Intracellular Trafficking of Cationic Carbon Dots in Cancer Cell Lines MCF-7 and HeLa—Time Lapse Microscopy, Concentration-Dependent Uptake, Viability, DNA Damage, and Cell Cycle Profile
Abstract
1. Introduction
2. Results
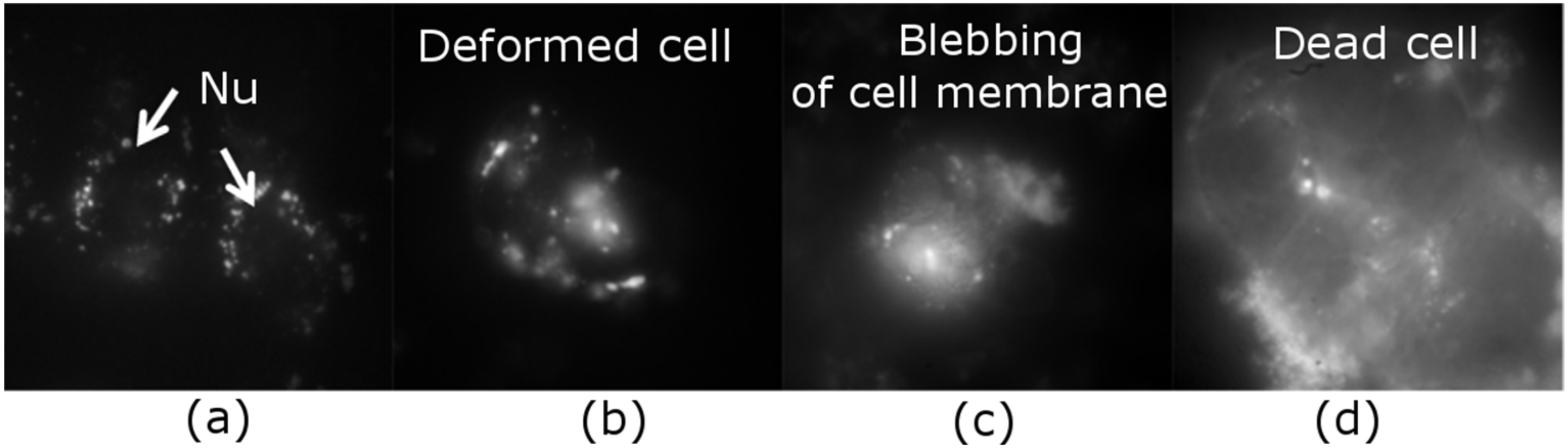
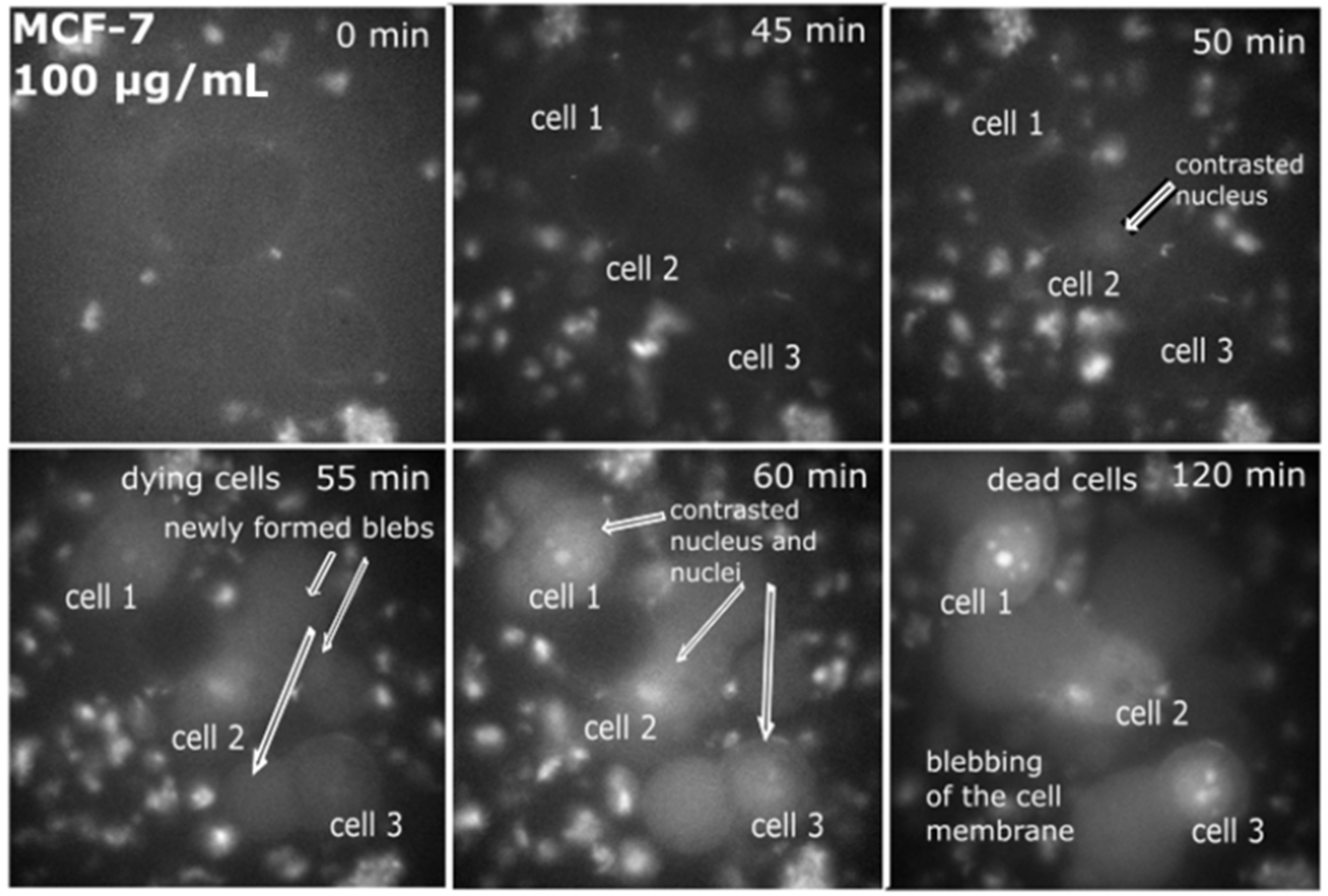
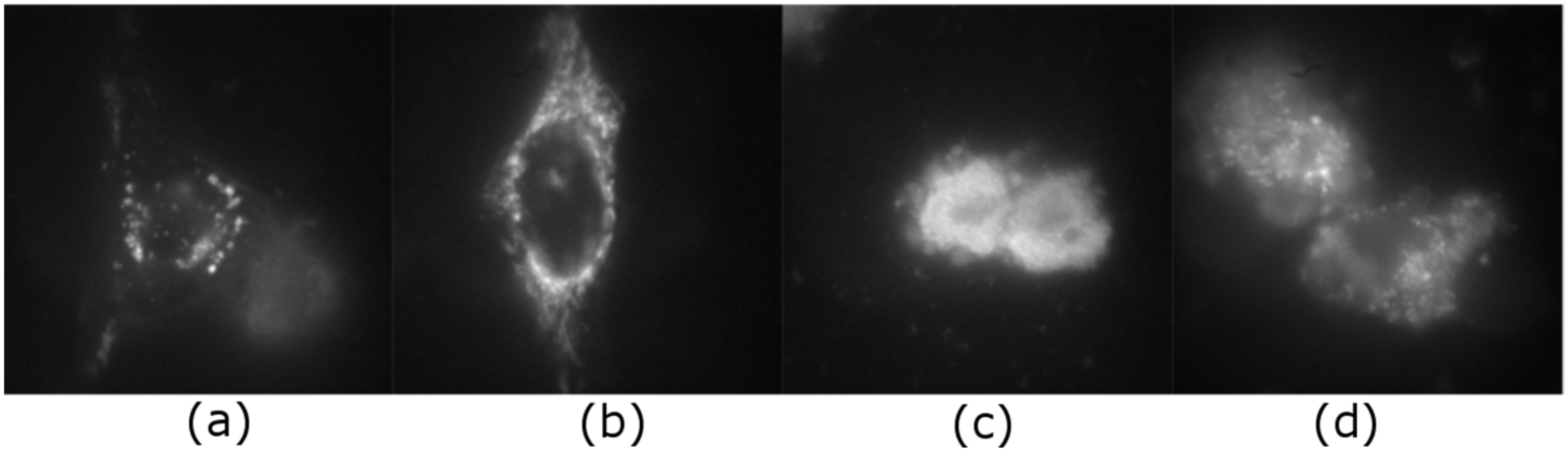
2.1. Intracellular Observation of QCDs-Labeled MCF-7 Cells
2.2. Intracellular Observation of QCDs-Labeled HeLa Cells
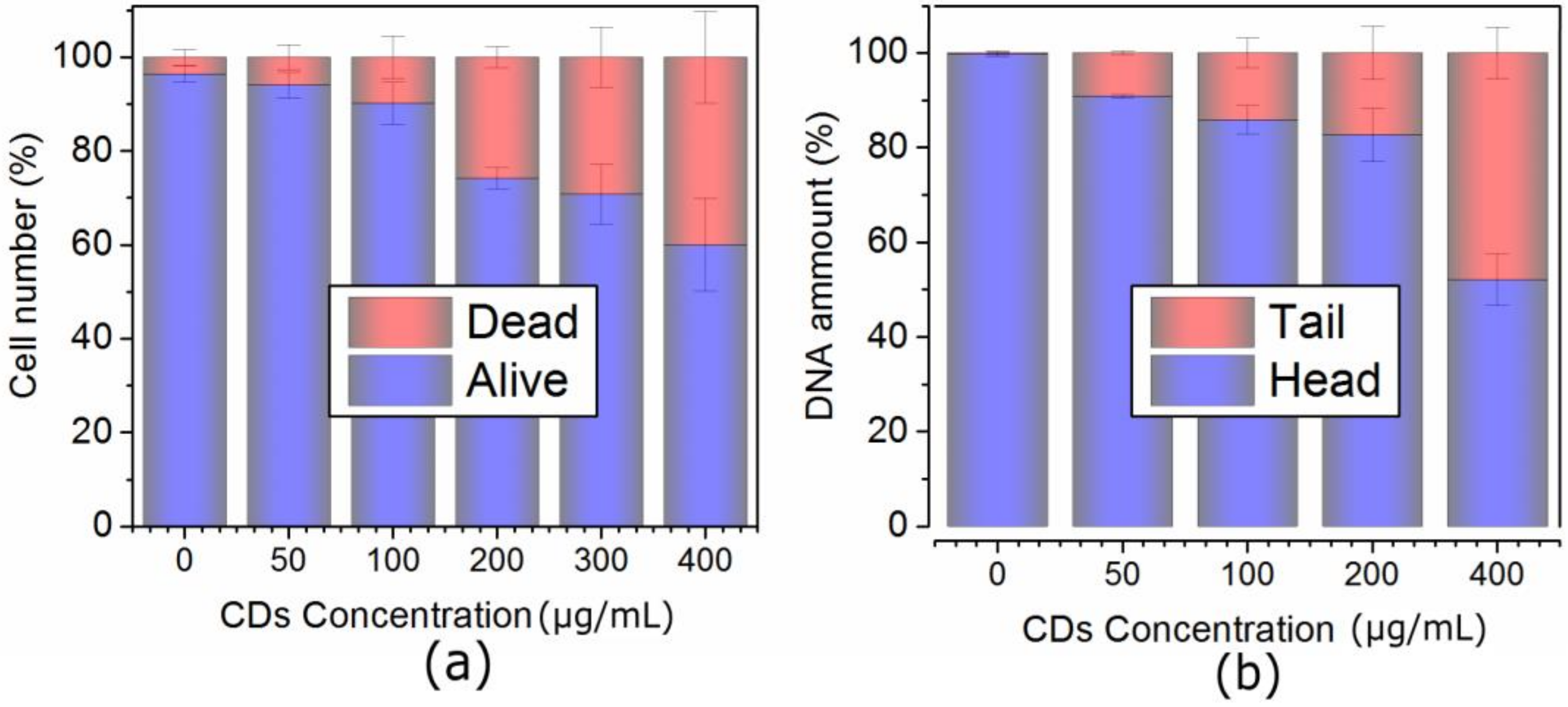
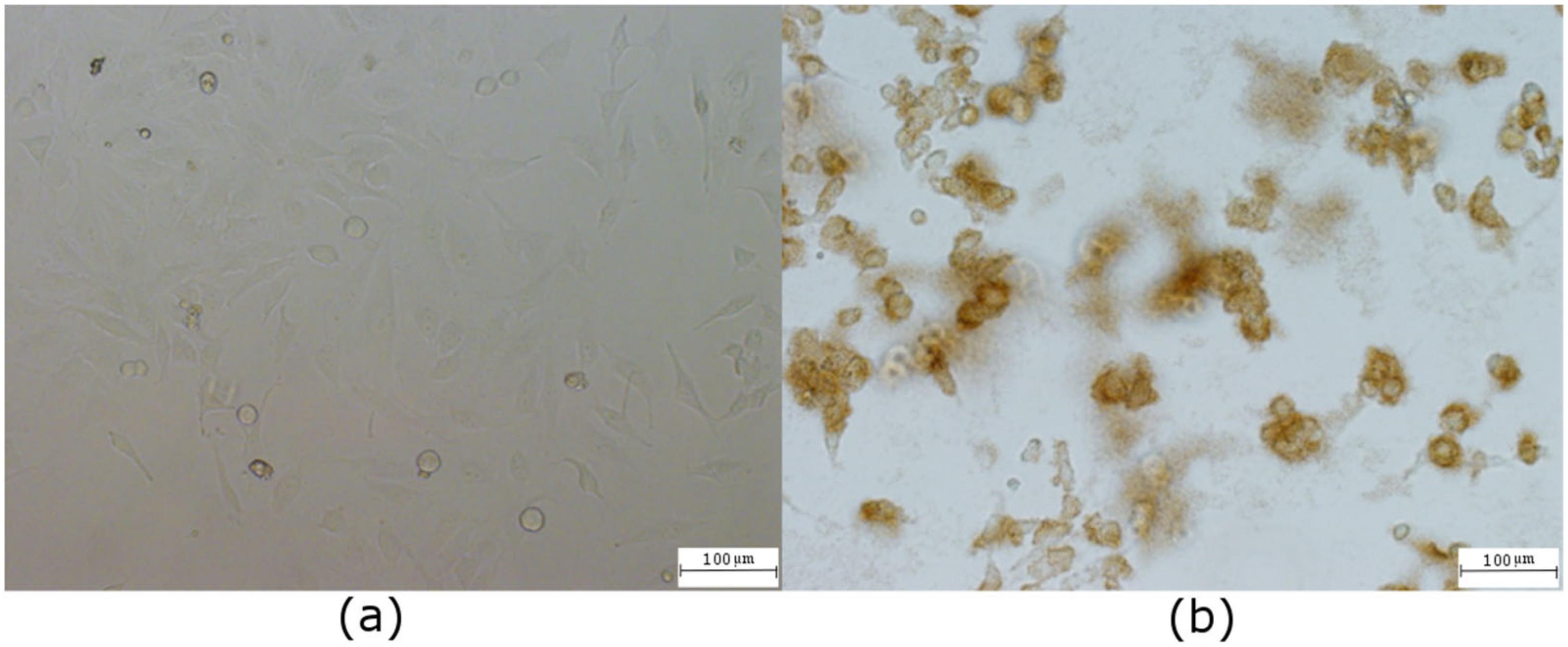
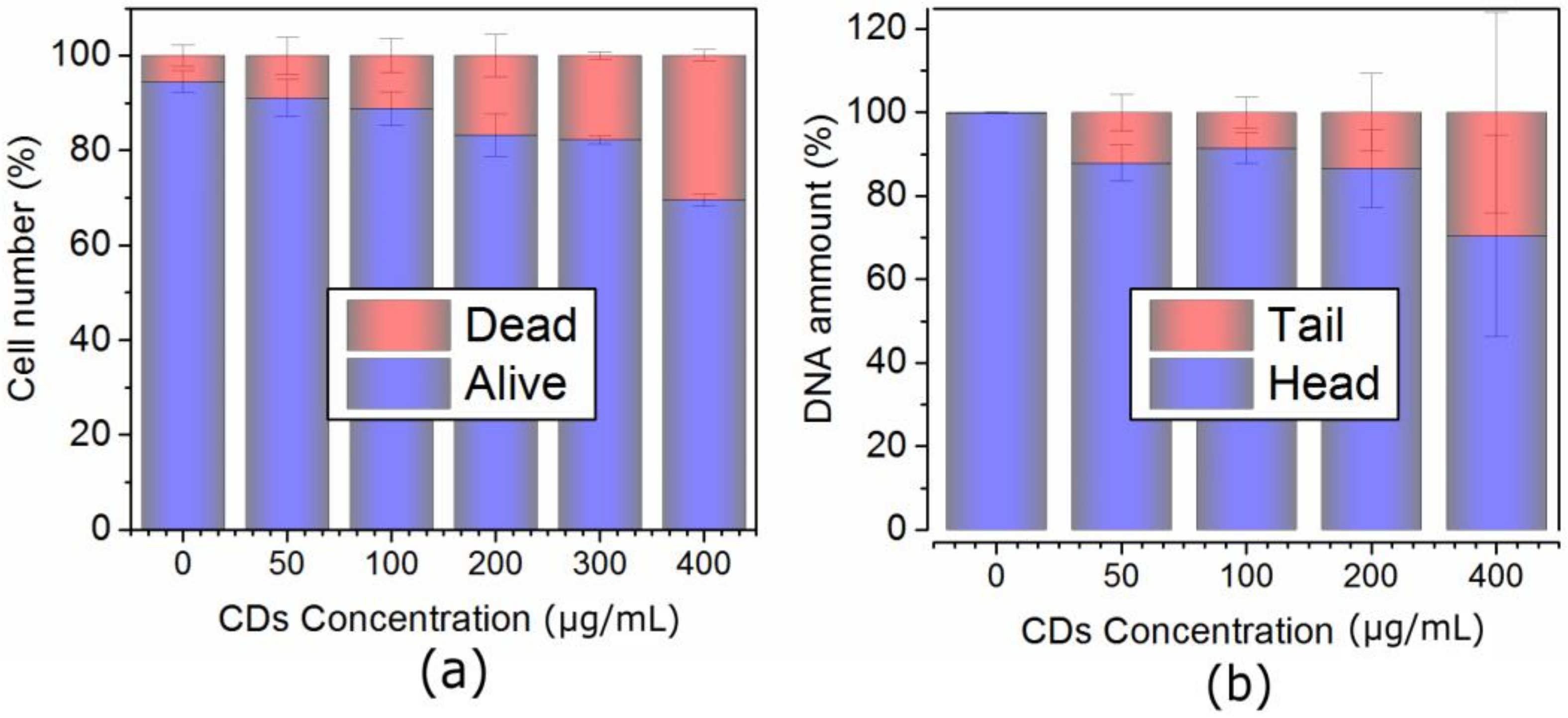
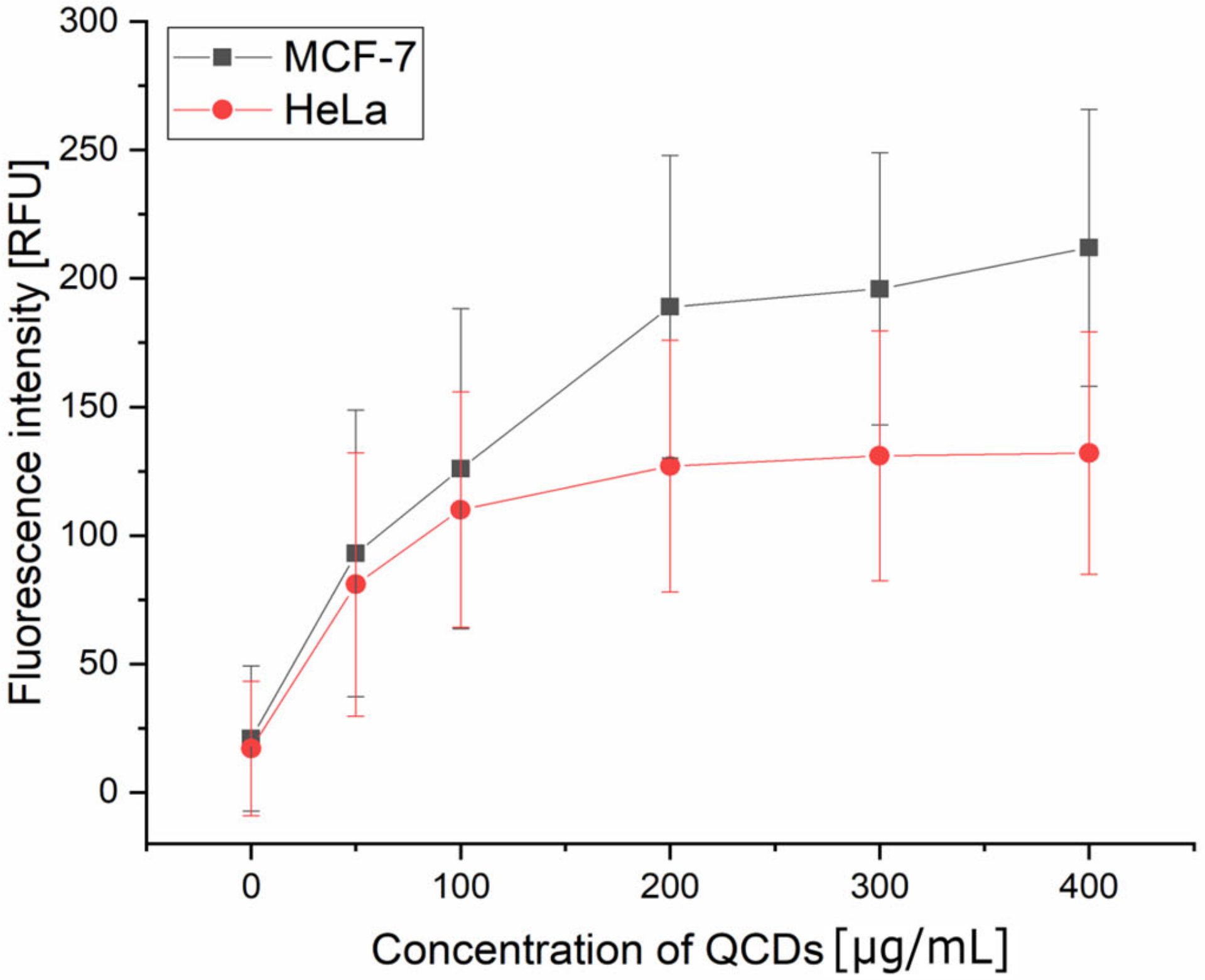
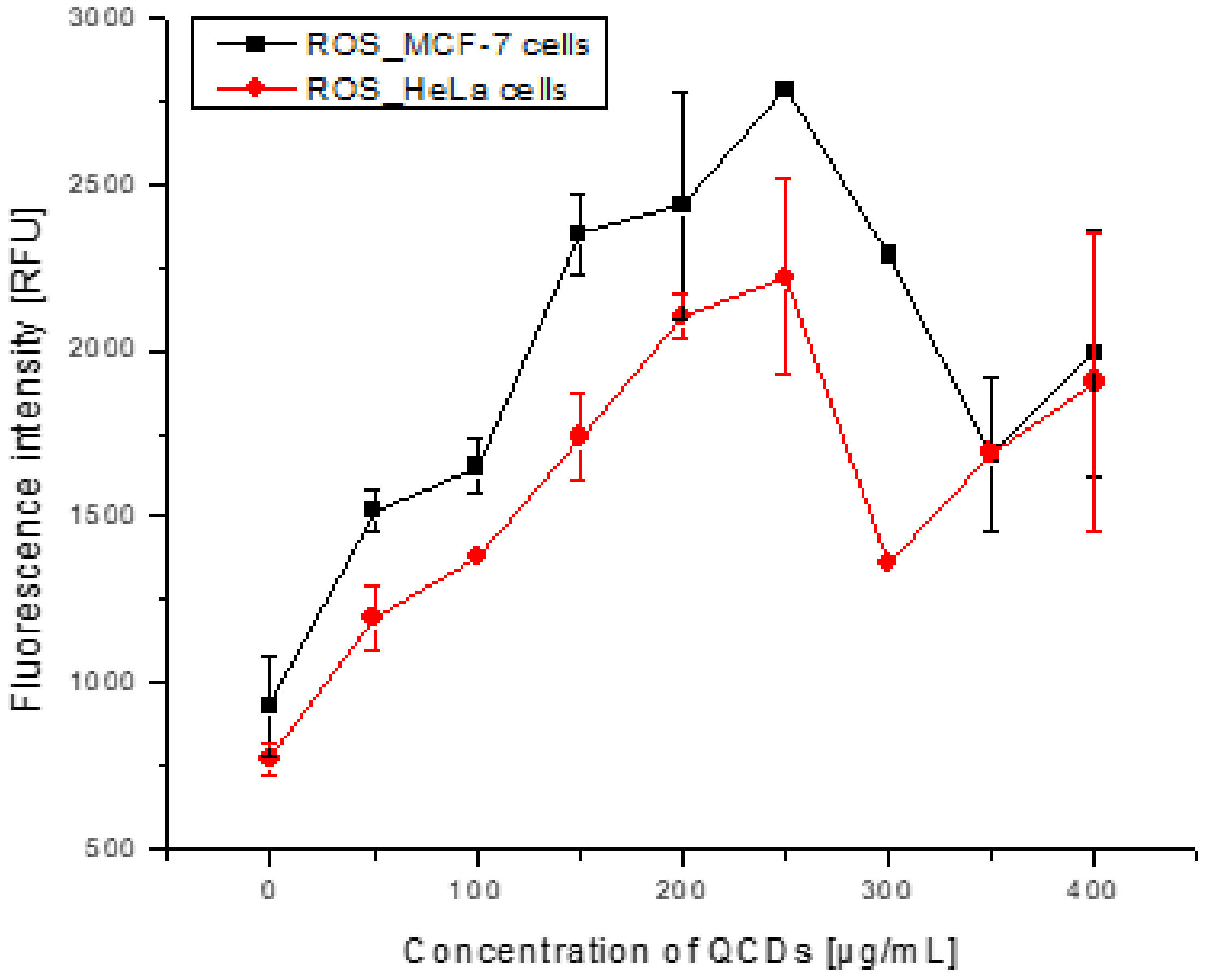
2.3. Concentration-Dependent Uptake and Oxidative Stress in Both Cell Lines
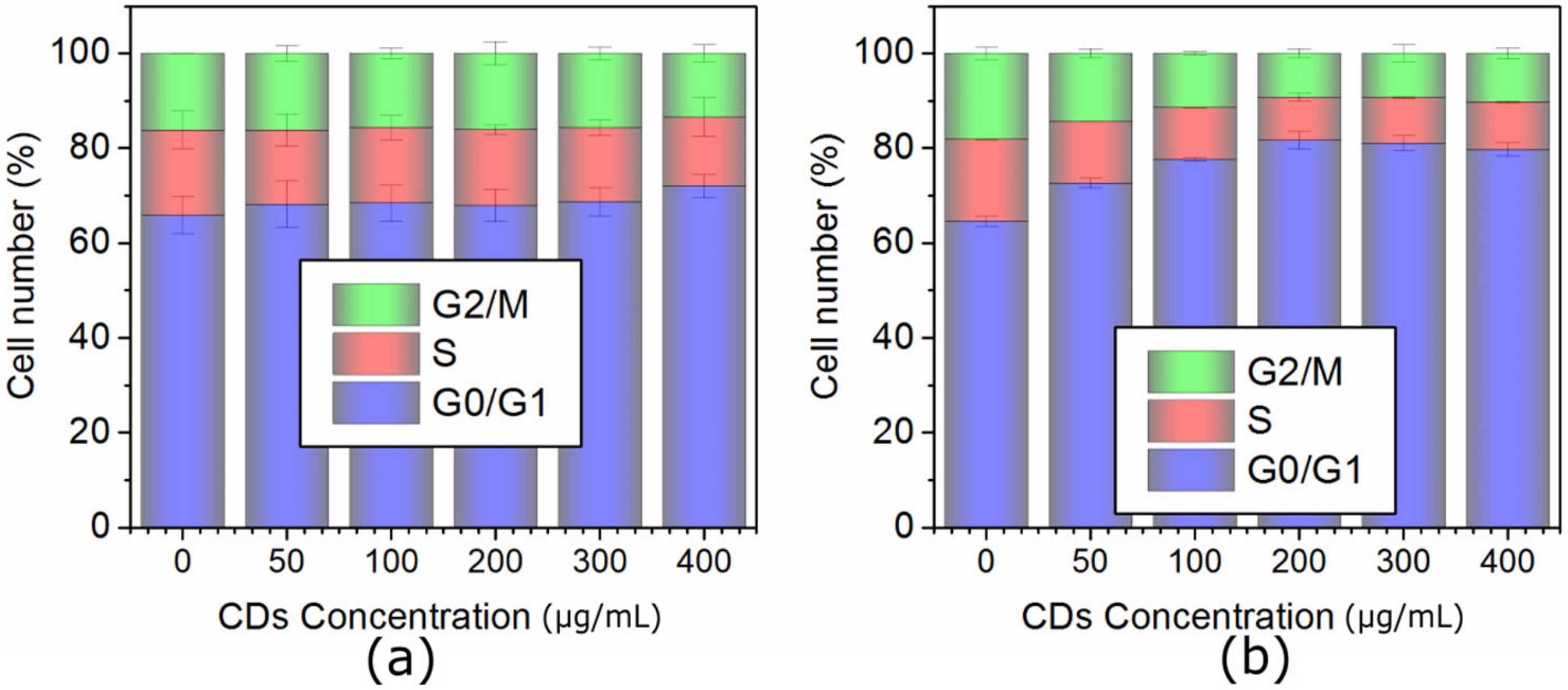
2.4. Cell Cycle Analysis of MCF-7 and HeLa Cells
3. Materials and Methods
3.1. Carbon Dots
3.2. Cell Cultivation, Microscopy
3.3. Fluorescence Microspectroscopy
3.4. Cell Cycle and Concentration-Dependent Uptake
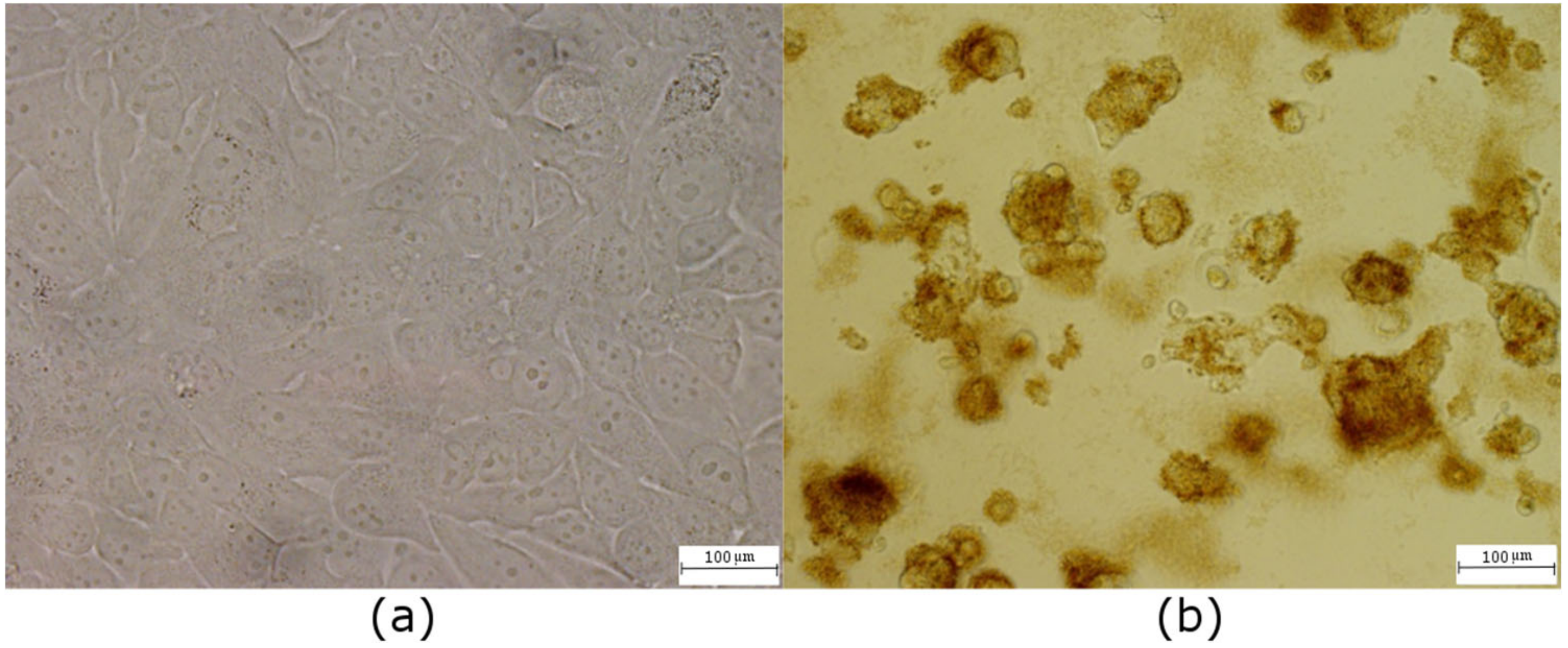
3.5. Viability
3.6. Comet Assay
3.7. Reactive Oxygen Species
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baker, S.N.; Baker, G.A. Luminescent carbon nanodots: Emergent Nanolights. Angew. Chem. Int. Ed. 2010, 49, 6726–6744. [Google Scholar] [CrossRef]
- Li, H.; Kang, Z.; Liu, Y.; Lee, S.-T. Carbon nanodots: Synthesis, properties and applications. J. Mater. Chem. 2012, 22, 24230–24253. [Google Scholar] [CrossRef]
- Song, Y.; Shi, W.; Chen, W.; Li, X.; Ma, H. Fluorescent carbon nanodots conjugated with folic acid for distinguishing folate-receptor-positive cancer cells from normal cells. J. Mater. Chem. 2012, 22, 12568–12573. [Google Scholar] [CrossRef]
- Ruan, S.; Qian, J.; Shen, S.; Zhu, J.; Jiang, X.; He, Q.; Gao, H. A simple one-step method to prepare fluorescent carbon dots and their potential application in non-invasive glioma imaging. Nanoscale 2014, 6, 10040–10047. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-M.; Lin, L.-P.; Wang, X.-X.; Lin, S.-Q.; Cai, W.-L.; Zhang, L.-H.; Zheng, Z.-Y. Highly selective and sensitive detection of Cu2+ with lysine enhancing bovine serum albumin modified-carbon dots fluorescent probe. Analyst 2012, 137, 2637–2642. [Google Scholar] [CrossRef]
- Kuo, T.R.; Sung, S.Y.; Hsu, C.W.; Chang, C.J.; Chiu, T.C.; Hu, C.C. One-pot green hydrothermal synthesis of fluorescent nitrogen-doped carbon nanodots for in vivo bioimaging. Anal. Bioanal. Chem. 2016, 408, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, M.; Czarnecka, J.; Bolibok, P.; Swidzinski, M.; Roszek, K. New insight into the fluorescence quenching of nitrogen-containing carbonaceous quantum dots—From surface chemistry to biomedical applications. Materials 2021, 14, 2454. [Google Scholar] [CrossRef]
- Havrdova, M.; Hola, K.; Skopalik, J.; Tomankova, K.; Petr, M.; Cepe, K.; Polakova, K.; Tucek, J.; Bourlinos, A.B.; Zboril, R. Toxicity of carbon dots—Effect of surface functionalization on the cell viability, reactive oxygen species generation and cell cycle. Carbon. 2016, 99, 238–248. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, Q.; Li, P.; Xun, X.; Zheng, L.; Ning, D.; Su, M. Surface charge controlled nucleoli selective staining with nanoscale carbon dots. PLoS ONE 2019, 14, e0216230. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 2012, 7, 5577–5591. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Li, Z.; Liu, Z.; Ren, J.; Qu, X. Luminescent carbon dot-gated nanovehicles for pH-triggered intracellular controlled release and imaging. Langmuir 2013, 29, 6396–6403. [Google Scholar] [CrossRef]
- Patra, S.; Roy, E.; Madhuri, R.; Sharma, P.K. The next generation cell-penetrating peptide and carbon dot conjugated nano-liposome for transdermal delivery of curcumin. Biomater. Sci. 2016, 4, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Fahmi, M.Z.; Chen, J.K.; Huang, C.C.; Ling, Y.C.; Chang, J.Y. Phenylboronic acid-modified magnetic nanoparticles as a platform for carbon dot conjugation and doxorubicin delivery. J. Mater. Chem. B 2015, 3, 5532–5543. [Google Scholar] [CrossRef]
- Misra, S.K.; Ohoka, A.; Kolmodin, N.J.; Pan, D. Next generation carbon nanoparticles for efficient gene therapy. Mol. Pharm. 2015, 12, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Pierrat, P.; Wang, R.; Kereselidze, D.; Lux, M.; Didier, P.; Kichler, A.; Pons, F.; Lebeau, L. Efficient in vitro and in vivo pulmonary delivery of nucleic acid by carbon dot-based nanocarriers. Biomaterials 2015, 51, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, X.; Bhirde, A.; Cao, J.; Zeng, Y.; Huang, X.; Sun, Y.; Liu, G.; Chen, X. Carbon-dot-based two-photon visible nanocarriers for safe and highly efficient delivery of siRNA and DNA. Adv. Healthc. Mater. 2014, 3, 1203–1209. [Google Scholar] [CrossRef]
- Das, S.; Debnath, N.; Cui, Y.; Unrine, J.; Palli, S.R. Chitosan, Carbon Quantum Dot, and Silica Nanoparticle Mediated dsRNA Delivery for Gene Silencing in Aedes aegypti: A Comparative Analysis. ACS Appl. Mater. Interfaces 2015, 7, 19530–19535. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; Qiang, L.; Chen, X.; Meng, X. Fluorescent carbon dots for bioimaging and biosensing applications. J. Biomed. Nanotech. 2014, 10, 2677–2699. [Google Scholar] [CrossRef]
- Yang, W.; Ni, J. Cationic carbon dots for modification-free detection of hyaluronidase. Anal. Chem. 2017, 89, 8384–8390. [Google Scholar] [CrossRef]
- Feng, F.; Miao, C. Positively charged and pH/sensitive carbon dots for fluorescence detection of copper ion. Bull. Korean Chem. Soc. 2020, 42, 227–234. [Google Scholar] [CrossRef]
- Fang, H.Y.; Huang, W.M.; Chen, D.H. One step synthesis of positively charged bifunctional carbon dot/silver composite. Nanotechnology 2019, 30, 365603. [Google Scholar] [CrossRef]
- Wang, H.; Lu, F.; Ma, C.; Ma, Y.; Zhang, M.; Wang, B.; Zhang, Y.; Liu, Y.; Huang, H.; Kang, Z. Carbon dots with positive surface charge from teraric acid and m-aminophenol for selective killing of Gram-positive bacteria. J. Mater. Chem. B 2021, 9, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Bourlinos, A.B.; Zboril, R.; Petr, J.; Bakandritsos, A.; Krysmann, M.; Giannelis, E.P. Luminescent surface quaternized carbon dots. Chem. Mater. 2012, 24, 6–8. [Google Scholar] [CrossRef]
- Song, Y.; Wang, Y.; Zhang, N.; Li, X.; Bai, X.; Li, T. Quaternized carbon-based nanoparticles embedded positively charged composite membranes towards efficient removal of cationic small-sized contaminants. J. Membr. Sci. 2021, 630, 119332. [Google Scholar] [CrossRef]
- Hao, X.; Huang, L.; Zhao, C.; Chen, S.; Lin, W.; Lin, Y.; Zhang, L.; Sun, A.; Miao, C.; Lin, X.; et al. Antibacterial activity of positively charged carbon quantum dots without detectable resistance for wound healing with mixed bacteria infection. Mater. Sci. Eng. C 2021, 123, 111971. [Google Scholar] [CrossRef]
- Zuo, G.; Xie, A.; Pan, X.; Su, T.; Li, J.; Dong, W. Fluorine-Doped Cationic Carbon Dots for Efficient Gene Delivery. ACS Appl. Nano Mater. 2018, 1, 2376–2385. [Google Scholar] [CrossRef]
- Guo, R.B.; Chen, B.; Li, F.L.; Weng, S.H.; Zheng, Z.F.; Chen, M.; Wu, W.; Lin, X.H.; Yang, C.Y. Positive carbon dots with dual roles of nanoquencher and reference signal for the ratiometric fluorescence sensing of DNA. Sens. Actuators B Chem. 2018, 264, 193–201. [Google Scholar] [CrossRef]
- Yue, L.J.; Wei, Y.Y.; Fan, J.B.; Chen, L.; Li, Q.; Du, J.L.; Yu, S.P.; Yang, Y.Z. Research progress in the use of cationic carbon dots for the integration of cancer diagnosis with gene treatment. New Carbon Mater. 2021, 36, 373–389. [Google Scholar] [CrossRef]
- Havrdova, M.; Urbancic, I.; Barton Tomankova, K.; Malina, L.; Strancar, J.; Bourlinos, A.B. Self-targeting of carbon dots into the cell nucleus: Diverse mechanisms of toxicity in NIH/3T3 and L929 cells. Int. J. Mol. Sci. 2021, 22, 5608. [Google Scholar] [CrossRef]
- Malina, T.; Polakova, K.; Skopalik, J.; Milotova, V.; Hola, K.; Havrdova, M.; Tomankova, K.B.; Cmiel, V.; Sefc, L.; Zboril, R. Carbon dots for in vivo fluorescence imaging of adipose tissue-derived mesenchymal stromal cells. Carbon 2019, 152, 434–443. [Google Scholar] [CrossRef]
- Unnikrishnan, B.; Wu, R.S.; Wei, S.C.; Huang, C.C.; Chang, H.; Chang, H.T. Fluorescent carbon dots for selective labeling of subcellular organelles. ACS Omega 2020, 5, 11248–11261. [Google Scholar] [CrossRef]
- Jung, Y.K.; Shin, E.; Kim, B.-S. Cell nucleus-targeting zwitterionic carbon dots. Sci. Rep. 2015, 5, 18807. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.A.; Sheikh, S.; Zhang, Q.; Sueiro Ballesteros, L.; Herman, A.; Davis, S.A.; Morgan, D.J.; Berry, M.; Benito-Alifonso, D.; Galan, M.C. Selective photothermal killing of cancer cells using LED-activated nucleus targeting fluorescent carbon dots. Nanoscale Adv. 2019, 1, 2840–2846. [Google Scholar] [CrossRef]
- Yang, L.; Jiang, W.; Qiu, L.; Jiang, X.; Zuo, D.; Wang, D.; Yang, L. One pot synthesis of highly luminescent polyethylene glycol anchored carbon dots functionalized with a nuclear localization signal peptide for cell nucleus imaging. Nanoscale 2015, 7, 6104–6113. [Google Scholar] [CrossRef]
- Ci, J.; Tian, Y.; Kuga, S.; Niu, Z.; Wu, M.; Huang, Y. One-pot green synthesis of nitrogen-doped carbon quantum dots for cell nucleus labeling and copper(II) detection. Chem.—Asian J. 2017, 12, 2916–2921. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Guo, B.; Hao, L.; Liu, N.; Lin, Y.; Guo, W.; Li, X.; Gu, B. Doxorubicin-loaded environmentally friendly carbon dots as a novel drug delivery system for nucleus targeted cancer therapy. Colloids Surf. B 2017, 159, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Şimşek, S.; Şüküroğlu, A.A.; Yetkin, D.; Özbek, B.; Battal, D.; Genç, R. DNA-damage and cell cycle arrest initiated anti-cancer potency of super tiny carbon dots on MCF7 cell line. Sci Rep. 2020, 10, 13880. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, X.; Xian, M.; Dong, C.; Shuang, S. Folic acid-conjugated green luminescent carbon dots as a nanoprobe for identifying folate receptor-positive cancer cells. Talanta 2018, 183, 39–47. [Google Scholar] [CrossRef]
- Wang, J.; Liu, S.; Chang, Y.; Fang, L.; Han, K.; Li, M. High efficient delivery of siRNA into tumor cells by positively charged carbon dots. J. Macromol. Sci. A 2018, 55, 770–774. [Google Scholar] [CrossRef]
- Abe, J.; Yamada, Y.; Harashima, H. Validation of a Strategy for Cancer Therapy: Delivering Aminoglycoside Drugs to Mitochondria in HeLa Cells. J. Pharm. Sci. 2016, 105, 734–740. [Google Scholar] [CrossRef]
- Sima, M.; Vrbova, K.; Zavodna, T.; Honkova, K.; Chvojkova, I.; Ambroz, A.; Klema, J.; Rossnerova, A.; Polakova, K.; Malina, T.; et al. The Differential Effect of Carbon Dots on Gene Expression and DNA Methylation of Human Embryonic Lung Fibroblasts as a Function of Surface Charge and Dose. Int. J. Mol. Sci. 2020, 21, 4763. [Google Scholar] [CrossRef]
- Singh, S.; Singh, M.K.; Das, P. Biosensing of solitary and clustered abasic site DNA damage lesions with copper nanoclusters and carbon dots. Sens. Actuators B Chem. 2018, 255, 763–774. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, S.; Ge, S.; Wang, S.; Yan, M.; Zang, D.; Yu, J. Facile and sensitive paper-based chemiluminescence DNA biosensor using carbon dots dotted nanoporous gold signal amplification label. Anal. Methods 2013, 5, 1328–1336. [Google Scholar] [CrossRef]
- Huang, Q.T.; Lin, X.F.; Zhu, J.J.; Tong, Q.X. Pd-Au@carbon dots nanocomposite: Facile synthesis and application as an ultrasensitive electrochemical biosensor for determination of colitoxin DNA in human serum. Biosens. Bioelectron. 2017, 94, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Kudr, J.; Richtera, L.; Xhaxhiu, K.; Hynek, D.; Heger, Z.; Zitka, O.; Adam, V. Carbon dots based FRET for the detection of DNA damage. Biosens. Bioelectron. 2017, 92, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.W.; Monteiro-Riviere, N.A. Mechanisms of Quantum Dot Nanoparticle Cellular Uptake. Toxicol. Sci. 2009, 110, 138–155. [Google Scholar] [CrossRef] [PubMed]
- Czarnecka, J.; Kwiatkowski, M.; Wisniewski, M.; Roszek, K. Protein corona hinders N-CQDs oxidative potential and favors their application as nanobiocatalytic system. Int. J. Mol. Sci. 2021, 22, 8136. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Berndt, C.; Jones, D.J. Oxidative Stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef]
- Zhou, Y.; Sun, H.; Wang, F.; Ren, J.; Qu, X. How functional groups influence the ROS generation and cytotoxicity of graphene quantum dots. Chem. Commun. 2017, 53, 10588–10591. [Google Scholar] [CrossRef] [PubMed]
- Tomankova, K.; Horakova, J.; Harvanova, M.; Malina, L.; Soukupova, J.; Hradilova, S.; Kejlova, K.; Malohlava, J.; Licman, L.; Dvorakova, M.; et al. Cytotoxicity, cell uptake and microscopic analysis of titanium dioxide and silver nanoparticles in vitro. Food Chem. Toxicol. 2015, 82, 106–115. [Google Scholar] [CrossRef]
- Periasamy, V.S.; Athinarayanan, J.; Alfawaz, M.A.; Alshatwi, A.A. Carbon nanoparticle induced cytotoxicity in human mesenchymal stem cells through upregulation of TNF3, NFKBIA and BCL2L1 genes. Chemosphere 2016, 144, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Foster, I. Cancer: A cell cycle defect. Radiography 2008, 14, 144–149. [Google Scholar] [CrossRef]
- DiPaola, R.S. To arrest or not to G2-M cell-cycle arrest. Clin. Cancer Res. 2002, 8, 3512–3519. [Google Scholar]
- Essner, J.B.; Kist, J.A.; Polo-Parada, L.; Baker, G.A. Artifacts and errors associated with the ubiquitous presence of fluorescent impurities in carbon nanodots. Chem. Mater. 2018, 30, 1878–1887. [Google Scholar] [CrossRef]
- Krysmann, M.J.; Kelarakis, A.; Dallas, P.; Giannelis, E.P. Formation mechanism of carbogenic nanoparticles with dual photoluminescence emission. J. Am. Chem. Soc. 2012, 134, 747–750. [Google Scholar] [CrossRef] [PubMed]
- Arsov, Z.; Urbančič, I.; Garvas, M.; Biglino, D.; Ljubetič, A.; Koklič, T.; Štrancar, J. Fluorescence microspectroscopy as a tool to study mechanism of nanoparticles delivery into living cancer cells. Biomed. Opt. Express 2011, 2, 2083–2095. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Urbančič, I.; Arsov, Z.; Ljubetič, A.; Biglino, D.; Štrancar, J. Bleaching-corrected fluorescence microspectroscopy with nanometer peak position resolution. Opt. Express 2013, 21, 25291–25306. [Google Scholar] [CrossRef]
- Collins, A.R. The comet assay for DNA damage and repair: Principles, applications, and limitations. Mol. Biotechnol. 2004, 26, 249–261. [Google Scholar] [CrossRef]
- Tomankova, K.; Kejlova, K.; Binder, S.; Daskova, A.; Zapletalova, J.; Bendova, H.; Kolarova, H.; Jirova, D. In vitro cytotoxicity and phototoxicity study of cosmetics colorants. Toxicol Vitr. 2011, 25, 1242–1250. [Google Scholar] [CrossRef]
- Tomankova, K.; Kolarova, H.; Bajgar, R.; Jirova, D.; Kejlova, K.; Mosinger, J. Study of the photodyamic effect on the A549 cell line by atomic force microscopy and the influence of greeen tea extract on the production of reactive oxygen species. Ann. N. Y. Acad. Sci. 2009, 1171, 549–558. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Havrdová, M.; Urbančič, I.; Tománková, K.B.; Malina, L.; Poláková, K.; Štrancar, J.; Bourlinos, A.B. Intracellular Trafficking of Cationic Carbon Dots in Cancer Cell Lines MCF-7 and HeLa—Time Lapse Microscopy, Concentration-Dependent Uptake, Viability, DNA Damage, and Cell Cycle Profile. Int. J. Mol. Sci. 2022, 23, 1077. https://doi.org/10.3390/ijms23031077
Havrdová M, Urbančič I, Tománková KB, Malina L, Poláková K, Štrancar J, Bourlinos AB. Intracellular Trafficking of Cationic Carbon Dots in Cancer Cell Lines MCF-7 and HeLa—Time Lapse Microscopy, Concentration-Dependent Uptake, Viability, DNA Damage, and Cell Cycle Profile. International Journal of Molecular Sciences. 2022; 23(3):1077. https://doi.org/10.3390/ijms23031077
Chicago/Turabian StyleHavrdová, Markéta, Iztok Urbančič, Kateřina Bartoň Tománková, Lukáš Malina, Kateřina Poláková, Janez Štrancar, and Athanasios B. Bourlinos. 2022. "Intracellular Trafficking of Cationic Carbon Dots in Cancer Cell Lines MCF-7 and HeLa—Time Lapse Microscopy, Concentration-Dependent Uptake, Viability, DNA Damage, and Cell Cycle Profile" International Journal of Molecular Sciences 23, no. 3: 1077. https://doi.org/10.3390/ijms23031077
APA StyleHavrdová, M., Urbančič, I., Tománková, K. B., Malina, L., Poláková, K., Štrancar, J., & Bourlinos, A. B. (2022). Intracellular Trafficking of Cationic Carbon Dots in Cancer Cell Lines MCF-7 and HeLa—Time Lapse Microscopy, Concentration-Dependent Uptake, Viability, DNA Damage, and Cell Cycle Profile. International Journal of Molecular Sciences, 23(3), 1077. https://doi.org/10.3390/ijms23031077






