathy, S.P.; Stevens, J.; Schultz, D.C. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell. Biol. 2006, 26, 8623–8638. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold-Making protein folding accessible to all. bioRxiv 2021, arXiv:2021.2008.2015.456425. [Google Scholar]
- Stoll, G.A.; Oda, S.I.; Chong, Z.S.; Yu, M.; McLaughlin, S.H.; Modis, Y. Structure of KAP1 tripartite motif identifies molecular interfaces required for retroelement silencing. Proc. Natl. Acad. Sci. USA 2019, 116, 15042–15051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Keown, J.R.; Black, M.M.; Raclot, C.; Demarais, N.; Trono, D.; Turelli, P.; Goldstone, D.C. A Dissection of Oligomerization by the TRIM28 Tripartite Motif and the Interaction with Members of the Krab-ZFP Family. J. Mol. Biol. 2019, 431, 2511–2527. [Google Scholar] [CrossRef] [PubMed]
- Mannini, R.; Rivieccio, V.; D’Auria, S.; Tanfani, F.; Ausili, A.; Facchiano, A.; Pedone, C.; Grimaldi, G. Structure/function of KRAB repression domains: Structural properties of KRAB modules inferred from hydrodynamic, circular dichroism, and FTIR spectroscopic analyses. Proteins 2006, 62, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.E.; Shylo, N.A.; Alexander, K.A.; Churchill, A.J.; Copperman, C.; Garcia-Garcia, M.J. The Transcriptional Repressive Activity of KRAB Zinc Finger Proteins Does Not Correlate with Their Ability to Recruit TRIM28. PLoS ONE 2016, 11, e0163555. [Google Scholar] [CrossRef] [PubMed]
- Staby, L.; O’Shea, C.; Willemoes, M.; Theisen, F.; Kragelund, B.B.; Skriver, K. Eukaryotic transcription factors: Paradigms of protein intrinsic disorder. Biochem. J. 2017, 474, 2509–2532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tycko, J.; del Rosso, N.; Hess, G.T.; Aradhana, A.; Banerjee, A.; Mukund, A.; Van, M.V.; Ego, B.K.; Yao, D.; Spees, K.; et al. High-Throughput Discovery and Characterization of Human Transcriptional Effectors. Cell 2020, 183, 2020–2035.e16. [Google Scholar] [CrossRef]
- Amemiya, C.T.; Alfoldi, J.; Lee, A.P.; Fan, S.; Philippe, H.; Maccallum, I.; Braasch, I.; Manousaki, T.; Schneider, I.; Rohner, N.; et al. The African coelacanth genome provides insights into tetrapod evolution. Nature 2013, 496, 311–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biscotti, M.A.; Adolfi, M.C.; Barucca, M.; Forconi, M.; Pallavicini, A.; Gerdol, M.; Canapa, A.; Schartl, M. A Comparative View on Sex Differentiation and Gametogenesis Genes in Lungfish and Coelacanths. Genome Biol. Evol. 2018, 10, 1430–1444. [Google Scholar] [CrossRef]
- Parvanov, E.D.; Tian, H.; Billings, T.; Saxl, R.L.; Spruce, C.; Aithal, R.; Krejci, L.; Paigen, K.; Petkov, P.M. PRDM9 interactions with other proteins provide a link between recombination hotspots and the chromosomal axis in meiosis. Mol. Biol. Cell 2017, 28, 488–499. [Google Scholar] [CrossRef]
- de Bruijn, D.R.; dos Santos, N.R.; Kater-Baats, E.; Thijssen, J.; van den Berk, L.; Stap, J.; Balemans, M.; Schepens, M.; Merkx, G.; van Kessel, A.G. The cancer-related protein SSX2 interacts with the human homologue of a Ras-like GTPase interactor, RAB3IP, and a novel nuclear protein, SSX2IP. Genes Chromosomes Cancer 2002, 34, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Schmitges, F.W.; Radovani, E.; Najafabadi, H.S.; Barazandeh, M.; Campitelli, L.F.; Yin, Y.; Jolma, A.; Zhong, G.; Guo, H.; Kanagalingam, T.; et al. Multiparameter functional diversity of human C2H2 zinc finger proteins. Genome Res. 2016, 26, 1742–1752. [Google Scholar] [CrossRef] [PubMed]
- Biscotti, M.A.; Gerdol, M.; Canapa, A.; Forconi, M.; Olmo, E.; Pallavicini, A.; Barucca, M.; Schartl, M. The Lungfish Transcriptome: A Glimpse into Molecular Evolution Events at the Transition from Water to Land. Sci. Rep. 2016, 6, 21571. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, T.J.; Clements, J.; Finn, R.D. Skylign: A tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinform. 2014, 15, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Remboutsika, E.; Lutz, Y.; Gansmuller, A.; Vonesch, J.L.; Losson, R.; Chambon, P. The putative nuclear receptor mediator TIF1alpha is tightly associated with euchromatin. J. Cell Sci. 1999, 112, 1671–1683. [Google Scholar] [CrossRef] [PubMed]
- Alerasool, N.; Segal, D.; Lee, H.; Taipale, M. An efficient KRAB domain for CRISPRi applications in human cells. Nat. Methods 2020, 17, 1093–1096. [Google Scholar] [CrossRef]
- Lau, C.H.; Suh, Y. In vivo epigenome editing and transcriptional modulation using CRISPR technology. Transgenic Res. 2018, 27, 489–509. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Bojar, D.; Fussenegger, M. A CRISPR/Cas9-based central processing unit to program complex logic computation in human cells. Proc. Natl. Acad. Sci. USA 2019, 116, 7214–7219. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Subfamily | Group | Count | HMM hsKRAB-A | HMM PRDM9 Ortho | HMM SSX Ortho | HMM Lcha ZNF |
|---|---|---|---|---|---|---|
| mKRAB-A | hsKRAB-A | 416 | 26.50 [3.57–31.20] | 4.20 [<2–14.89] | 4.31 [<2–10.57] | 5.23 [<2–16.59] |
| aKRAB-A | PRDM9 ortho | 183 | 8.59 [<2–10.20] | 29.72 [13.54–31.31] | 13.92 [4.52–16.55] | 13.51 [3.28–14.85] |
| aKRAB-A | SSX ortho | 224 | 5.64 [<2–7.70] | 14.69 [3.64–20.89] | 22.12 [14.68–24.17] | 8.74 [2.74–11.23] |
| aKRAB-A | lcha ZNF | 257 | 7.01 [4.59–10.64] | 15.19 [8.48–18.31] | 9.89 [6.89–13.33] | 22.68 [16.51–24.14] |
| Group | hsKRAB-A | PRDM9 Ortho | SSX Ortho | Lcha ZNF |
|---|---|---|---|---|
| hsKRAB-A | - | 6.77 × 10−132 | 2.65 × 10−132 | 2.04 × 10−129 |
| PRDM9 ortho | 1.65 × 10−61 | - | 1.38 × 10−59 | 1.40 × 10−60 |
| SSX ortho | 6.85 × 10−75 | 4.18 × 10−71 | - | 6.83 × 10−75 |
| lcha ZNF | 1.15 × 10-085 | 2.73 × 10−85 | 1.15 × 10−85 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lorenz, P.; Steinbeck, F.; Krause, L.; Thiesen, H.-J. The KRAB Domain of ZNF10 Guides the Identification of Specific Amino Acids That Transform the Ancestral KRAB-A-Related Domain Present in Human PRDM9 into a Canonical Modern KRAB-A Domain. Int. J. Mol. Sci. 2022, 23, 1072. https://doi.org/10.3390/ijms23031072
Lorenz P, Steinbeck F, Krause L, Thiesen H-J. The KRAB Domain of ZNF10 Guides the Identification of Specific Amino Acids That Transform the Ancestral KRAB-A-Related Domain Present in Human PRDM9 into a Canonical Modern KRAB-A Domain. International Journal of Molecular Sciences. 2022; 23(3):1072. https://doi.org/10.3390/ijms23031072
Chicago/Turabian StyleLorenz, Peter, Felix Steinbeck, Ludwig Krause, and Hans-Jürgen Thiesen. 2022. "The KRAB Domain of ZNF10 Guides the Identification of Specific Amino Acids That Transform the Ancestral KRAB-A-Related Domain Present in Human PRDM9 into a Canonical Modern KRAB-A Domain" International Journal of Molecular Sciences 23, no. 3: 1072. https://doi.org/10.3390/ijms23031072
APA StyleLorenz, P., Steinbeck, F., Krause, L., & Thiesen, H.-J. (2022). The KRAB Domain of ZNF10 Guides the Identification of Specific Amino Acids That Transform the Ancestral KRAB-A-Related Domain Present in Human PRDM9 into a Canonical Modern KRAB-A Domain. International Journal of Molecular Sciences, 23(3), 1072. https://doi.org/10.3390/ijms23031072






